Abstract
Mating and sexual development have been associated with virulence in various fungal pathogens including Cryptococcus neoformans. This fungus is a significant pathogen of humans because it causes life-threatening cryptococcal meningitis in immunocompromised people such as AIDS patients. The virulence of C. neoformans is known to be associated with the mating type of the cells (α or a), with the α mating type being predominant among clinical isolates. However, the mechanisms by which mating and sexual development are controlled by environmental conditions and their relationship with virulence require further investigation. Cir1 is a GATA-type transcription factor that regulates the expression of genes required for utilization of essential metals such as iron and copper, and also genes required for major virulence factors including the polysaccharide capsule and melanin. Here we investigated the role of Cir1 in the mating of C. neoformans. Our results demonstrate that mutants lacking CIR1 are defective in mating, and that Cir1 contributes to copper mediated enhancement of sexual filamentation. Furthermore, we found that Cir1 influences the expression of mating pheromone genes suggesting that this protein plays a role in the early phase of sexual development on V8 mating medium.
Keywords: CIR1, copper, Cryptococcus neoformans, iron, mating
INTRODUCTION
The process of sexual development has been well characterized in many fungi including the model yeast Saccharomyces cerevisiae, several pathogens of plants and animals and commonly studied filamentous fungi such as Neurospora crassa (Lee et al., 2010). In the fungal pathogens, sexual development is often associated with virulence by mechanisms that are poorly understood (Hsueh and Heitman, 2008). The plant pathogens, Ustilago maydis and U. hordei, are exceptions because the formation of a filamentous, dikaryotic cell type by fusion of mating partners in these fungi is essential for host infection and subsequent completion of the coincident infection and sexual cycle (Hsueh and Heitman, 2008). Ustilago mutants that are incapable of mating and sexual development are known to be avirulent (Feldbrugge et al., 2004). Human fungal pathogens, such as Cryptococcus neoformans, Candida albicans and Aspergillus fumigatus, also possess the machinery for sexual development, but the role of mating in pathogenesis has yet to be clarified.
C. neoformans is a basidiomycete fungal pathogen that causes life-threatening cryptococcal meningitis in immunocompromised people such as AIDS patients (Bicanic and Harrison, 2004). The fungus is yeast-like and generally haploid, and it possesses a bipolar mating system in which a single MAT locus determines the mating type of cells (MATα or MATa). Previous studies suggested that mating-type and sexual development may be associated with virulence in C. neoformans. For example, it has been found that strains of the MATα mating type are predominant in the environment, and that a MATα strain was more virulent than a MATa strain in a congenic mating pair of capsular serotype D strains (Kwon-Chung et al., 1992). The MAT locus in C. neoformans is somewhat unusual because it spans a region of more than 100 kb and contains ~20 genes encoding pheromones (MFα or MFa), pheromone receptors (STE3α or STE3a), components of the MAP kinase signaling pathway for pheromone perception, a homeodomain transcription factor (SXI1α or SXI2a), and proteins not clearly related to mating (Hsueh and Heitman, 2008; Hull et al., 2005). The mating pheromones of C. neoformans are short peptides that mediate initial signaling events via MAP kinase pathway components including the pheromone receptor Ste3, a heterotrimeric G protein β subunit Gpb1, a MAPKK kinase Ste11, a MAPK kinase Ste7, a MAP kinase Cpk1 and a transcription factor Ste12 (Lengeler et al., 2000; Wang and Heitman, 1999). In addition to the MAP kinase pathway, the cAMP/protein kinase A pathway also regulates sexual development in C. neoformans. The Gpa1 protein is a key upstream component of the cAMP pathway that participates in the sensing of nutritional signals and that encodes a conserved G-protein α subunit. The gpa1 mutant has been shown to be defective not only in the expression of virulence factors (melanin formation and capsule induction), but also in mating; these results support a role in sexual development and further link this process with virulence (Alspaugh et al., 1997).
In the laboratory, sexual development in C. neoformans can be induced by co-culturing both MATα and MATa cells on V8 agar, a solid medium containing 5% V8 juice. A recent study by Kent et al. (2008) revealed that no single factor in V8 medium induces sexual development, but it appears, instead, that multiple factors coordinately contribute to the process. Moreover, it was discovered that one of the essential metals, copper, plays an important role in the induction of sexual development. It was also shown that copper increases the transcript levels for the pheromone genes (MFα or MFa), but the underlying mechanism for this influence remains to be determined.
Previously, we identified the GATA-type transcription factor Cir1 as a major regulator of the expression of the iron regulon and all of the known major virulence factors. These include the polysaccharide capsule, melanin deposition in the cell wall and the ability to grow at mammalian body temperature. Global transcriptome analysis using microarrays identified downstream target genes of Cir1 and revealed that Cir1 also regulates genes associated with the MAT locus (e.g., STE11α and MYO2). For example, STE11α was 2.28-fold and 2.42-fold upregulated in the cir1 mutant in low-iron and high-iron medium, respectively (Jung et al., 2006). Moreover, genes in the signaling path-ways that influence sexual development in C. neoformans, such as the cAMP pathway and the MAP kinase pathway, were also differentially expressed in the cir1 mutants (Idnurm et al., 2005; Jung et al., 2006). Cir1 may also regulate copper uptake and homeostasis because the cir1 mutant showed differential expression of the copper exporting ATPase Ccc2 compared to the wild-type strain, as well as differential expression of laccase, which requires copper and catalyzes melanin formation.
Overall, the discovery of connections between Cir1, signaling components and copper led us to investigate the phenotypic characteristics of the cir1 mutants in relation to sexual development. In the present study, we constructed and employed cir1 mutants of both mating types to investigate the influence of Cir1 on the initial stages of sexual development: fusion and filament formation. The results reveal that Cir1 plays a role in mating and filament formation on V8 medium and that this protein contributes to the influence of copper on mating.
MATERIALS AND METHODS
Strains, growth conditions and mating assays
All strains used in this study have the D capsular serotype background and their genotypes are listed in Table 1. Strains were maintained in yeast extract, bacto-peptone medium with 2.0% glucose (YPD, Difco). To evaluate the initial stages of sexual development, mating crosses involving mixtures of strains were conducted on solid V8 medium (Erke, 1976). Briefly, cells were grown in YPD at 30℃ overnight and washed twice with phosphate buffered saline (PBS). Cell number was determined with a hemocytometer and 1 × 108 MATα or MATa cells were withdrawn from the cell suspensions. These cells were mixed by pipetting and 10 μl of each was spotted on V8 medium. Plates were incubated at room temperature in the dark for two to seven days, and the periphery of each mixed colony was observed microscopically.
Table 1.
Strains used in this study
| Name | Genotype | Reference |
|---|---|---|
| JEC21 | MATα | Kwon-Chung et al. (1992) |
| JEC21cir1Δ#46 | MATα, cir1Δ::NAT | Jung et al. (2006) |
| JEC21cir1Δ#57 | MATα, cir1Δ::NAT | Jung et al. (2006) |
| JEC20 | MATa | Kwon-Chung et al. (1992) |
| JEC20cir1Δ#7 | MATa, cir1Δ::NAT | This study |
| JEC20cir1Δ#37 | MATa, cir1Δ::NAT | This study |
RNA extraction and Northern blotting
Petri plates of solid V8 medium containing mating cultures or control cultures of single mating type cells were incubated at room temperature in the dark for 24 h. Cells were scraped off the plates and lyophilized for RNA extraction. Trizol (Invitrogen) was used for total RNA extraction following the manufacturer’s recommendations. Northern blotting was performed as described by Sambrook et al. (1989) with five μg of total RNA from each strain. Hybridization probes were designed for genes from each mating type and were amplified separately by PCR with the primers listed in Table 2. A Rediprime II random prime labeling system (GE Healthcare) was used for probe labeling, and the membrane was exposed to a Phosphor Screen (GE Healthcare) for 16 h, followed by scanning using a Pharos FX™ Plus Molecular Imager (Bio-Rad).
Table 2.
Primers used in this study
| Name | Sequence |
|---|---|
| J2CIR-KO-F | GCGCAGTTCTGTCGATCGTCCCGAATTG |
| J2CIR-KO-R | CCGATTTTCGAACACTTCCAGTACATCC |
| MFα-F | TCACTGCCATCTTCACCACCTTCA |
| MFα-R | GATGACACAAAGGGTCATGCCACC |
| MFa-F | TCACTGCTACCTTCTCAACCCTTT |
| MFa-R | AACGCAAGAGTAAGTCGGGCCCT |
| J2GPD1-F | ACACATGGTCGCTTCAAGGGCTCCG |
| J2GPD1-R | ACATAGGAGCATCAGCAGAAGGAGCG |
Mutant construction
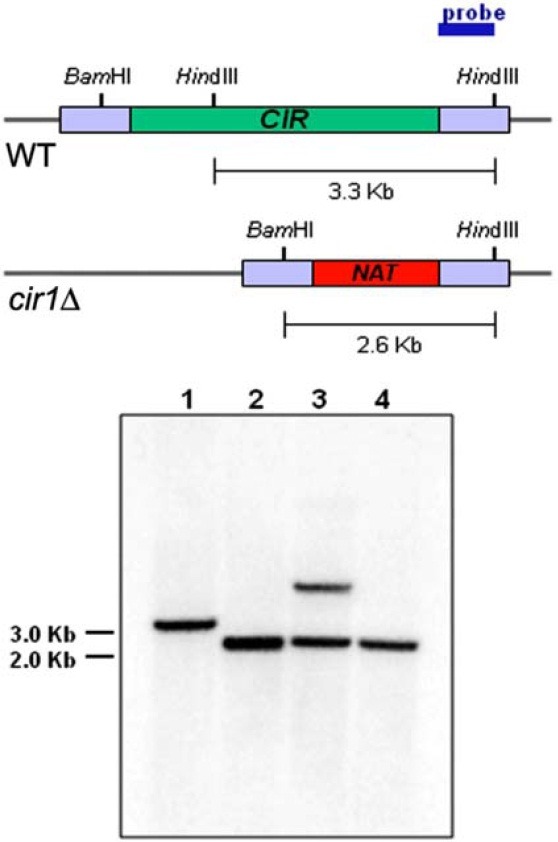
A disruption cassette was employed to construct cir1 mutants of the MATa strain. The cassette contained the nourseothricin acetyltransferase gene (NAT) and 5′ and 3′ flanking sequences of CIR1 amplified by PCR using primers J2CIR-KO-F and J2CIR-KO-R (Table 2), and the plasmid pWH008 as a template (Jung et al., 2006). The disruption cassette was biolistically transformed into the JEC20 wild-type strain (Table 1) (Toffaletti et al., 1993). Positive transformants were identified by PCR and confirmed by Southern blot analysis (Fig. 1). Two independent mutants were selected and used throughout the study. Surface reductase activity of the mutant cells was evaluated by the 2,3,5-triphenyltetrazolium chloride (TTC, Sigma) overlay method, as described previously (Hassett and Kosman, 1995; Ogur et al., 1957).
Fig. 1. Disruption of CIR1 in the MATa strain was confirmed by Southern blot analysis. Restriction maps of the genomic regions containing the wild-type or the disrupted CIR1 allele are shown. Genomic DNA of the serotype D MATa strain (JEC20) was digested with BamHI/HindIII and hybridized with the probe indicated. Lane 1 contains DNA from the serotype D wild-type strain JEC20. Lanes 2 and 4 contain DNA from the cir1 mutant strains and show that the disruption cassette integrated at the CIR1 locus. The mutants represented by lanes 2 and 3 were designated as cir1Δ#7 and cir1Δ#37, respectively. Lane 3 contains DNA from a transformant that has an ectopic integration of the disruption cassette.
RESULTS AND DISCUSSION
Construction of a serotype D MATa cir1 mutant
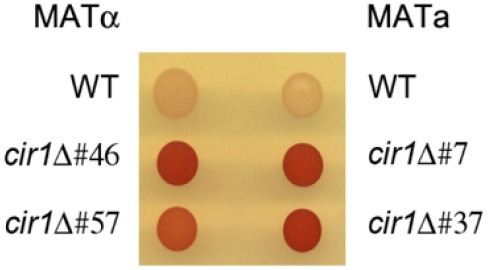
We previously constructed the cir1 mutation in MATα cells to study the iron regulatory roles of Cir1 in the serotype D background of C. neoformans (Jung et al., 2006). In this paper, we designate this mutant as cir1αΔ to indicate that the mutation is in the MATα mating type. In the current study, we also deleted CIR1 in a MATa strain to investigate the influence of Cir1 on mating. Two independent cir1 mutants were constructed in MATa cells (designated cir1aΔ) (Materials and Methods), and confirmed by Southern blot analysis (Fig. 1). These cir1aΔ mutants (JEC20cir1Δ#7 and JEC20cir1Δ#37) and two independent cir1αΔ mutants (JEC21cir1Δ#46 and JEC21cir1Δ#57) from our previous study were used in all subsequent mating experiments (Jung et al., 2006). Initially, we confirmed that deletion of CIR1 in MATa cells resulted in iron-related phenotypes by performing a 2, 3, 5-triphenyltetrazolium chloride (TTC) overlay assay to assess surface ferric reductase activity. We previously found that this activity is higher in the cir1αΔ mutants compared to the wild-type strain (Jung et al., 2006). As with the cir1αΔ mutants, we found elevated reductase activity for the cir1aΔ mutants, thus confirming loss of Cir1 function in MATa cells (Fig. 2).
Fig. 2. Disruption of CIR1 in the MATa strain caused elevated cell surface reductase activity. Both MATα cir1 mutants and MATa cir1 mutants displayed increased cell surface reductase activity, as indicated by the red colony color in the presence of TTC (Materials and Methods). The two independent mutants in each mating type displayed identical phenotypes.
Influence of Cir1 on mating in the presence and absence of copper
The ability of cir1 mutants to mate was tested on V8 medium and we found that mating mixtures involving cir1αΔ (αΔ × a, αΔ × aΔ) showed significantly reduced mating filamentation when compared with mixtures of wild-type strains (Figs. 3A and 3B). To rule out the possibility of a growth deficiency of the cir1αΔ or cir1aΔ mutants on V8 medium, the same assay plates were incubated up to seven days and the mixtures were again observed for mating filaments. Prolonged incubation did not remedy the filamentation defect observed in mixtures containing the cir1αΔ mutants (αΔ × a, αΔ × aΔ), suggesting that Cir1 indeed plays a role in mating in MATα cells. The cir1aΔ mutants showed no distinguishable mating phenotype, in contrast with the cir1αΔ mutants. That is, the mixtures containing the cir1aΔ mutants and the wild-type MATα cel ls (α × aΔ) showed wildtype levels of mating filamentation on V8 medium (Fig. 3A). Taken together, these results indicated that loss of Cir1 caused a unilateral mating defect in MATα cells, but not in MATa cells. Unilateral mating responses and difference between cells of opposite mating type have been reported previously for C. neoformans. For example, overexpression of the G-protein β subunit Gpb1 triggers the formation of more mating conjugation tubes in MATa cells than in MATα cells (Wang et al., 2000). Additionally, Wickes et al. (1996) found that only MATα strains display filamentous growth associated with monokaryotic fruiting. Therefore, we hypothesize that Cir1 is one of a number of regulatory functions that govern mating filamentation in a cell type-specific manner in C. neoformans.
Fig. 3. Cir1 in the MATα strain influences mating filament formation. Mixtures of cultures from the following strains were carried out on solid V8 medium, V8 medium containing 100 μM FeCl3 or V8 medium containing 100 μM CuSO4: MATα wild-type and MATa wild-type (α × a), MATα cir1Δ#46 and MATa wild-type (αΔ × a), MATα wild-type and MATa cir1Δ#7 (α × aΔ), MATα cir1Δ#46 and MATa cir1Δ#7 (αΔ × aΔ). (A) Each panel represents the periphery of a mixed colony. (B) Higher magnification (× 400) revealed significantly reduced levels of filamentation from a mixture containing MATα cir1Δ. (C) The mixture containing independently generated MATα cir1 mutants (46 and 57) and MAT a cir1 mutants (37) displayed a lack of filamentation at the colony periphery.
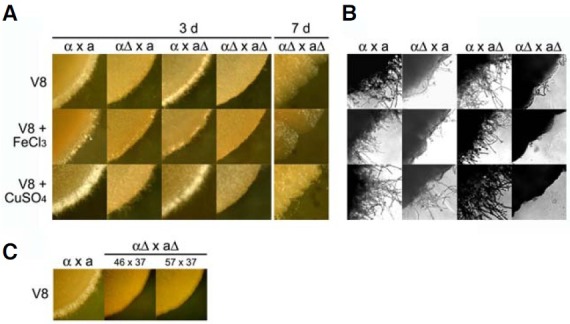
Kent et al. (2008) previously developed a defined V8 medium to examine the role of different components, and they demonstrated that the inclusion of copper is important to achieve robust mating. We therefore tested the effect of copper on mating mixtures with the cir1 mutants to investigate whether Cir1 mediates the response. We also tested the influence of iron because of the prominent role of Cir1 in iron homeostasis (Jung et al., 2006). Iron or copper was added to V8 medium and mating was assayed by observing filamentation. As expected from the work of Kent et al. (2008), we found an overall enhancement of mating filamentation by exogenously added copper, but there was no significant influence of iron (Fig. 3A). The level of filamentation for a unilateral mating mixture containing the cir1αΔ mutants (αΔ × a) was also enhanced to a small extent by copper addition (Figs. 3A and 3B). In contrast, the mixture with a bilateral defect in Cir1 (αΔ × aΔ) was not responsive to copper addition, thus indicating a role for Cir1.
It is possible that differential expression of genes required for copper homeostasis in the cir1 mutant background may explain the influence of Cir1 on copper stimulation of filamentation. Previous transcriptome analysis with the cir1αΔ mutant demonstrated differential regulation of genes involved in copper uptake (Jung et al., 2006). For example, the transcript for the copper exporting ATPase Ccc2 was 8.82-fold up-regulated in the mutant compared to the wild-type in the low-iron condition. The transcript for the copper uptake transporter Ctr4 was 4.86-fold up-regulated under this condition. The V8 mating medium contains a relatively low level of iron (~1 μM) (Kent et al., 2008). It is therefore possible that the regulatory influence of Cir1 on copper homeostasis functions reduces the intracellular accumulation that would otherwise stimulate mating. The absence of an influence of iron is consistent with the findings of Kent et al. (2008) that iron was not a key component of their defined V8 mating medium.
Cir1 regulates genes involved in mating and sexual development
To further investigate the role of Cir1 in the early stages of sexual development, we determined the expression of genes encoding the mating pheromones (MFα and MFa) in cir1 mutants. Transcript levels of the genes in mating mixtures containing wild-type cells and/or cir1 mutants were analyzed by Northern hybridization (Fig. 4). Our results showed that the transcript level of MFα was increased on V8 medium in mixtures of the cir1Δα mutant with the wild-type MATa strain or the cir1Δa mutant (αΔ × a and αΔ × aΔ), compared with cultures of single wild-type or mutant cells (Fig. 4). Interestingly, the mixture containing the cir1Δa mutant also displayed increased expression of MFα (α × aΔ). This result suggests that deletion of CIR1 in the MATa cells may increase the expression of secreted pheromone (or another unidentified factor) to induce transcription of MFα in MATα cells. The involvement of Cir1 in expression of MFa was also observed in that the mixtures containing the cir1Δα mutants showed increased transcript levels of MFa (αΔ × a and αΔ × aΔ). This result indicated that deletion of CIR1 in the MATα cells triggers increased transcription of MFa in MATa cells. We did note that, in contrast to the situation with MFα upon deletion of CIR1 in MATα cells, elevated MFa transcription was not observed in the mixture containing the cir1Δa mutant (α × aΔ). This result further reinforces the difference in the relative influence of the cir1 mutation for MATα and MATa cells, as observed for mating filament formation (Fig. 3).
Fig. 4. Cir1 influences of the transcript levels for pheromone genes. RNA was extracted from either mixtures of cells undergoing mating or haploid cells on V8 medium or V8 medium containing 100 μM CuSO4. Northern blot analysis was performed to assess the transcript levels of genes encoding the mating pheromones MFα and MFa.
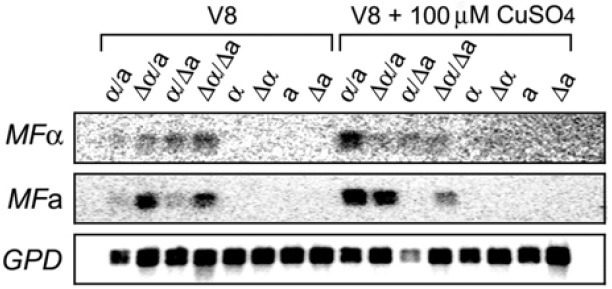
As observed by Kent et al. (2008) our analysis also revealed that addition of copper significantly increased expression of MFα in mixtures containing wild-type cells (Fig. 4, α × a in V8 + CuSO4 versus V8). However, the same was not true in mixtures containing cir1Δα and/or cir1Δa mutants (αΔ × a,215; aΔ or αΔ × aΔ) thus indicating that Cir1 is required for the elevated transcript level of MFα that results from copper addition. As mentioned above for the previous transcriptome analysis (Jung et al., 2006), this result could represent an indirect influence on the expression of genes required for copper homeostasis in the cir1 mutants. Addition of copper increased the transcript level of MFa in mixtures with wild-type strains (α × a, in the presence of CuSO4), as was found with MFα. However, the copper response was not observed for the MFa transcript in the cir1Δa mutant mixed with a wild-type partner (α × aΔ, in the presence of CuSO4 versus V8 alone). This result suggested that the cir1Δa mutants may be deficient in copper uptake or perception. Taken together, our data indicated that Cir1 generally has a negative influence on the expression of the pheromone transcripts in both mating types in V8 medium. The situation appears to be more complex in V8+ CuSO4 where Cir1 may make a positive contribution to the influence of copper. Overall, the reduction in mating filamentation in the cir1 mutants suggested that Cir1 is required for the morphological transition during early sexual development in both MATα and MATa cells. Our hypothesis is well supported by recent findings that suggest temporally distinct pathways during mating filamentation in C. neoformans. Specifically, Stanton et al. (2010) reported that pheromones and pheromone receptors play a major role in mating partner recognition during the initial period of sexual development, and that the transcription factor Sxi2a is required for subsequent filamentation and spore formation.
Our data are consistent with the view that Cir1 influences sexual development via regulation of mating pheromone expression, and it may also contribute to the morphological transition resulting in filamentation. Moreover, we found that Cir1 plays a role in the influence of copper on mating by a mechanism that likely involves regulation of metal homeostasis. The influence of Cir1 appears to be more substantial in MATα cells than in MATa cells, for unknown reasons. One possibility is that Cir1 may differentially influence the expression or the activity of the a- and α-specific homeodomain transcription factors (Sxi2a and Sxi1α) that negatively regulate the pheromone genes (Hull et al., 2005). As suggested by our previous study, differential expression of various components in the cAMP pathway and the MAP kinase pathway may also contribute to the deficiency of the cir1 mutant in sexual development. It is possible that the influence of signaling components, copper, and Cir1 on mating are interconnected. Overall, these studies provide new insights into environmental factors (such as copper) that influence mating in C. neoformans by revealing coordination with the extensive Cir1 regulatory network for metal homeostasis and virulence.
Acknowledgments
The authors thank Guanggan Hu, Cletus D’Souza and Joyce Wang for critical reading of the manuscript. This work was supported by awards from the National Institutes of Health R01 AI053721 (JWK), the Canadian Institutes of Health Research (JWK) and Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (2010-0004226) (WHJ). JWK is a Burroughs Wellcome Fund Scholar in Molecular Pathogenic Mycology.
References
- 1.Alspaugh J.A., Perfect J.R., Heitman J. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev. (1997);11:3206–3217. doi: 10.1101/gad.11.23.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bicanic T., Harrison T. S. Cryptococcal meningitis. Br. Med. Bull. (2004);72:99–118. doi: 10.1093/bmb/ldh043. [DOI] [PubMed] [Google Scholar]
- 3.Erke K.H. Light microscopy of basidia, basidiospores, and nuclei in spores and hyphae of Filobasidiella neoformans (Cryptococcus neoformans) J. Bacteriol. (1976);128:445–455. doi: 10.1128/jb.128.1.445-455.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feldbrugge M., Kamper J., Steinberg G., Kahmann R. Regulation of mating and pathogenic development in Ustilago maydis. Curr. Opin. Microbiol. (2004);7:666–672. doi: 10.1016/j.mib.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 5.Hassett R., Kosman D.J. Evidence for Cu(II) reduction as a component of copper uptake by Saccharomyces cerevisiae. J. Biol. Chem. (1995);270:128–134. doi: 10.1074/jbc.270.1.128. [DOI] [PubMed] [Google Scholar]
- 6.Hsueh Y.P., Heitman J. Orchestration of sexual reproduction and virulence by the fungal mating-type locus. Curr. Opin. Microbiol. (2008);11:517–524. doi: 10.1016/j.mib.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hull C.M., Boily M.J., Heitman J. Sex-specific homeodomain proteins Sxi1alpha and Sxi2a coordinately regulate sexual development in Cryptococcus neoformans. Eukaryot. Cell. (2005);4:526–535. doi: 10.1128/EC.4.3.526-535.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Idnurm A., Bahn Y.S., Nielsen K., Lin X., Fraser J.A., Heitman J. Deciphering the model pathogenic fungus Cryptococcus neoformans. Nat. Rev. Microbiol. (2005);3:753–764. doi: 10.1038/nrmicro1245. [DOI] [PubMed] [Google Scholar]
- 9.Jung W.H., Sham A., White R., Kronstad J.W. Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans. PLoS Biol. (2006);4:e410. doi: 10.1371/journal.pbio.0040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kent C.R., Ortiz-Bermudez P., Giles S.S., Hull C.M. Formulation of a defined V8 medium for induction of sexual development of Cryptococcus neoformans. Appl. Environ. Microbiol. (2008);74:6248–6253. doi: 10.1128/AEM.00970-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwon-Chung K.J., Edman J.C., Wickes B.L. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect Immun. (1992);60:602–605. doi: 10.1128/iai.60.2.602-605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee S.C., Ni M., Li W., Shertz C., Heitman J. The evolution of sex: a perspective from the fungal kingdom. Microbiol. Mol. Biol. Rev. (2010);74:298–340. doi: 10.1128/MMBR.00005-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lengeler K.B., Davidson R.C., D’Souza C., Harashima T., Shen W.C., Wang P., Pan X., Waugh M., Heitman J. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. (2000);64:746–785. doi: 10.1128/mmbr.64.4.746-785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ogur M., St. John R., Nagai S. Tetrazolium overlay technique for population studies of respiration deficiency in yeast. Science. (1957);125:928–929. doi: 10.1126/science.125.3254.928. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J., Fritch E.F., Maniatis T. Molecular Cloning, a laboratory manual. Cold Spring Harbor Laboratory Press; New York, USA: (1989). [Google Scholar]
- 16.Stanton B.C., Giles S.S., Staudt M.W., Kruzel E.K., Hull C.M. Allelic exchange of pheromones and their receptors reprograms sexual identity in Cryptococcus neoformans. PLoS Genet. (2010);6:e1000860. doi: 10.1371/journal.pgen.1000860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toffaletti D.L., Rude T.H., Johnston S.A., Durack D.T., Perfect J.R. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. (1993);175:1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang P., Heitman J. Signal transduction cascades regulating mating, filamentation, and virulence in Cryptococcus neoformans. Curr. Opin. Microbiol. (1999);2:358–362. doi: 10.1016/S1369-5274(99)80063-0. [DOI] [PubMed] [Google Scholar]
- 19.Wang P., Perfect J.R., Heitman J. The G-protein beta subunit GPB1 is required for mating and haploid fruiting in Cryptococcus neoformans. Mol. Cell. Biol. (2000);20:352–362. doi: 10.1128/mcb.20.1.352-362.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wickes B.L., Mayorga M.E., Edman U., Edman J.C. Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the alpha-mating type. Proc. Natl. Acad. Sci. USA. (1996);93:7327–7331. doi: 10.1073/pnas.93.14.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]


