Abstract
Cell death is an important process of plant responses to development and biotic/abiotic stresses. In rice plants, PBZ1, a PR10 family protein, has been shown to accumulate in tissues undergoing cell death. However, the function of PBZ1 in cell death remains yet to be demonstrated. Here, we report that exogenous recombinant PBZ1 protein induces cell death in rice suspension-cultured cells (SCCs) and also in leaves of Nicotiana tabacum in a dosedependent manner. This finding was confirmed in vivo in transgenic Arabidopsis lines harboring the PBZ1 gene under the control of a dexamethasone (DEX)-inducible promoter. The DEX-treated leaves of transgenic Arabidopsis induced expression of PBZ1 at transcript and protein levels and showed cell death morphology. TUNEL analysis detected DNA fragmentation, a hallmark of programmed cell death, in rice SCCs treated with the PBZ1 protein. Recombinant PBZ1 protein also exhibited RNase activity and exhibited internalization inside BY-2 cells. Taken together, PBZ1 induces cell death not only in rice, but also in tobacco and Arabidopsis via its RNase activity inside the cell. PBZ1 could be used as a marker to understand the mechanism by which PBZ1 confers the cell death morphology in rice and other model plants.
Keywords: dexamethasone, PBZ1, PR-10 protein family, programmed cell death, RNase activity, TUNEL
INTRODUCTION
Probenazole (Oryzemate®, 3-allyloxy-l,2-benzisothiazole-1,1- dioxide), a well known effective fungicide, incites plants to resist rice blast fungus by accumulating salicylic acid (SA) and inducing pathogen-related (PR) proteins in plants (Nakasita et al., 2002; Takayoshi et al., 2007). The rice PBZ1 was the first identified as probenazole-inducible gene (Midoh and Iwata, 1996). The PBZ1 protein belongs to the PR-10 family, proteins of which are highly conserved across the plant kingdoms (Liu and Ekramoddoullah, 2006). Sequence homology between ginseng ribonuclease (RNase) and PR-10 proteins could be used to predict RNase activity for a PR-10 protein (Moiseyev et al., 1997). To date, a few PR-10 proteins have been shown to possess the RNase activity as demonstrated in birch pollen (Bufe et al., 1996; Swoboda et al., 1996), white lupin (Bantignies et al., 2000), cotton (Zhou et al., 2002) and pepper (Park et al., 2004). More recently, the rice pathogen-related protein 10 (termed JIOsPR10; Jwa et al., 2001) was found to have the RNase activity (Kim et al., 2008b).
PBZ1 was highly responsive to rice blast fungus and Xanthomonas oryzae infection in rice suspension-cultured cells (SCCs) and leaves, and that expressed earlier in an incompatible than compatible interaction (Kim et al., 2003; 2004). Furthermore, predominantly expressed multiple isoforms of the PBZ1 protein were detected in response to rice blast fungus and elicitors using a gel-based proteomics approach (Kim et al., 2003). Although identity and function of these isoforms have yet to be determined, it is likely that PBZ1 is a target of posttranslational modification (Kim et al., 2003).
The PBZ1 gene was also shown to be highly responsive to abiotic stresses in rice, such as cold stress and phytohormones (reviewed in Jwa et al., 2006). Using the gibberellin (GA)- insensitive dwarf mutant (gid1), PBZ1 was revealed to have increased response to exogenous GA, which helps tolerance to cold stress and pathogen attack (Tanaka et al., 2006). Jasmonic acid, protein phosphatase 2A inhibitors, cantharidin, and abscisic acid induced the PBZ1 expression in rice (Rakwal et al., 2001). Interestingly, SA activated the PBZ1 expression in rice leaves in the light but suppressed by dark in rice SCCs (Hwang et al., 2008; Kim et al., 2003; 2004; Rakwal et al., 2001).
In addition to induced accumulation of PBZ1 in rice and its SCCs in response to various biotic and abiotic factors, proteomics analysis of the rice spl1 lesion mimic mutant demonstrated a tight correlation between the expressed PBZ1 protein and programmed cell death (PCD) (Kim et al., 2008a). In that study, the PBZ1 protein expressed and accumulated in leaf senescence, root caps, aleurone layers, coleoptiles, and hypersensitive tissues infected with pathogen. PBZ1 tightly colocalized in every PCD tissues, and has been proposed as a PCD marker protein in rice. Despite these findings, very few biochemical studies have been conducted for understanding the function of PBZ1 in plant defense system. In this study, we provide a number of convincing evidences to demonstrate that PBZ1 induces cell death in rice and other model plants possibly via its RNase activity.
MATERIALS AND METHODS
Plant materials and growth conditions
Mature rice seeds (cv. Jinheung) were obtained from the National Yeongnam Agricultural Experimentation Station. Dehulled seeds were sterilized with 70% ethanol for 10 min, followed by 3% sodium hypochlorite for 30 min. Seeds were then rinsed extensively with sterile water. Seeds (4℃ for 48 h) were placed on the N6 medium containing 0.2% phytagel to induce callus formation (Ohira et al., 1973). The embryonic calli were used to establish SCCs in the N6 medium with constant shaking (120 rpm at 25℃) on a rotary shaker in the dark and subcultured in the same medium at weekly interval unless stated otherwise. Wild type tobacco plants (Nicotiana tabacum) were grown in soil in a growth chamber under a 16-h-light/8-h-dark photoperiod at 25℃. The tobacco BY-2 (N. tabacum cv bright yellow-2) suspension cell line was maintained according to the method described by Yang et al. (2004). Arabidopsis (Arabidopsis thaliana Col-0) plants carrying the PBZ1 gene (pTA7001:: PBZ1) and its wild type (control) were grown in soil in a growth chamber (120 μmol m-2 s-1 white light intensity, 16-h-light/8-hdark photoperiod and 22℃). Sterilized seeds of Arabidopsis were spread on the basal MS solid medium (1X Murashige and Skoog basal salt mixture, 2% Sucrose, 2.5 mM MES, pH 5.7, and 0.25% phytagel) and plates were incubated at 22℃ under 16-h-light/8-h-dark photoperiod.
Production of recombinant PBZ1 protein
The PBZ1 cDNA was amplified by PCR using a pair of sense primers (5′-CGGATCCATGGCTCCGGCCTGCGTC-3′) and antisense primers (5′-CATCTTAGGCGTATTCGGCAGGG-3′), which contained BamHI restriction sites (underline). The PCR product was subcloned into the pGEM-T vector (Promega, USA). The BamHI and Pst1 fragment of pGEM-T, possessing the open reading frame of PBZ1, was cloned in-frame into the pQE30 vector (Qiagen, USA) at the C-terminal 6xHis tag to produce recombinant 6xHis-tagged PBZ1 protein. E. coli cells harboring the pQE30::PBZ1 plasmid were grown in the LB medium at 30℃. Once the cell density at OD600 reached to 0.6, IPTG was added to a final concentration of 1 mM and continued to culture for another 5 to 6 h. Recombinant PBZ1 protein was purified from E. coli cells using a Ni2+-NTA purification kit according to the supplier’s instructions (Qiagen, USA).
Production of Arabidopsis transgenic lines
The 477-bp PBZ1 full-length cDNA was inserted into the binary transformation plasmid pTA7001, which contains the twocomponent glucocorticoid inducible system (Aoyama and Chua 1997). The constructed binary plasmid, termed pTA7001::PBZ1, was used to transform Arabidopsis (A. thaliana Col-0) using the Agrobacterium-mediated transformation method as described previously (Clough and Bent, 1998). Generated transgenic plants were screened for both hygromycin-resistant and the presence of PBZ1 gene. Northern and Western blot analyses of transgenic and wild type plants were also performed as mentioned above.
Evans and trypan blue staining
Leaves of Arabidopsis and its transgenic plants (four-week-old) were submerged in 0.05% Evans blue containing 50 mM HEPES-KOH (pH 7.2) for 15 min and then washed extensively with the same buffer to remove excess dye. Incorporated dye with dead cells was extracted with 50% ethanol containing 0.1% SDS at RT for 12 h and quantified spectrophotometrically at A595 (Turner and Novacky, 1974). To detect dead and dying cells, stained leaves were washed with 70% ethanol to remove Evans blue and chloroplast, followed by staining of leaves with trypan blue (Dangl et al., 1991).
Leaves of dexamethasone (DEX)-inducible PBZ1 transgenic and its wild type Arabidopsis plants (four-week-old) were also stained with lactophenol-trypan blue (10 ml of lactic acid, 10 ml of glycerol, 10 g of phenol and 10 mg of trypan blue dissolved in 10 ml of distilled water) by boiling them in the same staining solution for approximately 1 min (Keogh et al., 1980). Stained leaves were decolorized with chloral hydrate solution (2.5 g of chloral hydrate dissolved in 1 ml of distilled water) for at least 30 min, mounted and viewed under the microscope equipped with phase-contrast optics.
Northern blot analysis
Total RNA (20 μg) was separated on 0.8% formaldehydedenaturing agarose gels and blotted onto nylon membranes. The PBZ1 cDNA was labeled with [α-32P] dCTP using the Primer-a-Gene labeling system (Pomega, USA). The nylon membrane was hybridized with the PBZ1 probe overnight at 65℃, and washed once each with 2× SSC (300 mm NaCl, 30 mM Na citrate, pH 7.0) and 0.2× SSC solutions containing 0.1% SDS for 15 min at RT and at 65℃, respectively. Membranes were exposed to an X-ray film for 3 days at -80℃.
Western blot analysis
Total protein (20 μg) was separated by 12% SDS-PAGE and transferred to a PVDF membrane using a semidry electrophoretic apparatus (Hoefer, USA). The blotted membrane was incubated in 1× TTBs buffer (50 mM Tris-HCl, pH 8.2, 0.1% v/v Tween 20, and 150 mM NaCl) containing 7% skim milk at RT for 4 h to block non-specific binding, followed by incubation in the same buffer containing the purified PBZ1 polyclonal antibody (1:1000) for another 2 h. The membrane was washed three times (15 min each) with TTBs at RT. A secondary antirabbit IgG antibody conjugated with horseradish peroxidase (1:10,000 dilution in TTBs) was added and signals were detected using the ECL system (Perkin Elmer Life Sciences, USA).
DNA fragmentation assay
The rice SCCs were treated with the recombinant PBZ1 protein (10 μM), BSA (100 μg/ml), PBZ1 antibody (100 μl) or PBZ1 protein (10 μM) together with PBZ1 antibody (100 μl) for 24 h. Cultured cells were collected, frozen with liquid nitrogen and grounded to fine powder again with the help of liquid nitrogen using mortar and pestle. Extraction buffer (7 M Urea, 350 mM NaCl, 10 mM Tris-Cl pH 7.6, 20 mM EDTA, and 2% SDS) was added and incubated at 65℃ for 10 min to extract genomic DNA. Extracted genomic DNA was treated with phenol/chloroform/ isoamylalcohol (25:24:1 volume) and precipitated with isopropanol. Pellet thus obtained was washed with 70% ethanol, briefly incubated at room temperature (RT) to remove any residual ethanol and finally dissolved in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.4). Genomic DNA (each 10 μg) was separated on 1.5% agarose gel, stained with ethidium bromide and photographed under UV illumination.
TUNEL assay for nuclear DNA fragmentation
Rice cultured cells treated with the recombinant PBZ1 protein (10 μM) for 24 h were fixed with 2.5% paraformaldehyde in PBS and sectioned using a razor blade. TUNEL assay for nuclear DNA fragmentation was performed using the DIG DNA labeling and detection kit according to the manufacturer’s instructions (Roche Diagnostics, Germany). Stained sections were observed under a bright-field light microscope and immediately photographed. For nuclear staining, the fluorescent dye DAPI (100 μg/ml) was used. Samples were examined using Laser-Scanning Confocal Microscopy (Olympus Co. Ltd., Japan) equipped with GFP and DAPI filter.
Internalization and localization of the FITC-labeled PBZ1 protein
The BSA and recombinant PBZ1 proteins were labeled with FITC (fluorescein isothiocyanate) using a labeling kit as per the manufacturer’s instructions (Peirce, USA). Tobacco BY-2 SCCs were co-cultured with FITC-labeled protein for its internalization and localization analyses. For localization, co-cultured cells were stained with 4 μM endocytic tracer FM4-64. An Olmypus confocal laser-scanning microscopy was used to observe and record fluorescence signals. The excitation wavelength was 488 nm; emission was detected for GFP between 500 and 530 nm and for FM4-64 between 620 and 680 nm.
RNase activity assay
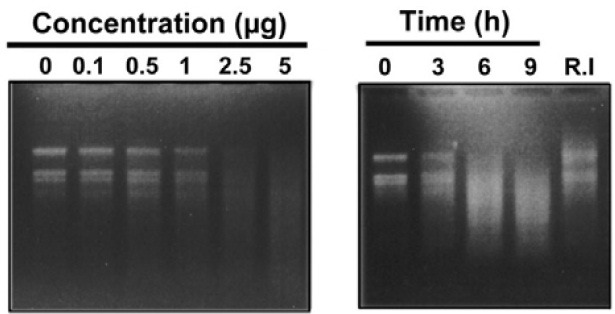
RNase activity assay of the recombinant PBZ1 protein was performed using the total RNA (5 μg) isolated from rice leaf as described previously (Kim et al., 2008b). Briefly, different concentrations of the PBZ1 protein (0.1, 0.5, 1, 2.5 and 5 μg) were incubated with total RNA at RT for 4 h in case of dosedependent experiment. For time-course experiment, the PBZ1 protein (2.5 μg) was incubated with total RNA at RT for 3, 6 and 9 h. BSA (10 μg/ml) and the protein purified using Ni2+-NTA resin fractions from a culture of E. coli lacking the PBZ1 cDNA clone were separately incubated with total RNA to serve as appropriate controls. RNase inhibitor (50 units) was also used as a control to monitor its effect on the activity of PBZ1 protein. For this purpose, the PBZ1 protein was first incubated with RNase inhibitor, followed by addition of total RNA. These incubated samples were extracted with phenol-chloroform to remove protein. Samples thus prepared were loaded and separated on 0.8% formaldehyde-denaturing agarose gel. Gels were stained with ethidium bromide, exposed to UV light and photographed.
RESULTS AND DISCUSSION
Recombinant PBZ1 protein causes cell death in rice SCCs and tobacco leaves
The recombinant PBZ1 protein was over-expressed in E. coli and purified using the Ni2+ affinity column (Supplementary Fig. 1). The purified recombinant PBZ1 protein was used to raise PBZ1 polyclonal antibody for conducting all biochemical studies, unless otherwise stated. The purity and sensitivity of PBZ1 antibody were also confirmed by Western blot analysis (data not shown).
To investigate whether the PBZ1 protein induces cell death, rice SCCs were first stained with Evans blue (Fig. 1). For this purpose, rice SCCs were co-incubated for 24 h with PBZ1 protein, PBZ1 protein with purified PBZ1 antibodies, or PBZ1 antibodies alone, respectively. Results showed that suspended cells treated with PBZ1 protein turned into the dark brown color (i.e. cell death), whereas cells treated with others remained light yellow, suggesting that PBZ1 protein induces cell death in rice SCCs (Fig. 1A). The pre-immune serum did not cause any cell death (Fig. 1A). To know the sensitivity of PBZ1 protein in inducing cell death, dose- and time-dependent experiments were performed (Figs. 1B and 1C). Cell death sensitivity was determined by staining the treated cells with Evans blue. Compared to control (BSA), cell death increased almost exponentially with increased concentration of the PBZ1 protein (Fig. 1B). Moreover, increase in cell death was also dependent on posttreatment time, where cell death started to increase after 6 h and continued exponentially till 24 h. Hence, PBZ1 induces cell death in a dose- and time-dependent manner.
Fig. 1. Rice suspension-cultured cells exhibit cell death upon treatment with recombinant PBZ1 protein. Degree of cell death-induced by the PBZ1 protein was determined by staining them with Evans blue for 24 h, followed by spectrophotometric quantification at A595. (A) Morphology of cell death. Dark brown color serves as an indication of cell death. (B) Effect of increasing concentration of PBZ1 protein (1, 2.5, 5 and 10 μM) on cell death. BSA (100 μg/ml) represents appropriate control. (C) Effect of prolonged incubation (0, 6, 12, 18 and 24 h) of cultured cells with PBZ1 protein (100 μg/ml) on cell death. (D) Cell death blocking assay. Effect of increasing amount of PBZ1 antibody (0, 5, 10, 50 and 100 μl) on cell deathinduced by PBZ1 protein (100 μg/ml) was monitored. Each bar represents S.E. of three independent experiments.
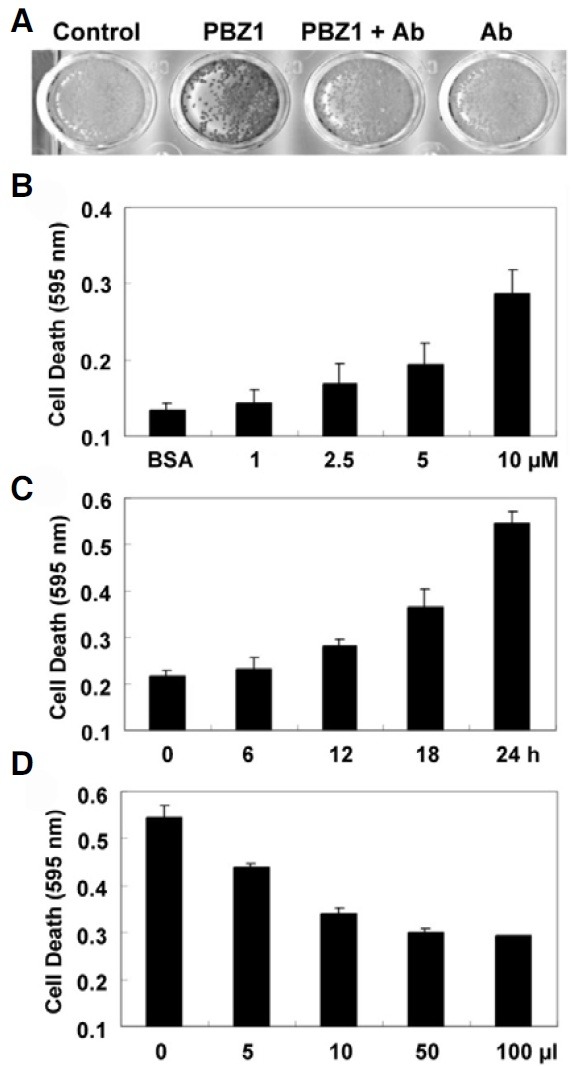
Cell death blocking assay was carried out using PBZ1 antibody. In such a situation, interactions between recombinant PBZ1 protein and PBZ1 antibody will result in reduced amount of the free PBZ1 protein in the medium, and hence show a decrease in cell death. To do this, increasing amount of PBZ1 antibody (0, 5, 10, 50 and 100 μl) was first co-incubated with recombinant PBZ1 proteins (100 μg/ml) and then added into rice SCCs. As depicted in Fig. 1D, PBZ1 antibody strongly inhibited phenomenon of the PBZ1-induced cell death in a dosedependent manner (Fig. 1D).
To confirm above results in planta, leaves of tobacco (Nicotiana tabacum) were infiltrated with various concentrations of recombinant PBZ1 protein alone or together with PBZ1 antibody (Fig. 2). Cell death morphology was clearly visible within 72 h post application (Fig. 2A). The amount of PBZ1 as low as 50 μg/ml was effective enough to cause apparent cell death (Fig. 2A; Number 4). No cell death was detected with application of two different controls [HEPES buffer (1) or 100 μg/ml BSA (2) of Fig. 2A]. Interestingly, co-infiltration of the recombinant PBZ1 protein (100 μg/ml) along with PBZ1 antibody (10 μl) dramatically reduced the size of cell death (number 4 of Fig. 2B) compared to infiltration of the PBZ1 protein alone (number 3 of Fig. 2B). These results indicate that the recombinant PBZ1 protein induces cell death in rice SSCs and in planta in leaves of tobacco.
Fig. 2. Infiltration of the recombinant PBZ1 protein causes cell death in tobacco leaves. Tobacco leaves were infiltrated on an abaxial side using a needless syringe and morphological observation was recorded at 72 h post application. (A) Doesdependent effect of PBZ1 protein on cell death. 1, 10 mM HEPES buffer (pH 7.0); 2, acetylated BSA (100 μg/ml); 3 through 6 are recombinant PBZ1 protein at the concentration of 10, 25, 50 and 100 μg/ml, respectively. (B) Cell death blocking experiment. 1, acetylated BSA (100 μg/mL); 2, PBZ1 antibody (10 μl); 3, PBZ1 protein (100 μg/ml); and 4, PBZ1 protein (100 μg/ml) plus PBZ1 antibody (10 μl). Numbers 1 and 2 serve as appropriate controls.

Enhanced expression of PBZ1 triggers cell death also in Arabidopsis
To investigate whether PBZ1 induces cell death in vivo, transgenic Arabidopsis plants over-expressing the PBZ1 gene under the control of a constitutive promoter were generated. Harvested seeds were selected on kanamycin selection medium. However, none of the seeds was survived after germination, suggesting that constitutive expression of PBZ1 might have a detrimental effect on seedling establishment. This result led us to use an inducible promoter to generate transgenic Arabidopsis plants.
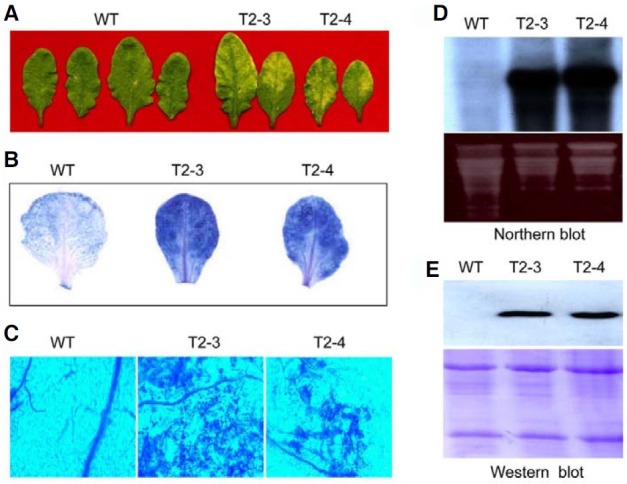
To do so, independent transgenic lines of Arabidopsis were generated that harbored the PBZ1 gene under the control of DEX-inducible promoter as a strong synthetic glucocorticoid, which can be induced by application of DEX solution. Twenty μM of DEX solution was applied to the leaf surface of both wild type and transgenic plants. As shown in Fig. 3A, two independent transgenic lines (T2-3 and T2-4) manifested a cell death-like symptoms (leaf senescence as yellow color), but not the wild type. Even DEX application had no effect on wild type leaves (Fig. 3A). Staining of these leaves with trypan blue displayed occurrence of cell death in the mesophyll cells of transgenic leaves (Figs. 3B and 3C). Northern and Western blot analyses confirmed strong accumulation of PBZ1 at RNA and protein levels in DEX-treated leaves of transgenic lines but not wild type (Figs. 3D and 3E). Hence, in vivo accumulation of PBZ1 causes cell death in addition to its similar role in rice and tobacco.
Fig. 3. Induced expression of rice PBZ1 in Arabidopsis causes cell death. Leaves of transgenic (T2-3 and T2-4) and wild type Arabidopsis were infiltrated with DEX (20 μM) for 72 h; T2 stands for 2nd generation whereas the number 3 and 4 are two independent lines. Treated leaves were used for observing cell death morphology and confirmation of enhanced expression of PBZ1. (A) Cell death symptom on leaves as senescence. (B) Binding of trypan blue with cell death tissue of leaves. Senescence leaves of T2-3 and T2-4 stain strongly with trypan blue compared to wild type (WT). (C) Observation of leaves stained with trypan blue under microscopy. (D) Northern blot analysis of total RNA isolated from leaves. (E) Western blot analysis of total protein isolated from leaves.
Rice cultured cells-treated with the PBZ1 protein reveal DNA fragmentation – a sign of PCD
Since DNA fragmentation is a hallmark of PCD (Bröker et al., 2005), it was monitored on genomic DNA isolated from suspended cells treated with BSA, PBZ1 antibody, PBZ1 protein together with anti-PBZ1 or PBZ1 protein for 24 h (Fig. 4). Results showed clear DNA fragmentation by the PBZ1 protein treatment (Fig. 4A; lane 5), and that was dramatically suppressed due to co-treatment with PBZ1 antibody (Fig. 4A; lane 4). No DNA fragmentation was observed with BSA or PBZ1 antibody (Fig. 4A; lanes 2 and 3). DNA fragmentation was originally described during apoptosis in animal cells (Wyllie et al., 1980). In plant cells, DNA fragmentation was documented in tobacco BY-2 cells undergoing PCD in response to abiotic stress (Koukalová et al., 1997) including CdSO4 treatment (Fojtová and Kovařík, 2000).
Fig. 4. DNA fragmentation and TUNEL analyses of rice SCCs-treated with recombinant PBZ1 protein. (A) DNA fragmentation assay. Rice SCCs were treated with BSA (control; 100 μg/ml), antibody (100 μl anti- PBZ1; control), PBZ1 + antibody [the PBZ1 protein (10 μM) and anti-PBZ1 (100 μl)] or PBZ1 [the PBZ1 protein (10 μM)]. M stands for DNA marker (1.5 kb DNA ladder). (B) TUNEL assay to determine nuclear DNA fragmentation. Rice cultured cells treated for 24 h were used for TUNEL assay. The fluorescent dye DAPI was used for nuclear staining. Control represents the PBZ1 protein (10 μM) plus PBZ1 antibody (100 μl).
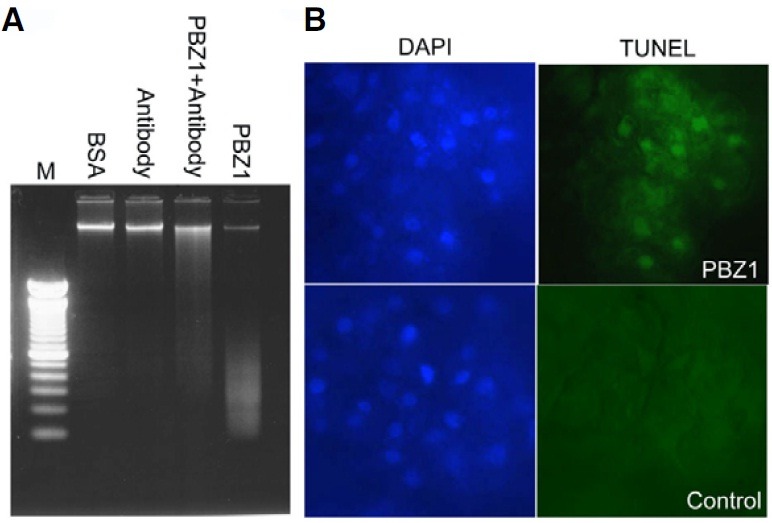
The TUNEL assay was also applied to independently determine PBZ1-induced nuclear DNA fragmentation in rice SCCs. As shown in Fig. 4B, results of TUNEL-positive signals correlated well with that of DNA fragmentation obtained from electrophoretic analysis. Again, PBZ1 antibody used as a negative control showed no positive signal for DNA fragmentation (data not shown). Previously, accumulation of the PBZ1 protein was reported to coincide well with the seed aleurone layer showing TUNEL signal (Kim et al., 2008). Therefore, the PBZ1 protein causes DNA fragmentation in rice and that is a recognized sign of PCD also in plants.
Internalization of the FITC-labeled recombinant PBZ1 protein
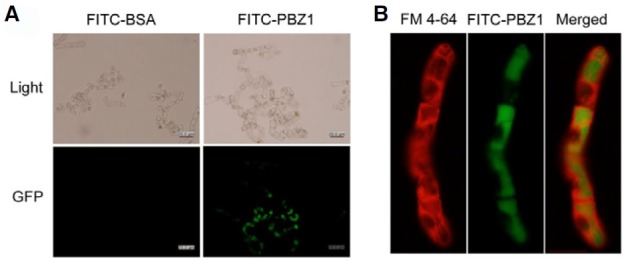
To investigate how exogenous PBZ1 induces cell death, we carried out the internalization of FITC-labeled recombinant PBZ1 protein (FITC-PBZ1) using the tobacco BY-2 cell by confocal microscopy. When FITC-PBZ1 was incubated with BY-2 cells for 12 h, an intense fluorescent signal was observed inside in almost all cells (Fig. 5A; FITC-PBZ1). The fluorescent signal became more intense as the time passed (data not shown). No fluorescent signal was detected in control (Fig. 5A; FITC-BSA). This result indicates that FITC-PBZ1 is effectively translocated inside into the intact BY-2 cells. To confirm the internalization of FITC-PBZ1, the plasma membrane of BY-2 cells were further labeled with an endocytic tracer fluorescent dye FM4-64, which showed red fluorescent signal (Fig. 5B). PBZ1 protein labeled with FITC was detected as green fluorescent (Fig. 5B). Merged data showed that PBZ1 was internalized into the cytoplasm, indicating that PBZ1 is likely to function inside the cell.
Fig. 5. Internalization and localizatio n analyses of BY-2 cells incubated with FITC-PBZ1. (A) Internalization analysis of FITC-PBZ1 or FITC-BSA (control). Images of light and fluore scence signals were observed by Co nfocal microscopy. Scale bars are 20 μm. (B) Localization of FITC-PBZ 1. Treated cells were stained with plasma-membrane fluorescence dy e FM 4-64 in red (FM 4-64). FITC signal of FITC-PBZ1 was detected under green filter (FITC-PBZ1). FM 4-64 and FITC-PBZ1 signals were merged to observe the localization of PBZ1 (Merged).
Recombinant PBZ1 protein possesses the RNase activity
PR-10 proteins found in other plants have been shown to possess RNase activity (Bantignies et al., 2000; Bufe et al., 1996; Park et al., 2004; Swoboda et al., 1996; Zhou et al., 2002). As PBZ1 belongs to the PR-10 protein family, it is likely that PBZ1 possesses RNase activity. In the RNA degradation assay, the recombinant PBZ1 protein showed significant ribonucleolytic activity at the concentration of 2.5 μg/ml, where total RNA was almost degraded within 3 h of incubation (Fig. 6A). Furthermore, when incubation time was prolonged (3, 6 and 9 h) with the same concentration (2.5 μg/ml), the extent of RNA degradation also increased (Fig. 6B). Appropriate controls, including Ni2+- NTA resin fractions from a culture of E. coli lacking the PBZ1 cDNA clone and 10 μg/ml of BSA, failed to degrade total RNA. On the other hand, RNase inhibitor (R.I), specific to pancreatic-type RNase, largely blocked the RNA degradation caused by the recombinant PBZ1 protein. These data indicate that PBZ1 protein possess RNase activity.
Fig. 6. RNase activity assay of recombinant PBZ1 protein. Total RNA (5 μg) used for this assay was isolated from rice leaf. (A) Dosage- dependent effect of the PBZ1 protein on RNA degradation. (B) Effect of incubation time on RNA degradation using 2.5 μg PBZ1 protein. R.I (50 units) represents RNase inhibitor. Prepared samples were loaded and separated on 0.8% formaldehyde-denatured agarose gel.
Although previous studies have shown RNase activity in the PR-10 proteins (Kim et al., 2008b; Moiseyev et al., 1997; Park et al., 2004), their function remains largely uninvestigated. In this study, a role for PBZ1 in cell death is supported by DNA fragmentation, internalization into the cytoplasm via plasma membrane and RNA degradation. In our previous report, we showed that localizations of PBZ1 dramatically coincided with tissues undergoing PCD, such as leaf senescence, root aerenchyma formation, coleoptiles senescence, root cap and seed aleurone layer (Kim et al., 2008a). These results suggest that PBZ1 may function in intracellular RNA degradation, potentially leading to the PCD in rice and perhaps in tobacco and Arabidopsis. Walter et al. (1996) have proposed that cytosolic RNases may be involved in selective and/or highly regulated degradation of existing RNAs during stress or pathogen attack.
RNase-mediated cell death in animal cells consists of cytosolic internalization and RNA cleavage (Haigis and Ranines, 2002). Studies of several ribonucleases have focused largely on the role of intracellular ribonucleolytic activity to cytotoxicity (Leland and Raines, 2001). However, little is known about the internalization pathway of RNase. Previously, Makarov and Ilinskaya (2003) have proposed that RNase binds to glycolipids and glycoproteins on the outer part of the plasma membrane and promotes their internalization. Ribonucleases must reach to the cytosol to degrade cellular RNA, but little is known about internalization of RNase A, even in animal systems (Haigis and Ranines, 2002; Olmo et al., 2001).
In flowering plants, cytotoxic S-RNases secreted by the pistil to selectively inhibit self-pollen have been studied (Goldraij et al., 2006; Luu et al., 2000). Using microscopy and immunolabelling methods, it was suggested that S-RNase was sorted to a vacuolar compartment or accumulated to the cytoplasm of all pollen-tubes in compatible pollinations but it was indistinguishable from that found in fully incompatible crosses because S-RNase cytotoxicity causes RNA degradation and unstable compartment for self-incompatiblity in plants (Goldraij et al., 2006; Luu et al., 2000). These previous studies support that idea that some of the PBZ1 present in cytoplasm is there to function inside the cell, even though most of PBZ1 is localized into the vacuole. These results will be important to further our understanding on PBZ1-mediated cell death mechanism in rice.
CONCLUSION
We previously reported that the expression level and localization of PBZ1 dramatically coincided with tissues undergoing PCD (Kim et al., 2008a). Even though those findings implied a possible role for PBZ1 in cell death, lack of evidence failed to demonstrate whether PBZ1 does indeed induce cell death in plants. Therefore, biochemical studies were required for understating and demonstrating the function of PBZ1 in plant defense system.
We have applied biochemical and transgenic approaches to further our understanding on PBZ1 function in rice, and also in tobacco and Arabidopsis plants. We demonstrate that the recombinant PBZ1 protein causes cell death in vitro not only in rice SCCs but also in planta in leaves of tobacco. In addition, when the PBZ1 gene is introduced into Arabidopsis under the DEX-inducible promoter, it induces cell death in vivo in leavestreated with DEX. Therefore, PBZ1-induced cell death appears to be a general phenomenon in plants. We also provide a number of biochemical evidences showing that PBZ1 protein functions in an intracellular tissue with activities to degrade DNA and RNA, which are well-accepted hallmark features of the PCD in plants and animals. Hence, findings presented in this study establish a novel role for PBZ1 in plant cell death. Having this evidence, PBZ1 is possibly one of the key components of PCD. But the question arises, how does PBZ1 induces PCD? Use of PBZ1 as a marker of cell death could help in understanding the mechanism and identifying new components of PCD in rice and other plants.
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
Acknowledgments
This work was supported by Biogreen 21 (03-2008-0172), Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2009-0089251), and the 2010 Specialization Project Research Grant funded by the Pusan National University, and by scholarships from the Brain Korea 21 Program (Y. Wang).
References
- 1.Aoyama T., Chua N.H. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. (1997);11:605–612. doi: 10.1046/j.1365-313x.1997.11030605.x. [DOI] [PubMed] [Google Scholar]
- 2.Bantignies B., Seguin J., Muzac I., Dedaldechamp F., Gulick P., Ibrahim R. Direct evidence for ribonucleolytic activity of a PR-10-like protein from white lupin roots. Plant Mol. Biol. (2000);42:871–881. doi: 10.1023/a:1006475303115. [DOI] [PubMed] [Google Scholar]
- 3.Bröker L.E., Kruyt F.A.E., Giaccone G. Cell death independent of caspases: a review. Clin. Cancer Res. (2005);11:3155–3162. doi: 10.1158/1078-0432.CCR-04-2223. [DOI] [PubMed] [Google Scholar]
- 4.Bufe A., Spangfort M.D., Kahlert H., Schlaak M., Becker W.M. The major birch pollen allergen, bet V1, shows ribonuclease activity. Planta. (1996);199:413–415. doi: 10.1007/BF00195733. [DOI] [PubMed] [Google Scholar]
- 5.Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. (1998);16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 6.Dangl J.L., Lehnackers H., Kiedrowski S., Debener T., Rupprecht C., Arnold M., Somssich I.E. In Advances in Molecular Genetics of Plant-Microbe Interactions. H. Hennecke, and D.P.S. Verma, eds. Kluwer Academic; Dordrecht, The Netherlands: (1991). pp. 78–83. Vol. 1. [Google Scholar]
- 7.Fojtová M., Kovařík A. Genotoxic effect of cadmium is associated with apoptotic changes in tobacco cells. Plant Cell Environ. (2000);23:531–537. [Google Scholar]
- 8.Goldraij A., Kondo K., Lee C.B., Hancock C.N., Sivaguru M., Vazquez-Santana S., Kim S., Phillips T.E., Cruz-Garcia F., McClure B. Compartmentalization of S-RNase and HT-B degradation in self-incompatible Nicotiana. Nature. (2006);439:805–810. doi: 10.1038/nature04491. [DOI] [PubMed] [Google Scholar]
- 9.Haigis M.C., Raines R.T. Secretory ribonucleases are internalized by a dynamin-independent endocytic pathway. J. Cell Sci. (2002);116:313–324. doi: 10.1242/jcs.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hwang S.H., Lee I.A., Yie S.W., Hwang D.J. Identification of an OsPR10a promoter region responsive to salicylic acid. Planta. (2008);227:1141–1150. doi: 10.1007/s00425-007-0687-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jwa N.S, Agrawal G.K., Rakwal R., Park C.H., Agrawal V.P. Molecular cloning and characterization of a novel Jasmonate inducible pathogenesis-related class 10 protein gene, OsPR10, from rice (Oryza sativa L.) seedling leaves. Biochem. Biophys. Res. Commun. (2001);286:973–983. doi: 10.1006/bbrc.2001.5507. [DOI] [PubMed] [Google Scholar]
- 12.Jwa N.S., Agrawal G.K., Tamogami S., Yonekura M., Han O.S., Iwahashi H., Rakwal R. Role of defense/stressrelated marker genes, proteins and secondary metabolites in defining rice self-defense mechanisms. Plant Physiol. Biochem. (2006);44:261–273. doi: 10.1016/j.plaphy.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 13.Keogh R.C., Deverall B.J., McLeod S. Comparison of histological and physiological responses to Phakopsora pachyrhizi in resistant and susceptible soybean. Trans. Br. Mycol. Soc. (1980);74:329–333. [Google Scholar]
- 14.Kim S.T., Cho K.S., Yu S., Kim S.G., Hong J.C., Han C., Bae D.W., Nam M.H., Kang K.Y. Proteomic analysis of differentially expressed proteins induced by rice blast fungus and elicitor in suspension-cultured rice cells. Proteomics. (2003);3:2368–2378. doi: 10.1002/pmic.200300577. [DOI] [PubMed] [Google Scholar]
- 15.Kim S.T., Kim S.G., Hwang D.H., Kang S.Y., Kim H.J., Lee B.H., Lee J.J., Kang K.Y. Proteomic analysis of pathogen-responsive proteins from rice leaves induced by rice blast fungus, Magnaporthe grisea. Proteomics. (2004);4:3569–3578. doi: 10.1002/pmic.200400999. [DOI] [PubMed] [Google Scholar]
- 16.Kim S.T., Kim S.G., Kang Y.H., Wang Y., Kim J.Y., Yi N., Kim J.K., Rakwal R., Koh H.J., Kang K.Y. Proteomics analysis of rice lesion mimic mutant (spl1) reveals tightly localized probenazole-induced protein (PBZ1) in cells undergoing programmed cell death. J. Proteome Res. (2008a);7:1750–1760. doi: 10.1021/pr700878t. [DOI] [PubMed] [Google Scholar]
- 17.Kim S.T., Yu S., Kang Y.H., Kim S.G., Kim J.Y., Kim S.H., Kang Y.H. The rice pathogen-related protein 10 (JIOsPR10) is induced by abiotic and biotic stresses and exhibits ribonuclease activity. Plant Cell Rep. (2008b);27:593–603. doi: 10.1007/s00299-007-0485-6. [DOI] [PubMed] [Google Scholar]
- 18.KoukalováKovařík B., Fajkus A., Široký J. Chromatin fragmentation associated with apoptotic changes in tobacoo cells exposed to cold stress. FEBS Lett. (1997);414:289–292. doi: 10.1016/s0014-5793(97)01008-9. [DOI] [PubMed] [Google Scholar]
- 19.Leland P.A., Raines R.T. Cancer chemotherapy-ribonucleases to the rescue. Chem. Biol. (2001);8:405–413. doi: 10.1016/s1074-5521(01)00030-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J.J., Ekramoddoullah A.K.M. The family 10 of plant pathogenesis-related proteins: Their structure, regulation, and function in response to biotic and abiotic stresses. Physiol. Mol. Plant Pathol. (2006);68:3–13. [Google Scholar]
- 21.Luu D.T., Qin X., Morse D., Cappadocia M. S-RNase uptake by compatible pollen tubes in gametophytic selfincompatibility. Nature. (2000);407:649–650. doi: 10.1038/35036623. [DOI] [PubMed] [Google Scholar]
- 22.Makarov A.A., Ilinskaya O.N. Cytotoxic ribonucleases: molecular weapons and their targets. FEBS Lett. (2003);540:15–20. doi: 10.1016/s0014-5793(03)00225-4. [DOI] [PubMed] [Google Scholar]
- 23.Midoh N., Iwata M. Cloning and characterization of a probenazole-inducible gene for an intracellular pathogenesisrelated protein in rice. Plant Cell Physiol. (1996);37:9–18. doi: 10.1093/oxfordjournals.pcp.a028918. [DOI] [PubMed] [Google Scholar]
- 24.Moiseyev G.P., Fedoreyeva L.I., Zhuravlev Y.N., Yasnetslaya E., Jekel P.A., Beintema J.J. Primary structures of two ribonucleases from ginseng calluses new members of the PR-10 family of intracellular pathogenesis-related plant proteins. FEBS Lett. (1997);407:207–210. doi: 10.1016/s0014-5793(97)00337-2. [DOI] [PubMed] [Google Scholar]
- 25.Nakashita H., Yasuda M., Nishioka M., Hasegawa S., Arai Y., Uramoto M., Yoshida S., Yamaguchi I. Chloroisonicotinamide derivative induces a broad range of disease resistance in rice and tobacco. Plant Cell Physiol. (2002);43:823–831. doi: 10.1093/pcp/pcf097. [DOI] [PubMed] [Google Scholar]
- 26.Ohira K., Ojima K., Fujiwara A. Studies on the nutrition of rice cell culture. A simple, defined medium for rapid growth in suspension culture. Plant Cell Physiol. (1973);14:1113–1121. [Google Scholar]
- 27.Park C.J., Kim K.J., Shin R., Park J.M., Shin Y.C., Paek K.H. Pathogenesis-related protein 10 isolated from hot pepper functions as a ribonuclease in an antiviral pathway. Plant J. (2004);37:186–198. doi: 10.1046/j.1365-313x.2003.01951.x. [DOI] [PubMed] [Google Scholar]
- 28.Rakwal R., Agrawal G.K., Yonekura M. Lightdependent induction of OsPR10 in rice (Oryza sativa L.) seedlings by the global stress signaling molecule jasmonic acid and protein phosphatase 2A inhibitors. Plant Sci. (2001);161:469–479. [Google Scholar]
- 29.Swoboda I., Hoffmann-Sommergrube K., O’Riordain G., Scheiner O., Heberle-Bors E., Vicente O. Bet V1 proteins, the major birch pollen allergens and members of a family of conserved pathogenesis-related proteins, show ribonuclease activity in vitro. Physiol. Plant. (1996);96:433–438. [Google Scholar]
- 30.Takayoshi I., Shigemi S., Ichiro M., Yuko O. Probenazole-induced accumulation of salicylic acid confers resistance to Magnaporthe grisea in adult rice plants. Plant Cell Physiol. (2007);48:915–924. doi: 10.1093/pcp/pcm062. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka N., Matsuoka M., Kitano H., Asano T., Kaku H., Komatsu S. gid1, a gibberellin-insensitive dwarf mutant, shows altered regulation of probenazole-inducible protein (PBZ1) in response to cold stress and pathogen attack. Plant Cell Environ. (2006);29:619–631. doi: 10.1111/j.1365-3040.2005.01441.x. [DOI] [PubMed] [Google Scholar]
- 32.Turner T.G., Novacky A. The quantitative relation between plant and bacterial cells involved in the hypersensitive reaction. Phytopathology. (1974);64:885–890. [Google Scholar]
- 33.Walter M.H., Liu J.W., Wünn J., Hess D. Bean ribonuclease-like pathogenesis-related protein genes (Ypr10) display complex patterns of developmental, dark-induced and exogenous-stimulus-dependent expression. Eur. J. Biochem. (1996);239:281–293. doi: 10.1111/j.1432-1033.1996.0281u.x. [DOI] [PubMed] [Google Scholar]
- 34.Wyllie A.H., Kerr J.F., Currie A.R. Cell death: the significance of apoptosis. Int. Rev. Cytol. (1980);68:251–306. doi: 10.1016/s0074-7696(08)62312-8. [DOI] [PubMed] [Google Scholar]
- 35.Yang S.W., Kim S.K., Kim W.T. Perturbation of NgTRF1 expression induces apoptosis-like cell death in tobacco BY-2 cells and implicates NgTRF1 in the control of telomere length and stability. Plant Cell. (2004);16:3370–385. doi: 10.1105/tpc.104.026278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou X.J., Lu S., Xu Y.H., Wang J.W., Chen X.Y. A cotton cDNA (GaPR-10) encoding a pathogenesis-related 10 protein with in vitro ribonuclease activity. Plant Sci. (2002);162:629–636. [Google Scholar]


