Abstract
One of the most remarkable types of migration found in animals is diadromy, a life-history behaviour in which individuals move between oceans and freshwater habitats for feeding and reproduction. Diadromous fishes include iconic species such as salmon, eels and shad, and have long fascinated biologists because they undergo extraordinary physiological and behavioural modifications to survive in very different habitats. However, the evolutionary origins of diadromy remain poorly understood. Here, we examine the widely accepted productivity hypothesis, which states that differences in productivity between marine and freshwater biomes determine the origins of the different modes of diadromy. Specifically, the productivity hypothesis predicts that anadromous lineages should evolve in temperate areas from freshwater ancestors and catadromous lineages should evolve in tropical areas from marine ancestors. To test this, we generated a time-calibrated phylogeny for Clupeiformes (herrings, anchovies, sardines and allies), an ecologically and economically important group that includes high diversity of diadromous species. Our results do not support the productivity hypothesis. Instead we find that the different modes of diadromy do not have predictable ancestry based on latitude, and that predation, competition and geological history may be at least as important as productivity in determining the origins of diadromy.
Keywords: diadromy, anadromy, catadromy, habitat transitions, life history, migration
1. Introduction
Diadromy is a remarkable life-history behaviour in which individuals migrate between oceans and freshwater rivers during a predictable phase of their life cycle, typically for feeding and reproduction [1]. Diadromous fishes have been a long-standing fascination of biologists [1–4], both because of the impressive physiological and behavioural adaptations necessary for survival in different habitats, and because diadromy has significant implications for the ecology, evolution and biogeography of fishes [5–8]. For instance, diadromy can impact genetic diversity and amounts of gene flow [9], alter life-history traits [10,11], and have cascading effects between marine, freshwater and terrestrial food webs [12]. Moreover, many of the approximately 250 diadromous fish species found nearly worldwide [7,13,14] are heavily studied iconic food and sport fishes (e.g. salmon, sturgeon, shad, eel).
There are several outstanding questions concerning diadromy: (1) What factors promote these remarkable migrations that require energetically costly physiological and osmoregulatory changes to deal with salinity differences between environments? (2) Under what conditions do the different types of diadromy (see below) evolve? Why would some species migrate to oceans while other species migrate to freshwater habitats to reproduce or feed? (3) Does diadromy facilitate the evolution of long-term lineage transitions between marine and freshwater habitats? (4) Lastly, what are the macroevolutionary and biogeographic implications of diadromy? Here, we test hypotheses regarding the evolutionary origins of diadromous lineages (questions 1 and 2), and address the macroevolutionary patterns and implications of this life-history behaviour (questions 3 and 4).
Three types of diadromy have been described: catadromy, anadromy and amphidromy [1]. Catadromous fishes are born in marine biomes and migrate to freshwater, where they spend most of their lives feeding and growing before returning to the ocean to reproduce. Anadromous fishes do the opposite—they are born in freshwater and migrate to the ocean, where they feed and grow, before migrating to freshwater to reproduce. Amphidromous species migrate between marine and freshwaters at a particular life-cycle stage (often as juveniles), but not for the purpose of spawning. More detailed descriptions and definitions of diadromy have been reviewed elsewhere [1,14–17].
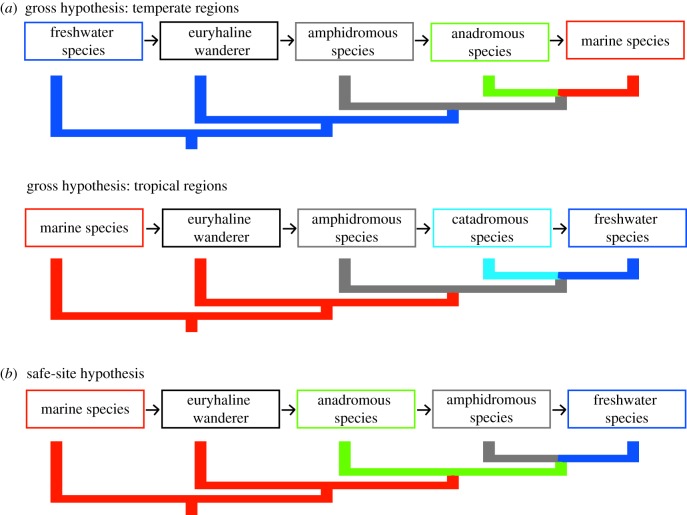
The evolutionary origins of diadromy are the subject of considerable theoretical discussion [1,14,18–23]. In an important contribution, Gross [22] and Gross et al. [23] proposed that differences in ecological productivity between marine and freshwater biomes determine the origins of the different modes of diadromy (herein referred to as the ‘productivity hypothesis’). Assuming the accuracy of Gross’s analyses of productivity data, oceans have higher productivity than freshwaters in temperate regions, and freshwaters have higher productivity than oceans in tropical regions. In turn, higher latitudes have more anadromous species and lower latitudes have more catadromous species (figure 1). This apparent geographical relationship between productivity and mode of diadromy provides some evidence that temperate anadromous fishes are derived from freshwater ancestors that began migrating to oceans to exploit the higher productivity, while tropical catadromous fishes are derived from marine ancestors that began migrating to rivers to exploit these higher productivity habitats [22,23]. Gross [22] further proposed that diadromy represents an intermediate condition between the evolution of fully freshwater to fully marine species (or vice versa). Gross's productivity model therefore makes the phylogenetic prediction (figure 2) that anadromous species have evolved from a freshwater ancestor, while catadromous species evolved from a marine ancestor.
Figure 1.
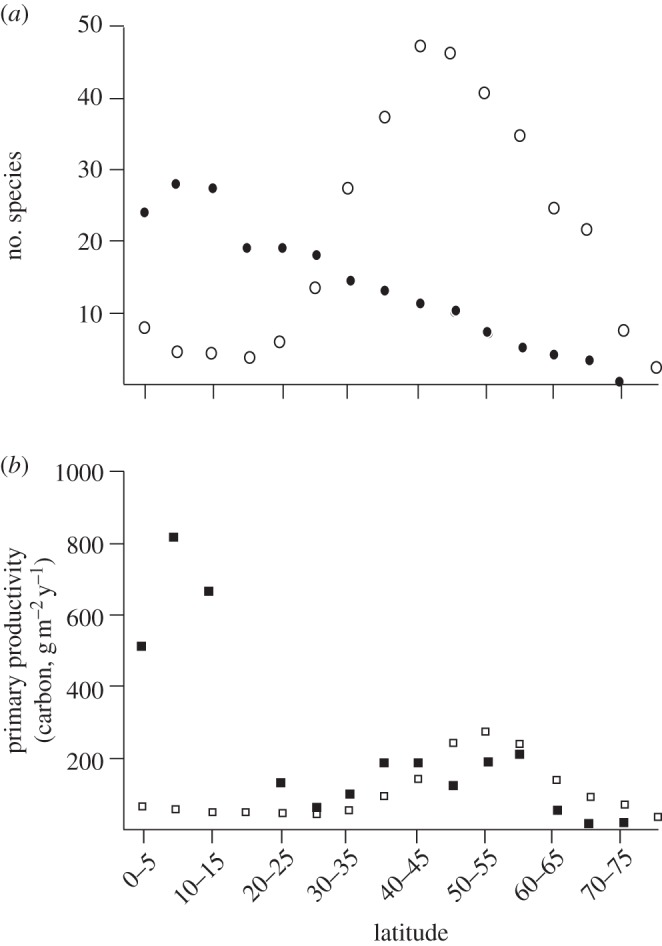
Evidence for the productivity hypothesis for the evolution of diadromy, adapted from Gross et al. [23]. (a) The number of all known anadromous (open circles) and catadromous (filled circles) fish species found across 5° latitudinal increments averaged from both Northern and Southern hemispheres, based on McDowall [18]. Diadromy occurs across the major craniate groups, from lampreys to percomorphs. (b) Annual primary productivity (measured in grams of carbon fixed per square metre per year) of aquatic biomes across different latitudes. Freshwater data (filled squares) averaged from 135 different water bodies; marine data (open squares) from Bunt [24]. See [23] for details on how species numbers and primary productivity were calculated.
Figure 2.
Hypotheses and phylogenetic predictions for the origins of diadromy based on (a) the productivity hypothesis [23] and (b) the safe-site hypothesis [25].
An alternative hypothesis for the origin of diadromy is the ‘safe-site hypothesis’, specifically proposed to explain the evolution of anadromy [25]. The safe-site hypothesis posits that freshwater habitats offer the eggs and larvae of marine fishes a sanctuary from marine predators. Thus, anadromy would be driven by the adaptive advantage offered by migration to freshwater habitats for reproduction. The safe-site hypothesis predicts that anadromous species evolved from a marine ancestor, and that freshwater species eventually evolve from anadromous lineages (figure 2).
Few studies have attempted to test hypotheses for the evolutionary origins of diadromy using a phylogenetic framework (but see [25,26]), and Gross's productivity hypothesis is based on species data that have not been examined in a phylogenetic framework. Here, we conduct a phylogenetic investigation of the evolution of diadromy in Clupeiformes, which includes some of the most important food fishes of all time (herring, sardines, anchovies and their allies). While most major fish clades are restricted to either marine or freshwater habitats [13,27], clupeiforms include marine, freshwater and diadromous species. In fact, Clupeidae alone has more diadromous species (approx. 30) than any other family of fishes except Gobiidae and Salmonidae, and the proportion of diadromous clupeids is about 10 times higher than in all other fishes [28]. Moreover, clupeiforms include both anadromous and catadromous species, making this group a superb model system for testing hypotheses on the evolution of diadromy and evolutionary transitions between aquatic biomes.
Using a time-calibrated phylogeny of clupeiforms, we reconstruct the evolution of marine, freshwater and diadromous lineages to explicitly test the productivity and the safe-site hypotheses for the origins of diadromous fishes. We also consider the role of diadromy in transitions between marine and continental freshwater biomes, and test whether diadromy facilitates evolutionary transitions between habitat types.
2. Material and methods
(a). Taxon sampling and molecular data
We assembled a data matrix consisting of four genes (rag1, rag2, cytb, 16 s) that included the broadest taxon sampling possible for clupeiforms by combining data from previously published studies [29–35] and newly generated sequence data for available taxa (see electronic supplementary material, table S1). The dataset includes 153 species from 64 of the 84 currently recognized genera and all major lineages of Clupeifomes.
Molecular laboratory protocols and primer information for newly generated sequences have previously been published [29,33]. Whenever possible, two or more specimens per species were sequenced for each gene and preliminary analyses were conducted as a measure of quality control. Duplicate species representatives were removed for all subsequent analyses. Sequences were edited using the computer software Geneious v. 5.4 [36] and aligned using the MUSCLE plugin [37] implemented in Geneious. For alignment of the 16 s data, we also used the Clustal X [38] plugin in Geneious to employ a range of gap opening and extension parameters to test the robustness of the MUSCLE alignment. Protein-coding genes were translated to amino acids to confirm open reading frames. Following alignment, we concatenated all four genes into a single matrix consisting of 5211 bp. The final matrix includes data for more than 70% of all possible cells, and comprises data from 152 species for 16 s (1352 bp), 155 species for cytb (1131 bp), 98 species for rag1 (1491 bp) and 102 species for rag2 (1237 bp).
(b). Data partitioning and model selection
We used the program PartitionFinder [39] to objectively determine the best-fit model of evolution and partitioning scheme simultaneously. Best-fit models were selected using Bayesian information criteria, selecting among the best models available under a ‘greedy’ search scheme using ‘models=mrbayes’ (see electronic supplementary material, table S2).
(c). Phylogeny and time-calibration
We conducted phylogenetic inference using maximum-likelihood (ML) and Bayesian methods. Our ML phylogenetic reconstructions were conducted using the program RAxML v. 7.3 [40]. For ML estimates, we used the best partitioning strategy chosen by PartitionFinder and a GTRGAMMA model for each partition (electronic supplementary material, table S2). We estimated support for nodes using the rapid-bootstrapping algorithm for 1000 non-parametric bootstrap replicates. We performed three separate ML searches to ensure that we searched tree space thoroughly and were not trapped on local optima.
We jointly estimated phylogeny and diversification times using a Bayesian relaxed clock method [41] in the program BEAST v. 1.7.2 [42]. We used an uncorrelated lognormal tree prior and a birth–death prior for rates of cladogenesis. Several initial BEAST runs showed that the partitioning strategy chosen by PartitionFinder resulted in over-parametrization, so the dataset was partitioned by gene with partitions unlinked and a GTR model with gamma-distributed rate heterogeneity used for each partition. We ran four independent analyses for 300 million generations, sampling every 1000th generation. We used Tracer v. 1.5 [43] to evaluate convergence of runs and to verify that effective sample sizes were more than 200 for all parameters. We determined that the first 50 million generations from the MCMC sample were a conservative burn-in. We combined runs using LogCombiner v. 1.6.1 [43] and the maximum credibility tree was generated in TreeAnnotator v. 1.6.1 [43].
To determine absolute divergence times, we used eight fossil and biogeographic age calibrations (see electronic supplementary material, text S3) with exponential priors to set a hard minimum and soft maximum bound [44]. Several of the fossil calibrations have been used in recent studies [34,45,46]; however, we included additional clupeiform fossils that have not yet been incorporated in diversification time analysis of this group.
(d). Ancestral character reconstruction
We used ancestral character reconstruction to determine the evolutionary history of diadromous, marine and freshwater lineages. Each species was coded for a discrete, unordered character (biome requirement) with four states: marine (0), freshwater (1), anadromous (3) or catadromous (4). Some species are thought to be facultatively diadromous, where not all individuals migrate (e.g. alewife) [28]. We coded facultative diadromous species as having the respective diadromous state (anadromous or catadromous).
We used the program Mesquite v. 2.73 [47] to conduct a maximum-parsimony (MP) and ML character reconstructions. For ML reconstructions, we used the Mk model [48]. By conducting these reconstructions on our time-calibrated phylogeny, we are able to determine the number, order and timing of transitions between marine, freshwater and diadromous life histories in clupeiforms.
3. Results
(a). Phylogeny
We recovered a well-resolved molecular phylogeny that is largely consistent with previous studies (figure 3; see also electronic supplementary material, figures S4 and S5). Our phylogenetic hypothesis includes several clupeiform lineages that have not previously been included in a molecular study. These results are presented and discussed in detail in electronic supplementary material, text S3.
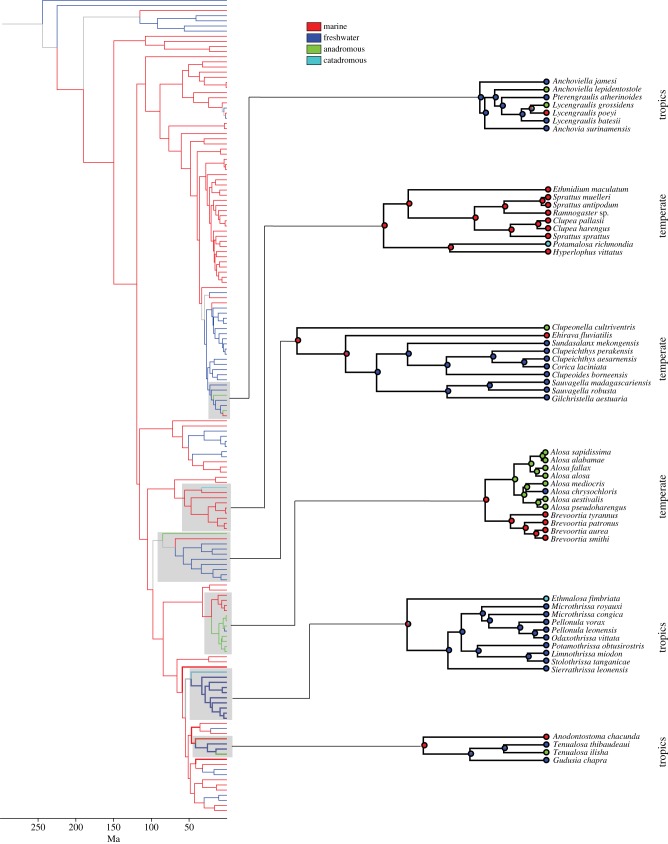
Figure 3.
Time-calibrated phylogeny of Clupeiformes showing ancestral character reconstructions of marine (red), freshwater (dark blue), anadromous (green) and catadromous (light blue) lineages. Branch colours indicate character states from maximum-likelihood reconstructions, with branch states considered unambiguous when the log-likelihood was 2.0 units higher than the alternative state.
(b). Ancestral character reconstructions: evolution of diadromy
Our ancestral character reconstructions indicate that catadromy evolved twice and anadromy evolved five times in Clupeiformes (figure 3). Amphidromy may have evolved once (see Discussion). In general, diadromous clupeiform lineages are monotypic and often placed in their own genus (Alosa is a notable exception).
The catadromous herring species Potamalosa richmondia was derived from a marine ancestor and occurs in the temperate regions of southeastern Australia. Ethmalosa fimbriata is a tropical catadromous herring species from the eastern Atlantic and western African rivers. This species was derived from a marine ancestor and is sister to a clade of freshwater herring from Africa.
The earliest instance of anadromy is Clupeonella cultriventris, which is a temperate anadromous lineage derived from a marine ancestor, although this is a tentative assessment because the phylogenetic placement of this species is not well supported. Our analyses indicate that seven anadromous species in the genus Alosa from temperate eastern North America and Eurasia were derived from a marine ancestor. Nested within the anadromous Alosa clade is a purely freshwater species (Alosa chrysochloris). Of the five anadromous lineages, there were three separate instances of an anadromous lineage evolving from a freshwater ancestor (Tenualosa ilisha, Anchoviella lepidentostole and Lycengraulis grossidens), all of which occurred in tropical regions.
(c). Ancestral character reconstructions: marine/freshwater transitions
Our analyses show that Clupeiformes is an ancestrally marine group that invaded continental freshwaters 12 times (excluding diadromous lineages; figure 3). There were only three reversals (freshwater to marine), all of which occurred in New World anchovy lineages distributed along the northeastern coast of South America. Lineages rarely invaded freshwaters in the same geographical area more than once. Freshwater lineages are generally much more diverse than diadromous lineages. In fact, Alosa is the only diadromous lineage that is not monotypic (see above). Freshwater clupeiform lineages are particularly diverse in South America (mostly anchovies), southeastern Asia and western Africa.
4. Discussion
(a). Evolution of catadromy: testing the productivity hypothesis
As a scenario for the origin of catadromy, Gross's productivity hypothesis predicts that in the tropics, marine (euryhaline) ancestors would undergo excursions to freshwaters to feed in these high-productivity environments (figures 1 and 2). The fitness gain from these freshwater excursions would lead to annual migration to freshwaters for an extended period of growth, yielding catadromous lineages. Over time, migration back to the natal marine habitat could cease, severing the link to the marine environment, and yielding an endemic, entirely freshwater lineage [22,23].
Our results show that of the two origins of catadromy we inferred one is consistent with the productivity hypothesis, while the other is not. The catadromous African bonga shad, E. fimbriata, was derived from a marine ancestor, a pattern consistent with Gross's model. Furthermore, E. fimbriata is sister to a clade of freshwater herring (Pellonulini) that has diversified in African rivers and lakes [34]. The pattern of a freshwater clade closely related to a tropical catadromous lineage is also consistent with Gross's model. However, the close affinity of Ethmalosa and Pellonulini needs to be further investigated because our results did not provide strong statistical support for this relationship (see the electronic supplementary material, text S3).
The second catadromous lineage shows a pattern that is inconsistent with the Gross hypothesis. The Australian catadromous herring P. richmondia was derived from a marine ancestor and is sister to Hyperlophus, a marine lineage that occurs along the southern and eastern coasts of Australia. However, P. richmondia occurs in temperate rather than tropical regions. Furthermore, P. richmondia does not have close phylogenetic affinity to freshwater lineages, despite having more than 30 million years to produce a freshwater descendant (figure 3; electronic supplementary material, figure S5). The only non-diadromous freshwater clupeid in Australia is Nematalosa erebi, which is phylogenetically distant from P. richmondia, and is related to a different set of marine species (figure 3).
Although the phylogenetics of other non-clupeiform catadromous lineages has not been studied in much detail, the pattern of marine ancestry for catadromous fishes may be fairly general [1]. Anguillid eels are the best-known and most species-rich catadromous group. These widespread fishes were probably derived from a deep-sea ancestor, and while they may have originated in the tropics [49], many catadromous eels migrate to feed and grow in temperate freshwaters, not tropical freshwaters. These migrations to temperate freshwater habitats contradict Gross's predictions because temperate freshwaters have relatively low productivity. Furthermore, catadromous eels have never yielded fully freshwater lineages, nor even produced landlocked forms [6], even though as a group they have a broad distribution, suggesting ample opportunity for landlocking. More recently, Feutry et al. [26] found that catadromous species in the tropical fish genus Kuhlia were derived from a marine ancestor, and that a catadromous lineage also gave rise to a marine lineage, a pattern that is also inconsistent with the productivity hypothesis.
(b). Evolution of anadromy: testing the productivity hypothesis
The productivity hypothesis posits a mirror opposite pattern for the origins of anadromous fishes (figures 1 and 2). In this scenario, a freshwater ancestor in temperate regions would start by making excursions into more productive temperate marine habitats for feeding and growth. Initially, this would involve a euryhaline wanderer, which eventually would give rise to an anadromous lineage. Over time, the annual migrations from freshwater to marine habitats would cease, resulting in an entirely marine descendant. We find no evidence in clupeiform lineages to support Gross's predictions for the origins of anadromy (figure 3). Three of the five anadromous lineages—T. ilisha, L. grossidens and An. lepidentostole—were each independently derived from a freshwater ancestor. However, these lineages all occur in tropical rather than temperate regions. The well-known anadromous Alosa spp. and the lesser-known Clupeonella occur in temperate regions, but these taxa were independently derived from marine ancestors. Thus, none of the anadromous clupeiform lineages fit the Gross model.
Other anadromous fish groups also fail to show evidence for the Gross hypothesis [1]. Some anadromous groups such as sticklebacks [50] and smelts [25] occur in temperate areas, but are derived from marine ancestors. Anadromous ariid catfishes [51,52] are derived from marine ancestors and occur in tropical regions. None of these lineages fit Gross's model. Salmonids have a complicated history, but anadromous lineages probably evolved from freshwater ancestors and occur in temperate regions [25,53]; thus, Salmonidae may be the only group that is consistent with Gross's model.
It is not immediately evident why phylogenetic analyses of diadromy paint a different picture for the evolution of anadromy and catadromy than the species count approach of Gross (figure 1). However, it is clear that any future tests for latitudinal gradients of both catadromy and anadromy should explicitly incorporate phylogenetic approaches that consider lineage evolution, rather than using phylogenetically uncorrected species counts. Current phylogenetic evidence indicates that other factors (see below) might be as important as productivity for the origins of diadromy.
(c). Evolution of diadromy: the roles of predation, competition and abiotic factors
The evolution of diadromy is probably affected by species interactions. Dodson et al.'s safe-site hypothesis proposes a marine ancestor migrating to freshwater to reproduce in habitats that have lower predation pressure (figure 2) [25]. Over time, such populations would stop returning to the sea, leaving entirely freshwater descendants [25]. Alosa shows a pattern that fits the safe-site hypothesis; an anadromous clade was derived from a marine ancestor and gave rise to a fully freshwater lineage (Al. chrysochloris). Clupeonella cultriventris was also derived from a marine ancestor, but is sister to a clade that includes both marine and freshwater members. This pattern does not fit the safe-site hypothesis because once a lineage has successfully established itself in freshwater, there is no predicted advantage to returning to the marine environment. The remaining anadromous lineages (T. ilisha, An. lepidentostole and L. grossidens) were derived from freshwater ancestors and thus do not support the safe-site hypothesis. Dodson et al. [25] found that phylogenetic analyses of smelts (Osmeroidei) supported the safe-site hypothesis, but that analyses of salmonids did not. More work is needed to determine whether clades with phylogenetic patterns consistent with the safe-site hypothesis show evidence for reduction in predation in freshwater habitats compared with marine habitats.
Competition is an important factor affecting macroevolutionary transitions between marine and freshwater lineages [29,52,54], and is also likely to play a role in determining which geographical regions diadromous species are able to invade [55]. This hypothesis is based on the concept that competition is strongest among closely related taxa [56–58]. In clupeiforms, we find that diadromy generally has not evolved multiple times in the same geographical area (figure 3), suggesting that once a diadromous lineage invades a region, it may prevent ecologically similar species from evolving the same migratory behaviour. Furthermore, diadromous species often occur in regions with low species diversity, which McDowall [55] argued was in part due to reduced competition from incumbent species. We suggest there is growing phylogenetic evidence that competition does play a role in the origins of diadromy.
One of the primary determinants of reduced competition in certain geographical areas is the palaeogeographical history of those areas. For example, northern latitudes have been subjected to repeated bouts of glacial advances and retreats, the most recent occurring only 10 000 years ago. These areas often have a high proportion of diadromous lineages (along with lineages of recent marine ancestry) [1,55]. Oceanic islands also have a high proportion of diadromous lineages in freshwater habitats [18,19]. McDowell [1,55] suggested that the high dispersal ability of diadromous lineages facilitates re-colonization of newly available, reduced competition habitats. While it is less clear what palaeogeographic events might favour the evolution of diadromy in continental rivers at lower latitudes, it is possible that marine incursions have played a role in reducing competition in some areas, thereby facilitating transitions between habitats [59,60]. Thus, the palaeogeography and geological history of certain areas may have played an important role in determining the evolutionary patterns of diadromy.
(d). Is diadromy a pathway for marine/freshwater evolutionary transitions?
An expected outcome in both the productivity and safe-site hypotheses is that diadromy is a precursor to permanent transitions between marine and freshwater biomes in fish lineages (although see [20,21,61,62]). This concept is intuitively appealing: a species that migrates between marine and freshwater environments could become isolated in one habitat and successfully establish a permanent population. Over time, this could result in speciation and subsequent diversification in the new habitat. The expected phylogenetic patterns from this process, assuming extinction has not erased ancestral species, are illustrated in figure 2. Clupeiformes was a likely candidate group for fulfilling these scenarios because the group includes many diadromous, marine and freshwater species. However, we find that diadromy has played a minimal role in transitions between marine and freshwater biomes (figure 3). Diadromy is only rarely an intermediate condition between freshwater and marine lineages. Furthermore, only one of the 12 marine-to-freshwater transitions we determined were associated with any diadromous species. To date, we lack an empirical example of a freshwater lineage evolving via catadromous ancestry (the Ethmalosa/Pellonulini clade is a possible exception), or an exclusively marine lineage evolving from an anadromous ancestor [6], which are the expected end points of the productivity and safe-site hypotheses [22].
The decoupled relationship between diadromy and biome transitions is a pattern that is repeated in other lineages [51,52]. There are instances of landlocked freshwater populations of anadromous marine lineages such as shads, salmon and sticklebacks, which in some cases have undergone morphological divergence [11,63]. Interestingly, these cases are all very recent (Pleistocene or younger), and generally neither diverse nor spread across large geographical areas, suggesting that there are constraints on diversification of freshwater populations derived from diadromous ancestors. It seems that diadromy is not a pathway for large-scale transitions between marine and freshwater habitats. It may instead be a macroevolutionary dead end, since most diadromous lineages are species-poor compared with their strictly marine or freshwater relatives.
Acknowledgements
For assistance with tissues and specimens, we thank the following people and institutions: Wayne Starnes (North Carolina Museum of Natural History), Andrew Bentley (University of Kansas Biodiversity Institute), Kevin Conway (Texas A&M University), Erling Holm (Royal Ontario Museum), Marisa Litz (National Marine Fisheries Service), Kelvin Lim and Tan Heok Hui (National University of Singapore), Kyle Piller (Southeastern Louisiana University), John Lyons (University of Wisconsin Zoological Museum), and Pablo Gesundheit. We thank the following people for assistance collecting specimens in the field: Mark Sabaj, Mike Littman, Hernán López-Fernández, Eric Lewallen, Don Taphorn and Calvin Bernard. Allan Baker, Hernán López-Fernández, Jason Weir, John Lundberg and Belinda Chang provided helpful feedback on this study. Hernán López-Fernández and Mark Holder graciously provided computer computation time for analyses.
Funding statement
Funding was provided by an NSERC Discovery Grant to N.R.L., and Centre for Global Change (University of Toronto) and Lerner Gray grants (AMNH) to D.D.B.
References
- 1.McDowall RM. 1988. Diadromy in fishes: migrations between freshwater and marine environments. London, UK: Croom Helm [Google Scholar]
- 2.Talbot GB, Sykers JE. 1958. Altantic coast migrations of American shad. Fish. Bull. US Fish Wildl. Serv. 58, 473–490 [Google Scholar]
- 3.Roule L. 1933. Fishes: their journeys and migrations, p. 270 London, UK: Rouledge [Google Scholar]
- 4.Myers GS. 1949. Salt-tolerance of fresh-water fish groups in relation to zoogeographical problems. Bijdragen tot de Dierkunde 28, 315–364 [Google Scholar]
- 5.McDowall RM. 1999. Diadromy and genetic diversity in Nearctic and Palearctic fishes. Mol. Ecol. 8, 527–528 (doi:10.1046/j.1365-294x.1999.00560.x) [Google Scholar]
- 6.McDowall RM. 2001. Diadromy, diversity and divergence: implications for speciation processes in fishes. Fish Fish. 2, 278–285 (doi:10.1046/j.1467-2960.2001.00050.x) [Google Scholar]
- 7.McDowall RM. 2008. Diadromy, history and ecology: a question of scale. Hydrobiologia 602, 5–14 (doi:10.1007/s10750-008-9290-7) [Google Scholar]
- 8.Waters JM, Wallis GP. 2001. Cladogenesis and loss of the marine life-history phase in freshwater galaxiid fishes (Osmeriformes: Galaxiidae). Evolution 55, 587–597 (doi:10.1554/0014-3820(2001)055[0587:calotm]2.0.co;2) [DOI] [PubMed] [Google Scholar]
- 9.Hasselman DJ, Ricard D, Bentzen P. 2013. Genetic diversity and differentiation in a wide ranging anadromous fish, American shad (Alosa sapidissima), is correlated with latitude. Mol. Ecol. 22, 1558–1573 (doi:10.1111/mec.12197) [DOI] [PubMed] [Google Scholar]
- 10.Closs GP, Hicks AS, Jellyman PG. 2013. Life histories of closely related amphidromous and non-migratory fish species: a trade-off between egg size and fecundity. Freshwater Biol. 58, 1162–1177 (doi:10.1111/fwb.12116) [Google Scholar]
- 11.Palkovacs EP, Dion KB, Post DM, Caccone A. 2008. Independent evolutionary origins of landlocked alewife populations and rapid parallel evolution of phenotypic traits. Mol. Ecol. 17, 582–597 (doi:10.1111/j.1365-294X.2007.03593.x) [DOI] [PubMed] [Google Scholar]
- 12.Schindler DE, Armstrong JB, Bentley KT, Jankowski K, Lisi PJ, Payne LX. 2013. Riding the crimson tide: mobile terrestrial consumers track phenological variation in spawning of an anadromous fish. Biol. Lett. 9, 20130048 (doi:10.1098/rsbl.2013.0048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson JS. 2006. Fishes of the World, 4th edn, p. 601 New York, NY: John Wiley & Sons [Google Scholar]
- 14.McDowall RM. 2009. Making the best of two worlds: diadromy in the evolution, ecology, and conservation of aquatic organisms. Am. Fish Soc. Symp. 69, 1–22 [Google Scholar]
- 15.McDowall RM. 1992. Diadromy: origins and definitions of terminology. Copeia 1992, 248–251 (doi:10.2307/1446563) [Google Scholar]
- 16.McDowall RM. 2007. On amphidromy, a distinct form of diadromy in aquatic organisms. Fish Fish. 8, 1–13 (doi:10.1111/j.1467-2979.2007.00232.x) [Google Scholar]
- 17.Myers GS. 1949. usage of anadromous, catadromous and allied terms for migratory fishes. Copeia 1949, 89–97 (doi:10.2307/1438482) [Google Scholar]
- 18.McDowall RM. 1987. The occurrence and distribution of diadromy among fishes. In Common strategies of anadromous and catadromous fishes (eds Dadswell M, Klauda R, Moffitt C, Saunders R.), pp. 1–13 Bethesda, MD: American Fisheries Society, Symposium 1 [Google Scholar]
- 19.McDowall RM. 1998. Driven by diadromy: its role in the historical and ecological biogeography of the New Zealand freshwater fish fauna. Ital. J. Zool. 65(Suppl. 1), 73–85 (doi:10.1080/11250009809386799) [Google Scholar]
- 20.McDowall RM. 1997. The evolution of diadromy in fishes (revisited) and its place in phylogenetic analysis. Rev. Fish. Biol. Fish 7, 443–462 (doi:10.1023/A:1018404331601) [Google Scholar]
- 21.Parenti LR. 2008. Life history patterns and biogeography: an interpretation of diadromy in fishes. Ann. Missouri Botanical Garden 95, 232–247 (doi:10.3417/2006051) [Google Scholar]
- 22.Gross M. 1987. Evolution of diadromy in fishes. In Common strategies of anadromous and catadromous fishes (eds Dadswell M, Klauda R, Moffitt C, Saunders R.), pp. 14–24 Bethesda, MD: American Fisheries Society, Symposium 1 [Google Scholar]
- 23.Gross MR, Coleman RM, McDowall RM. 1988. Aquatic productivity and the evolution of diadromous fish migration. Science 239, 1291–1293 (doi:10.1126/science.239.4845.1291) [DOI] [PubMed] [Google Scholar]
- 24.Bunt JS. 1975. Primary productivity of marine ecosystems. In Primary productivity of the biosphere (eds Leith H, Whittaker RH.), pp. 169–183 New York, NY: Springer [Google Scholar]
- 25.Dodson JJ, Laroche J, Lecomte F. 2009. Contrasting evolutionary pathways of anadromy in euteleostean fishes. Am. Fish Soc. Symp. 69, 63–77 [Google Scholar]
- 26.Feutry P, Castelin M, Ovenden JR, Dettaï A, Robinet T, Cruaud C, Keith P. 2013. Evolution of diadromy in fish: insights from a tropical genus (Kuhlia species). The American Naturalist 181, 52–63 (doi:10.1086/668593) [DOI] [PubMed] [Google Scholar]
- 27.Berra T. 2001. Freshwater fish distribution, p. 604 New York, NY: Academic Press [Google Scholar]
- 28.McDowall RM. 2003. Shads and diadromy: implications for ecology, evolution, and biogeography. Am. Fish Soc. Symp. 35, 11–23 [Google Scholar]
- 29.Bloom DD, Lovejoy NR. 2012. Molecular phylogenetics reveals a pattern of biome conservatism in New World anchovies (Family Engraulidae). J. Evol. Biol. 25, 701–715 (doi:10.1111/j.1420-9101.2012.02464.x) [DOI] [PubMed] [Google Scholar]
- 30.Lavoué S, Miya M, Kawaguchi A, Yoshino T, Nishida M. 2008. The phylogenetic position of an undescribed paedomorphic clupeiform taxon: mitogenomic evidence. Ichthyol. Res. 55, 328–334 (doi:10.1007/s10228-008-0044-3) [Google Scholar]
- 31.Lavoué S, Miya M, Nishida M. 2010. Mitochondrial phylogenomics of anchovies (family Engraulidae) and recurrent origins of pronounced miniaturization in the order Clupeiformes. Mol. Phylogenet. Evol. 56, 480–485 (doi:10.1016/j.ympev.2009.11.022) [DOI] [PubMed] [Google Scholar]
- 32.Lavoué S, Miya M, Saitoh K, Ishiguro NB, Nishida M. 2007. Phylogenetic relationships among anchovies, sardines, herrings and their relatives (Clupeiformes), inferred from whole mitogenome sequences. Mol. Phylogenet. Evol. 43, 1096–1105 (doi:10.1016/j.ympev.2006.09.018) [DOI] [PubMed] [Google Scholar]
- 33.Li CH, Orti G. 2007. Molecular phylogeny of clupeiformes (Actinopterygii) inferred from nuclear and mitochondrial DNA sequences. Mol. Phylogenet. Evol. 44, 386–398 (doi:10.1016/j.ympev.2006.10.030) [DOI] [PubMed] [Google Scholar]
- 34.Wilson AB, Teugels GG, Meyer A. 2008. Marine incursion: the freshwater herring of Lake Tanganyika are the product of a marine invasion into west Africa. PLoS ONE 3, e1979 (doi:10.1371/journal.pone.0001979) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lavoué S, Miya M, Musikasinthorn P, Chen W-J, Nishida M. 2013. Mitogenomic evidence for an indo-west pacific origin of the clupeoidei (Teleostei: Clupeiformes). PLoS ONE 8, e56485 (doi:10.1371/journal.pone.0056485) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drummond A, et al. 2010. Geneious. (v5.3 ed). See http://www.geneious.com .
- 37.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (doi:10.1093/nar/gkh340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 (doi:10.1093/nar/25.24.4876) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lanfear R, Calcott B, Ho SYW, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701 (doi:10.1093/molbev/mss020) [DOI] [PubMed] [Google Scholar]
- 40.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (doi:10.1093/bioinformatics/btl446) [DOI] [PubMed] [Google Scholar]
- 41.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4, e88 (doi:10.1371/journal.pbio.0040088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973 (doi:10.1093/molbev/mss075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 (doi:10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ho SYW, Phillips MJ. 2009. Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Syst. Biol. 58, 367–380 (doi:10.1093/sysbio/syp035) [DOI] [PubMed] [Google Scholar]
- 45.Alfaro ME, Santini F, Brock C, Alamillo H, Dornburg A, Rabosky DL, Carnevale G, Harmon LJ. 2009. Nine exceptional radiations plus high turnover explain species diversity in jawed vertebrates. Proc. Natl Acad. Sci. USA 106, 13 410–13 414 (doi:10.1073/pnas.0811087106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santini F, Harmon LJ, Carnevale G, Alfaro ME. 2009. Did genome duplication drive the origin of teleosts? A comparative study of diversification in ray-finned fishes. BMC Evol. Biol. 9, 194 (doi:10.1186/1471-2148-9-194) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maddison WP, Maddison DR. 2011 Mesquite: a modular system for evolutionary analysis. (2.75 ed.) [Google Scholar]
- 48.Pagel M. 1999. The maximum likelihood approach to reconstructing ancestral character states of discrete characters on phylogenies. Syst. Biol. 48, 612–622 (doi:10.1080/106351599260184) [Google Scholar]
- 49.Inoue JG, et al. 2010. Deep-ocean origin of the freshwater eels. Biol. Lett. 6, 363–366. (doi:10.1098/rsbl.2009.0989) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawahara R, Miya M, Mabuchi K, Near TJ, Nishida M. 2009. Stickleback phylogenies resolved: evidence from mitochondrial genomes and 11 nuclear genes. Mol. Phylogenet. Evol. 50, 401–404 (doi:10.1016/j.ympev.2008.10.014) [DOI] [PubMed] [Google Scholar]
- 51.Betancur-R R. 2010. Molecular phylogenetics supports multiple evolutionary transitions from marine to freshwater habitats in ariid catfishes. Mol. Phylogenet. Evol. 55, 249–258 (doi:10.1016/j.ympev.2009.12.018) [DOI] [PubMed] [Google Scholar]
- 52.Betancur-R R, Ortí G, Stein AM, Marceniuk AP, Alexander Pyron R. 2012. Apparent signal of competition limiting diversification after ecological transitions from marine to freshwater habitats. Ecol. Lett. 15, 822–830 (doi:10.1111/j.1461-0248.2012.01802.x) [DOI] [PubMed] [Google Scholar]
- 53.McLennan DA. 1994. A phylogenetic approach to the evolution of fish behavior. Rev. Fish Biol. Fish. 4, 430–460 (doi:10.1007/BF00042889) [Google Scholar]
- 54.Bloom DD, Weir JT, Piller KR, Lovejoy NR. 2013. Do freshwater fishes diversify faster than marine fishes? A test using state-dependent diversification analyses and molecular phylogenetics of New World Silversides (Atherinopsidae). Evolution 67, 2040–2057 (doi:10.1111/evo.12074) [DOI] [PubMed] [Google Scholar]
- 55.McDowall RM. 2008. Why are so many boreal freshwater fishes anadromous? Confronting ‘conventional wisdom’. Fish Fish. 9, 208–213 (doi:10.1111/j.1467-2979.2008.00271.x) [Google Scholar]
- 56.Violle C, Nemergut DR, Pu Z, Jiang L. 2011. Phylogenetic limiting similarity and competitive exclusion. Ecol. Lett. 14, 782–787 (doi:10.1111/j.1461-0248.2011.01644.x) [DOI] [PubMed] [Google Scholar]
- 57.Wiens JJ, et al. 2010. Niche conservatism as an emerging principle in ecology and conservation biology. Ecol. Lett. 13, 1310–1324 (doi:10.1111/j.1461-0248.2010.01515.x) [DOI] [PubMed] [Google Scholar]
- 58.Wiens JJ, Brandley MC, Reeder TW. 2006. Why does a trait evolve multiple times within a clade? Repeated evolution of snakelike body form in squamate reptiles. Evolution 60, 123–141 [PubMed] [Google Scholar]
- 59.Bloom DD, Lovejoy NR. 2011. The biogeography of marine incursions in South America. In Historical biogeograhpy of neotropical freshwater fishes (eds Albert JS, Reis RE.), pp. 137–144 Berkeley, CA: University of California Press [Google Scholar]
- 60.Lovejoy NR, Albert JS, Crampton WGR. 2006. Miocene marine incursions and marine/freshwater transitions: evidence from Neotropical fishes. J. South Am. Earth Sci. 21, 5–13 (doi:10.1016/j.jsames.2005.07.009) [Google Scholar]
- 61.McDowall RM. 1993. A recent marine ancestry for diadromous fishes—sometimes yes, but mostly no. Environ. Biol. Fishes 37, 329–335 (doi:10.1007/bf00005199) [Google Scholar]
- 62.McDowall RM. 2002. The origin of the salmonid fishes: marine, freshwater … or neither? Rev. Fish Biol. Fish. 11, 171–179 (doi:10.1023/A:1020370328194) [Google Scholar]
- 63.McKinnon JS, Rundle HD. 2002. Speciation in nature: the threespine stickleback model systems. Trends Ecol. Evol. 17, 480–488 (doi:10.1016/s0169-5347(02)02579-x) [Google Scholar]