Abstract
Experimental evolution provides a powerful manipulative tool for probing evolutionary process and mechanism. As this approach to hypothesis testing has taken purchase in biology, so too has the number of experimental systems that use it, each with its own unique strengths and weaknesses. The depth of biological knowledge about Caenorhabditis nematodes, combined with their laboratory tractability, positions them well for exploiting experimental evolution in animal systems to understand deep questions in evolution and ecology, as well as in molecular genetics and systems biology. To date, Caenorhabditis elegans and related species have proved themselves in experimental evolution studies of the process of mutation, host–pathogen coevolution, mating system evolution and life-history theory. Yet these organisms are not broadly recognized for their utility for evolution experiments and remain underexploited. Here, we outline this experimental evolution work undertaken so far in Caenorhabditis, detail simple methodological tricks that can be exploited and identify research areas that are ripe for future discovery.
Keywords: experimental evolution, Caenorhabditis, evolution
1. The tool of experimental evolution
Experimental evolution is the controlled study of evolutionary change as it occurs under experimenter-imposed conditions in the laboratory or field. At its most basic, experimental evolution combines two separate procedures: the multi-generation culturing of populations, and the quantification of change in those populations. Its great power is in being a broker between theory and nature. Much of evolutionary theory has been formalized since the early days of the great mathematical geneticists, and yet many aspects of theory have proved difficult to test with traditional experiments or comparative data. Experimental evolution provides a compelling methodological alternative. Despite experimental evolution being a relatively recent paradigm in evolutionary biology, it has now established itself as a powerful method for testing evolutionary theory [1,2].
The first evolution experiment, by Dallinger in 1878, described the adaptation to heat stress by ‘a minute septic organism’ [3]. In modern times, Lenski et al. [4] pioneered and popularized long-term experimental evolution with Escherichia coli, which has now been cultured for more than 55 000 generations [2]. This microbial beginning has since metastasized to test a broad variety of theories about the mechanisms of evolution [1,2,5].
Short generation time and laboratory tractability constitute key prerequisites for study organisms in carrying out long-term evolution in a compact period of time. Consequently, the majority of such experiments exploit microbes. Unfortunately, microbes cannot speak to the evolution of the many traits and properties that are unique to eukaryotes and metazoans, such as sexual selection, development and behaviour. Thus, key systems of study have expanded to include fruitflies, plants, fish and mice, among many others [6]. The nematode Caenorhabditis elegans and its relatives offer another powerful, but underexploited option for addressing ecological and evolutionary questions with experimental evolution. In this review, we synthesize the current state of what has been learned by applying experimental evolution to C. elegans with its potential for establishing new discoveries.
2. Virtues and vices of the Caenorhabditis elegans experimental system
Caenorhabditis elegans is one of the supermodels of modern biology. Since its debut in this capacity in 1974 [7], C. elegans became the first metazoan to have its genome sequenced [8], its complete cellular developmental pathway has been mapped [9] and its neural connection networks determined [10]. This focus of study has led to the invention and application of many molecular experimental methods, further accelerating C. elegans investigations; for example, RNA interference by feeding [11] and green-fluorescent protein expression reporters [12]. Thousands of laboratory mutants with a common genetic background confer experimentally useful phenotypes, each of which is publicly available and can be cryopreserved indefinitely [13]. With the wealth of knowledge and tools gleaned over nearly four decades of study, as well as intriguing organismal biology in its own right, the worm has now expanded into more diverse areas of biology, including evolution and ecology [14].
In the laboratory, the C. elegans life cycle can be as short as 50 h, with a single egg hatching into a larva that undergoes four moults before maturing into the adult hermaphrodite that lays around 300 self-fertilized eggs over a period of about a week (figure 1) [15]. C. elegans’s lifespan averages two weeks, but crowding and starvation induce a diapause-like ‘dauer’ stage, in which worms stop feeding and can live for several months. First-stage larvae survive cryopreservation in a glycerol solution, permitting indefinite storage of populations or isogenic strains. Worms grow readily to their adult length of approximately 1 mm when reared on a diet of E. coli or on many other bacterial species, or even in an axenic medium. Populations may be reared in liquid culture, on an agar substrate in Petri dishes or in three-dimensional environments.
Figure 1.
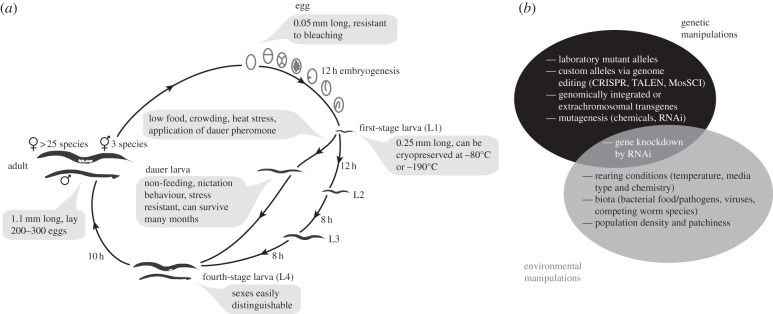
(a) Life cycle of C. elegans at 25°C, annotated with key life-history features pertinent to experimental study. (b) A schematic diagram indicating some of the many genetic and environmental manipulations possible in rearing worms. See main text for more details on methods and examples.
Caenorhabditis elegans is androdioecious, meaning that self-fertilizing hermaphrodites and rare males comprise natural and laboratory populations. Sex determination is chromosomal, with males being haploid for the X chromosome, but diploid for the five autosomes (hermaphrodites are diploid for all chromosomes). In N2, the commonly used reference strain, males occur in populations at the frequency at which they are expected to arise through X-chromosome non-disjunction in meiosis, although alternative genetic backgrounds and environmental conditions yield males with greater abundance [16]. When mated to a male, a hermaphrodite's sperm stores are supplemented by the male, so that she lays more eggs; outcrossed egg production occurs first owing to precedence of the larger male sperm in fertilization [17,18]. A drawback to this highly selfing sexual system for many purposes is that populations will inbreed at a high rate, creating extensive linkage disequilibrium and diminished heterozygosity in genetically variable populations. This issue can be circumvented by standard genetic modifications that transform hermaphrodites into females [19] or by using one of the many related species of Caenorhabditis that outcross obligatorily [20].
Indeed, C. elegans is no longer the only player for research in the Caenorhabditis genus, which contains at least 26 species in laboratory culture [21]. These other members of the genus have very similar life cycles to C. elegans (figure 1). Twelve of these species have had their genomes sequenced (see http://www.nematodes.org/nematodegenomes), permitting comparative methods to analyse development and genetics. The relatively compact size of Caenorhabditis genomes (100–150 Mb, approx. 25% of which is coding genes, approx. 17% repetitive DNA) also makes genome sequencing of experimental evolution populations a viable strategy with high-throughput sequencing [22,23]. Only Caenorhabditis briggsae, C. sp. 11 and C. elegans have self-fertilizing hermaphrodites. All other species in the genus share the ancestral gonochoristic (dioecious) reproductive habit with a 1 : 1 ratio of males and females, allowing standard crosses and experimental designs comparable with flies and beetles. Despite the many species, ecological understanding of this genus, as for many model organisms, is still in its infancy [14]. Nevertheless, cryopreserved strain collections from diverse populations around the world and numerous phylogeographic analyses provide the basis for relating experimental evolution to natural variation [24,25]. Ongoing advances in automation, image processing and microfluidics makes possible high-throughput studies for many traits [26]. Chemosensory, behavioural and fitness traits are particularly amenable to such automation. However, the streamlined morphology of all Caenorhabditis means that evolutionary study of form is a challenge in these organisms, excepting relatively subtle developmental phenotypes. The simplicity of form and behaviour may therefore be viewed as a drawback or as a benefit, depending on the focal question.
The molecular methods available to C. elegans researchers are unparalleled. Reversible gene knockdown by RNAi is straightforward in C. elegans by simply introducing or removing a plasmid into the food bacteria [11], and RNAi libraries are publicly available to target most genes in the genome. Publicly available gene deletion strains are plentiful, and recent work on TALENs and CRISPR/Cas9 has allowed targeted genome editing to engineer particular alleles in Caenorhabditis [27]. These molecular tools offer powerful means of interrogating the outcomes of experimental evolution in subsequent experiments.
3. Experimental evolution paradigms applied to Caenorhabditis elegans
Evolution, by definition, requires genetic change over time. Thus, there must be a source of genetic diversity for populations to undergo this change. In experimental evolution, genetic diversity is generally obtained in one of three ways: evolution from standing natural genetic variation, competition experiments between defined alternative alleles or evolution from new mutational input (naturally arising, or artificially elevated). Here, we describe these approaches as applied to Caenorhabditis and then point to specific example applications in the next section.
The large number of wild isolates of C. elegans available to researchers provides a cross-section of natural variation in the species as starting material with known genome sequence [24,28]. In order to generate diversity for selection to act upon quickly, an arbitrary number of strains can be crossed using a careful design to reduce linkage [19], to be used as a starting population [16,29]; established strain resources are available that were derived in this way [19,30]. Outbreeding species of Caenorhabditis harbour even greater genetic variation in traits and DNA, with Caenorhabditis brenneri having the highest molecular variation known for any eukaryote [31].
The starting genetic variation is more specific in experiments in which the effect of a single gene is quantified. To do this, an allele of a single gene (e.g. a knockout or gain of function laboratory mutation) with an interesting phenotypic effect can be tested in a common genetic background or introgressed into multiple backgrounds [18,32–34]. The strains can then be allowed to evolve together (figure 2). This approach allows testing of hypotheses about differences in fitness owing to single alleles under alternative controlled environmental conditions, but requires the genetic construction of appropriate tester strains.
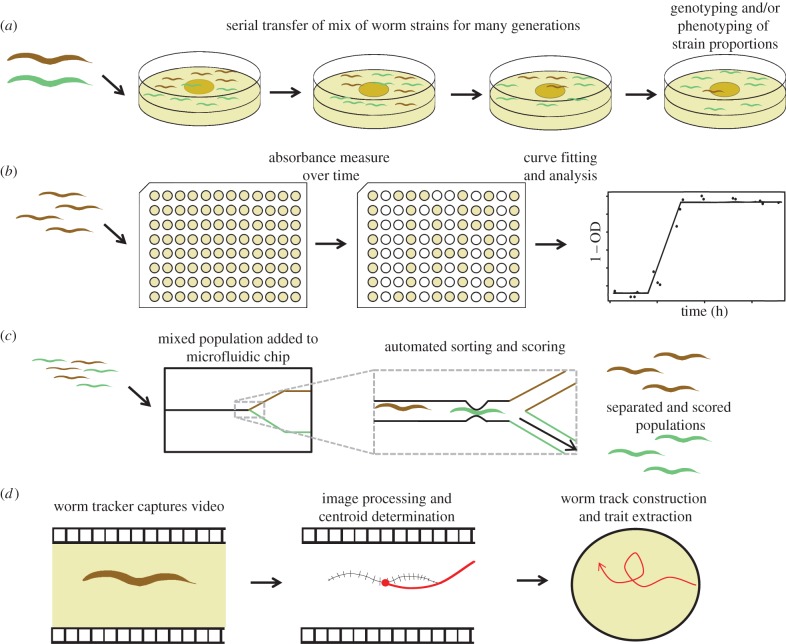
Figure 2.
Example fitness and phenotyping assays in Caenorhabditis. (a) Competitive fitness on Petri dishes (or in liquid). Two strains of worm are inoculated on each plate and serially transferred. Change in frequency of strains is determined over time by the use of a fluorescent transgenic marker [18,32]. (b) Non-competitive liquid fitness assays. Worms are inoculated into wells seeded with bacterial food in high-throughput microtitre plates. Absorbance change is measured over time and growth parameters determined for each well [35,36]. Alternatively, worms in liquid culture can be put through a biosorter to count, measure and sort the animals. (c) Microfluidic devices have been designed for worm sorting and phenotyping [37]. For example, worms are added to a microfluidic chip and stopped at a junction. Worms can be automatically scored for length, or sorted by fluorescence or another trait ranging from chemotaxis to fecundity [37]. (d) Animal behaviour also can be assessed in relatively high-throughput assays. For example, videos of worm movement when added to a plate with a chemoattractant or environmental gradient permit worm-tracking equipment or software [26], coupled with image processing, to determine and track the skeleton of the worm. Worm trajectories can then be quantified in terms of speed, curvature and other features. (Online version in colour.)
It is possible to wait for the slow response to de novo mutations from an isogenic ancestor [38], and this is exactly the approach taken by mutation accumulation (MA) experiments to measure the mutation process itself [39]. Alternatively, initially isogenic experimental populations can be stocked with new mutations by chemical mutagenesis [40–43] or with genetics, such as from a knockout of a mismatch repair gene [44,45]. RNAi knockdown of DNA repair genes provides an inducible means of introducing new mutations with an endogenous mutational spectrum [46]. These mutagenic approaches introduce many more new mutations than would occur naturally, allowing evolution to proceed more quickly.
4. Current contributions from Caenorhabditis elegans experimental evolution
Experimental evolution studies in Caenorhabditis have touched on diverse areas of biology. This work offers improved understandings of general problems in evolution, as well as of particulars about C. elegans biology. The first evolution experiment carried out with C. elegans, in fact, pre-dates its inception as a modern biological model [38]. Below, we discuss some of the key topics for which C. elegans experimental evolution has been important, including host–pathogen coevolution, fundamental mutational properties, mating systems and life-history theory.
(a). Pathogenesis, coevolution and ecological microcosms
To nematode worms, bacteria are simply food, but can be pathogenic upon ingestion. With the amenability of both microbes and worms to experimental evolution, coevolution of host–pathogen dynamics is a natural extension to pursue. Species interactions, environmental structure and multi-species systems are classic areas of theoretical study, and notoriously difficult to investigate from a microbial perspective. C. elegans, as a motile metazoan, allows a convenient inroad for these topics.
Different Caenorhabditis strains and species vary in their susceptibility to bacterial pathogens, including Serratia marcescens [47] and Bacillus thuringiensis [29]. This fact has been exploited to test models of host–pathogen dynamics by tracking coevolution between worm host and bacterial pathogen, and then quantifying responses to selection imposed on each of them. For example, trade-offs evolve between worm growth rates and resistance to pathogens [29,48], local adaptation occurs between pathogens and hosts [49,50], and outcrossing sex increases during host–pathogen coevolution [51–53]. With the recent discovery of viruses that infect natural populations of C. elegans and C. briggsae [54], viral coevolution experiments provide a further dimension to test these and related hypotheses.
In addition to coevolution, multi-species experiments can test ecologically motivated questions as well. A tri-trophic microcosm comprising C. elegans, Pseudomonas syringae and phage Φ6 has been developed to study simple ecosystem interactions [55]. Dispersal is an ecologically important trait, and a system has been developed and modelled in C. elegans to describe competition between strains that differ in dispersal rates under different ecological scenarios, made possible by virtue of specific alleles that affect motility and fertility [56,57]. By competing in a patchy environment, the experiments showed that environmental variation can favour the evolution of increased dispersal tendencies and that alternative dispersal strategies can coexist under intermediate rates of environmental disturbance [56,57]. Fragmented environments have also been used to test hypotheses about balancing selection for the maintenance of genetic variation within a population, specifically associated with genetic control of feeding strategies [33]. While Petri dishes provide a relatively uniform environment, imposing physical barriers to dispersal has allowed study of dispersal propensity [52] and construction of artificial dirt permits worms to perform behaviour in a more realistic three-dimensional environment [58]. The inclusion of multiple species and environmental heterogeneity into experimental designs suggests tantalizing opportunities to test theories about predator–prey dynamics, competition, biodiversity and ecosystem function.
(b). Understanding mutation
One of the most commonly undertaken applications in C. elegans experimental evolution has been for MA. This work has yielded great progress into characterizing the fundamental mutational parameters that underlie evolutionary theory to describe the rate, fitness effects, biases and types of mutation [39].
Keightley & Caballero [59] started MA in C. elegans by repeatedly bottlenecking to a single individual for 60 generations. They found that reproductive output declined by 0.03% per generation. Recent studies have expanded to use whole-genome sequencing of strains to quantify mutation rates directly after more than 300 generations of MA [22,23], as well as permutations on the MA scheme, including manipulations of mutation rate, the per generation bottleneck size, environmental conditions, natural genetic background of strain founders and focal species (table 1).
Table 1.
Major hypotheses tested with experimental evolution in Caenorhabditis.
| topic | key question or idea | finding | references |
|---|---|---|---|
| coevolution | is there genetic diversity for pathogen resistance? | found for resistance to S. marsecens, B. thuringensis, P. luminscens | [29,47,60] |
| does evolution of resistance to pathogens have trade-offs? | an increase in resistance, but reductions in growth and feeding rate | [29,48] | |
| does coevolution and local adaptation occur between host and pathogen? | populations showed higher resistance to their own pathogens and genetic diversity between populations increased | [49,50] | |
| red queen hypothesis | outcrossed sex allowed faster adaptation to parasites | [51–53] | |
| population structure and ecosystems | can ecosystems be constructed? | three species interactions and dependencies | [55] |
| is dispersal beneficial in varying environments? | dispersal is beneficial under random extinction, can be regulated by a single gene and can be selected for | [33,42,56,57] | |
| effects and accumulation of mutations | how do traits evolve with mutation accumulation (MA)? | fitness, body size, behaviour, oxidizing state and other traits degrade | [59,61–65] |
| does fitness recover after MA? | restoring selection, or greatly increasing mutation rate leads to fitness recovery | [43,66] | |
| what is the rate and spectrum of new mutations? | many mutations identified after 396 generations of MA by genome sequencing | [22,23,67] | |
| do mutation properties differ among genetic backgrounds, species or environments? | different strains, species and conditions do or do not have differing mutation profiles | [23,68–71] | |
| mating systems | are males evolutionary relics? | males reduce in frequency under the lack of selection, depending on the strain and genetic background | [16,41,72,73] |
| does outcrossing sex promote removal of detrimental mutations? | male frequencies increased under higher mutational loads | [41,44] | |
| does outcrossing sex accelerate adaptation? | male frequencies increased under directional selection | [42,51,52,74] | |
| does outcrossing sex help retain heterozygosity? | no difference between reproductive modes, balancing selection dominates | [30] | |
| do inbreeding and outbreeding depression depend on reproductive mode? | inbreeders showed outbreeding depression and vice versa | [75] | |
| how does sexual selection by sperm competition evolve? | competition led to larger sperm and restored male sexual function | [34,76,77] | |
| life history | does increased lifespan have pleiotropic costs? | fewer offspring for longer lived worms in one study, but not in another | [78,79] |
| is individual vigour linked to lifespan? | selection for good condition worms led to longer lifespans | [20] | |
| how does selection affect reproductive life-history trade-offs? | selection between faster generation times and offspring number changed the trade-off | [18] |
In addition to the accumulation of new mutations, some studies have explored the clearance of and interactions between mutations. For example, experiments using different population sizes confirmed theory that smaller populations fix more detrimental mutations [45]. The dynamics of recovery from MA have been explored, showing that epistatic compensatory mutations dominate [80]. Investigation of the clearance of mutations at differing mutation rates has found that an increase in mutation rate can lead to a paradoxical increase in fitness [43]. Some theory predicts gonochoristic species to have higher mutation rates than selfing species, and results appear to support this [68]. Different androdioecious species also have been tested for mutation rate differences, although the conclusions regarding species differences vary depending on how mutation rate is measured [23,69]. Further work with MA should focus on the distribution of selective effects of mutations, and the relative rate of beneficial and detrimental mutations.
(c). Mating systems
One especially appealing aspect of Caenorhabditis biology is the existence of distinct mating systems among species, and the ability to manipulate mating and sex determination systems in C. elegans using both genetics and exogenous treatments (e.g. RNAi, temperature) [81,82]. The evolution of sex is a long-standing problem in evolutionary biology [83], having attracted many experimental evolution studies in a variety of organisms [84]. While Caenorhabditis experiments cannot contrast sex versus asex, studies have investigated the related problem of selfing versus outcrossing [16]. This work also has attempted to explain the incidence of males in populations, which relates directly to the frequency of outcrossed reproduction, and the effect of those outcrossing males on adaptation.
Why do functional males still occur in C. elegans, given that they are not strictly necessary for reproduction? Males are rare in natural collections [85,86], and both modelling and multi-generation experiments based on the standard N2 laboratory strain indicate rapid loss of males from populations, suggesting that they are not necessary for population survival [72,86–88]. However, the males of the N2 strain seem to have particularly low sexual vigour compared with other wild isolates and males of other species [73,89,90]. Experimental evolution under elevated mutation rates, either endogenously via mismatch repair mutants or exogenously via chemical mutagenesis, results in males and outcrossing persisting for longer durations within populations over time [41,44]. Moreover, different genetic backgrounds allow greater male persistence and outcrossing across generations [16,41].
Theory also predicts that outcrossing sex will accelerate the rate of adaptation under directional selection [91]. Experimental evolution using starvation stress [92], directional selection [42,74] and coevolution with pathogens [51,52] all showed that outcrossing sex was favoured over selfing under these conditions. Experiments with alternative reproductive modes also suggest a role for balancing selection maintaining variation in an experimental setting [30].
The differing reproductive strategies among closely related species of Caenorhabditis allow comparative insights into reproductive behaviour. For example, wild isolates of Caenorhabditis remanei, a gonochoristic species, show strong inbreeding depression (and a lack of purging of deleterious mutations) when propagated over 13 generations, whereas the highly selfing C. elegans and C. sp. 11 yield outbreeding depression consistent with the presence of ‘coadapted gene complexes’ [75,93].
Sexual selection also is a long-standing area of study in evolutionary biology, and C. elegans provides a prime underexploited system for investigation. One study explored sperm competition: in C. elegans, large sperm are competitively superior [76]. When C. elegans populations were forced to reproduce by outcrossing, which imposed male–male sperm competition, selection appears to have driven the evolution of larger males and males making larger sperm [34,94]. Natural genetic variation exists for a variety of mating traits, upon which experimental selection pressures could act [18,90]. The sex determination pathway of C. elegans is well understood [95], and mutations in this pathway can produce intersex individuals. Worms which were intersex owing to mutations in tra-2 and xol-1 re-evolved high levels of sexual dimorphism in response to selection over the course of 50 generations, shedding light into the developmental evolution of sexual dimorphism [77]. Although species differ in attractiveness to female mating pheromones and mating propensity [89], many aspects of worm mating remain to be studied from an evolutionary perspective.
(d). Ageing and life history
Caenorhabditis elegans has become a model system for ageing research [96], and this is a fruitful target for selection experiments. Lifespan has long been predicted to show a trade-off between long life and faster reproduction [97], and much work attempts to determine whether this prediction holds true generally. Worms are particularly attractive for this, as they show ageing and senescence, unlike microbes, but have life cycles on the order of weeks (rather than years, as is common in many animals).
A strain of C. elegans carrying an allele of the age-1 gene—known to increase lifespan, but also leading to increased dauer formation—showed no cost of increased lifespan under benign conditions [78]. However, upon cyclical starvation, the long-lived worms rapidly declined in frequency in experimental populations owing to dauer formation, indicating that there was an antagonistic pleiotropic effect to the longer lifespan conferred by age-1. In other work, lifespan and reproduction were again probed by selecting for early offspring [79]. As expected, early offspring numbers increased and late offspring decreased. Yet lifespan did not show a corresponding decline, casting doubt on the idea of antagonistic pleiotropy between early- and late-acting genetic effects. Some recent theory posits that while high mortality will select for a decrease in lifespan, if mortality is condition-dependent, then longer lifespans will evolve [98]. To test this, C. remanei populations were subjected to condition-dependent selection, with worms dying either randomly or following heat stress, which preferentially kills low-condition worms [20]. The populations subjected to random mortality evolved a reduced lifespan, as expected from classic theory, but those with condition-dependent mortality evolved longer lifespans, consistent with the updated theory. The wealth of data on the molecular mechanisms of ageing in the worm offers a possibility of uniting both new and long-standing theory with evolutionary process and molecular function.
In C. elegans hermaphrodites, reproduction is often sperm-limited [17]. This is due to sperm being produced before a switch to oocyte production, leading to a trade-off between number of sperm produced and the earliest time at which fertilized eggs can be laid. Mutations known to influence sperm number have been competed in order to test theory about this fitness trade-off between early reproduction and total lifetime reproduction [18,99]. These examples illustrate the broad range of life-history trade-offs that are tractable for study by experimental evolution in the worm.
5. Prospects
Experimental evolution research in C. elegans has just scratched the surface of what is possible. Caenorhabditis elegans is an enviable research model in many respects, with the benefits of short life cycle and laboratory amenability combined with the trappings of a higher eukaryote, giving it a superb potential as an experimental system for evolutionary studies. Here, we highlight a few areas that are ripe for interrogation, or in need of development (table 2), in addition to the suite of topics commonly addressed by experimental evolution in other organisms [2].
Table 2.
Topics in need of development using Caenorhabditis experimental evolution.
| topic | comments |
|---|---|
| speciation | The recently discovered species in the genus that can be hybridized in the laboratory offer the opportunity for an experimental insight into speciation [100–103]. Laboratory experiments selecting for reproductive isolation and reinforcement have a long heritage in Drosophila, and work in Caenorhabditis can complement this. |
| repeatability of evolution | Caenorhabditis is a large genus and a large number of species allows a comparative approach. While recent studies have tested the repeatability of evolution [104], this has occurred in a single ancestral genotype. Analysis of the genetic change during adaptation in multiple closely related species would provide a fascinating insight into evolution in different genetic backgrounds. Comparative experimental evolution work on genomic mutation rates [23] and sexual systems [72,75,77] also will be extremely valuable. The diverse reproductive strategies have already been exploited for mutation studies, and adaptive evolution is an obvious next direction to explore in detail. |
| ecological theory | Host–parasite coevolution is, and will be, a prosperous area for C. elegans experimental evolution [49,52]. But this is just one possible ecosystem. The literature on ecosystem dynamics and predator prey interactions is vast, and the systems developed with nematodes offer a way to empirically test some of these models. Promising topics include dispersal, predator–prey/consumer–resource dynamics, environmental variability and maintenance of biodiversity. |
(a). Sexual selection and behavioural evolution
Tests of ideas about sexual selection remain largely unexplored in Caenorhabditis, despite the sperm competition work described above. Worms have many compelling features for investigating sexual selection and sexual conflict: sperm size differences in sperm competition, mating plugs, mating pheromones, plastic re-mating propensities. Mating is one of the most cognitively demanding procedures undertaken by male worms [105], and elucidating and modifying the mate recognition and decision systems by evolution would allow a fascinating insight into behaviour. Worms perform both learned and stereotyped behavioural responses, albeit simple from an anthropocentric view [106]. The complete neural network has been mapped, and worm behaviour on a two-dimensional plane (agar plates) offers a simple environment in which to quantify behavioural evolution via automated video image processing. In addition, there are many relevant C. elegans manipulative genetic tricks, and the transparent cuticle allows direct visualization of the reproductive tract contents [107], permitting a diversity of options for experimental evolution and follow-up study of molecular mechanism about evolutionary responses.
(b). Genotype–phenotype mapping
A persistent goal in evolutionary genetics is the mapping of natural phenotypic differences to their genetic causes. Methods such as bulk segregants analysis or X-QTL combine selection and evolution with genotyping, allowing the elucidation of this link. The recent construction and genome sequencing of more than 2000 mutagenized and wild strains of C. elegans provides a compelling substrate for experimental evolution and for connecting genotype to phenotype [28]. Until recent development of automated high-throughput phenotyping [35,36], fitness assays have not been as easy or as powerful as in many microbial systems. As these techniques are refined, more individuals and populations can be assayed for fitness, allowing better detection of phenotypic trait differences, fitness effects, adaptive trajectories and connection to real-world ecologies.
(c). Integration with systems biology
The developmental genetics of C. elegans has been exceptionally well characterized from traditional molecular genetic approaches. The work of Chandler et al. [77] shows the promise of evolutionary inroads into molecular and systems biology. By altering one or a few genes, and then allowing compensatory mutations to evolve through experimental evolution, one can generate targets for subsequent molecular genetic analysis. In addition to addressing intriguing hypotheses about the evolution of development and genetic networks, this approach provides a complementary method to quantitative genetics (e.g. QTL mapping, GWAS) and standard forward and reverse genetics (e.g. mutagenesis screens) to understand gene–phenotype mapping and molecular mechanisms.
6. Concluding remarks
Recent years have seen increasing adoption of Caenorhabditis in experimental evolution, but it is not yet mainstream. So far, the major thrusts in C. elegans experimental evolution have targeted understanding mutational properties, host–pathogen coevolution and mating system evolution. Given the experimental tractability and toolkit conferred on C. elegans by decades of development for biomedical research, and the compelling details of its organismal biology, the Caenorhabditis system is primed to address topical issues throughout the disciplines of evolution and ecology with experimental evolution (tables 1 and 2). The promise of high-throughput phenotyping and genome sequencing of experimental populations, coupled with multi-species systems, can rapidly help connect evolutionary process and mechanism.
Funding statement
A.D.C. is supported by funds from the Natural Sciences and Engineering Research Council of Canada, the United States’ National Institutes of Health, and a Canada Research Chair.
References
- 1.Buckling A, MacLean RC, Brockhurst MA, Colegrave N. 2009. The beagle in a bottle. Nature 457, 824–829 (doi:10.1038/nature07892) [DOI] [PubMed] [Google Scholar]
- 2.Kawecki TJ, Lenski RE, Ebert D, Hollis B, Olivieri I, Whitlock MC. 2012. Experimental evolution. Trends Ecol. Evol. 27, 547–560 (doi:10.1016/j.tree.2012.06.001) [DOI] [PubMed] [Google Scholar]
- 3.Dallinger WH. 1878. On the life-history of a minute septic organism: with an account of experiments made to determine its thermal death point. Proc. R. Soc. Lond. 27, 332–350 (doi:10.1098/rspl.1878.0055) [Google Scholar]
- 4.Lenski RE, Rose MR, Simpson SC, Tadler SC. 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 138, 1315–1341 (doi:10.1086/285289) [Google Scholar]
- 5.Garland TJ, Rose MR. 2009. Experimental evolution: concepts, methods, and applications of selection experiments. Berkeley, CA: University of California Press [Google Scholar]
- 6.Bell G. 2008. Selection: the mechanism of evolution, 2nd edn Oxford, UK: Oxford University Press [Google Scholar]
- 7.Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.C. elegans Sequencing Consortium 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012–2018 (doi:10.1126/science.282.5396.2012) [DOI] [PubMed] [Google Scholar]
- 9.Sulston JE, Horvitz HR. 1977. Post-embryonic cell lineages of the nematode Caenorhabditis elegans. Dev. Biol. 56, 110–156 (doi:10.1016/0012-1606(77)90158-0) [DOI] [PubMed] [Google Scholar]
- 10.White JG, Southgate E, Thomson JN, Brenner S. 1986. The structure of the nervous system of the nematode Caenorhabditis elegans. Phil. Trans. R. Soc. Lond. B 314, 1–340 (doi:10.1098/rstb.1986.0056) [DOI] [PubMed] [Google Scholar]
- 11.Timmons L, Fire A. 1998. Specific interference by ingested dsRNA. Nature 395, 854–854 (doi:10.1038/27579) [DOI] [PubMed] [Google Scholar]
- 12.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. 1994. Green fluorescent protein as a marker for gene expression. Science 263, 802–805 (doi:10.1126/science.8303295) [DOI] [PubMed] [Google Scholar]
- 13.Stiernagle T. 2006. Maintenance of C. elegans. WormBook, 11 February. (doi:10.1895/wormbook.1.101.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Félix M-A, Braendle C. 2010. The natural history of Caenorhabditis elegans. Curr. Biol. 20, R965–R969 (doi:10.1016/j.cub.2010.09.050) [DOI] [PubMed] [Google Scholar]
- 15.Wood WB. 1988. The nematode Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- 16.Anderson JL, Morran LT, Phillips PC. 2010. Outcrossing and the maintenance of males within C. elegans populations. J. Hered. 101, S62–S74 (doi:10.1093/jhered/esq003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ward S, Carrel JS. 1979. Fertilization and sperm competition in the nematode Caenorhabditis elegans. Dev. Biol. 73, 304–321 (doi:10.1016/0012-1606(79)90069-1) [DOI] [PubMed] [Google Scholar]
- 18.Murray RL, Cutter AD. 2011. Experimental evolution of sperm count in protandrous self-fertilizing hermaphrodites. J. Exp. Biol. 214, 1740–1747 (doi:10.1242/jeb.053181) [DOI] [PubMed] [Google Scholar]
- 19.Teotónio H, Carvalho S, Manoel D, Roque M, Chelo IM. 2012. Evolution of outcrossing in experimental populations of Caenorhabditis elegans. PLoS ONE 7, e35811 (doi:10.1371/journal.pone.0035811) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H, Maklakov AA. 2012. Longer life span evolves under high rates of condition-dependent mortality. Curr. Biol. 22, 2140–2143 (doi:10.1016/j.cub.2012.09.021) [DOI] [PubMed] [Google Scholar]
- 21.Kiontke KC, Félix M-A, Ailion M, Rockman MV, Braendle C, Pénigault J-B, Fitch DH. 2011. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol. Biol. 11, 339 (doi:10.1186/1471-2148-11-339) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denver DR, et al. 2009. A genome-wide view of Caenorhabditis elegans base-substitution mutation processes. Proc. Natl Acad. Sci. USA 106, 16 310–16 314 (doi:10.1073/pnas.0904895106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denver DR, Wilhelm LJ, Howe DK, Gafner K, Dolan PC, Baer CF. 2012. Variation in base-substitution mutation in experimental and natural lineages of Caenorhabditis nematodes. Genome Biol. Evol. 4, 513–522 (doi:10.1093/gbe/evs028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersen EC, Gerke JP, Shapiro JA, Crissman JR, Ghosh R, Bloom JS, Félix M-A, Kruglyak L. 2012. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat. Genet. 44, 285–290 (doi:10.1038/ng.1050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Félix M-A, Jovelin R, Ferrari C, Han S, Cho YR, Andersen EC, Cutter AD, Braendle C. 2013. Species richness, distribution and genetic diversity of Caenorhabditis nematodes in a remote tropical rainforest. BMC Evol. Biol. 13, 10 (doi:10.1186/1471-2148-13-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Husson SJ. 2012. Keeping track of worm trackers. WormBook, 10 September. (doi:10.1895/wormbook.1.156.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frøkjær-Jensen C. 2013. Exciting prospects for precise engineering of Caenorhabditis elegans genomes with CRISPR/Cas9. Genetics 195, 635–642 (doi:10.1534/genetics.113.156521) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson O, et al. 2013. The million mutation project: a new approach to genetics in Caenorhabditis elegans. Genome Res. 23, 1749–1762 (doi:10.1101/gr.157651.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schulte RD, Makus C, Hasert B, Michiels NK, Schulenburg H. 2010. Multiple reciprocal adaptations and rapid genetic change upon experimental coevolution of an animal host and its microbial parasite. Proc. Natl Acad. Sci. USA 107, 7359–7364 (doi:10.1073/pnas.1003113107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chelo IM, Teotónio H. 2013. The opportunity for balancing selection in experimental populations of Caenorhabditis elegans. Evolution 67, 142–156 (doi:10.1111/j.1558-5646.2012.01744.x) [DOI] [PubMed] [Google Scholar]
- 31.Cutter AD, Jovelin R, Dey A. 2013. Molecular hyperdiversity and evolution in very large populations. Mol. Ecol. 22, 2074–2095 (doi:10.1111/mec.12281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duveau F, Félix M-A. 2012. Role of pleiotropy in the evolution of a cryptic developmental variation in Caenorhabditis elegans. PLoS Biol. 10, e1001230 (doi:10.1371/journal.pbio.1001230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gloria-Soria A, Azevedo RBR. 2008. npr-1 regulates foraging and dispersal strategies in Caenorhabditis elegans. Curr. Biol. 18, 1694–1699 (doi:10.1016/j.cub.2008.09.043) [DOI] [PubMed] [Google Scholar]
- 34.LaMunyon CW, Ward S. 2002. Evolution of larger sperm in response to experimentally increased sperm competition in Caenorhabditis elegans. Proc. R. Soc. Lond. B 269, 1125–1128 (doi:10.1098/rspb.2002.1996) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elvin M, Snoek LB, Frejno M, Klemstein U, Kammenga JE, Poulin GB. 2011. A fitness assay for comparing RNAi effects across multiple C. elegans genotypes. BMC Genomics 12, 510 (doi:10.1186/1471-2164-12-510) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramani AK, Chuluunbaatar T, Verster AJ, Na H, Vu V, Pelte N, Wannissorn N, Jiao A, Fraser AG. 2012. The majority of animal genes are required for wild-type fitness. Cell 148, 792–802 (doi:10.1016/j.cell.2012.01.019) [DOI] [PubMed] [Google Scholar]
- 37.Shi W, Wen H, Lin B, Qin J. 2011. Microfluidic platform for the study of Caenorhabditis elegans. Top. Curr. Chem. 304, 323–338 (doi:10.1007/128_2011_145) [DOI] [PubMed] [Google Scholar]
- 38.Brun J. 1965. Genetic adaptation of Caenorhabditis elegans (Nematoda) to high temperatures. Science 150, 1467–1467 (doi:10.1126/science.150.3702.1467)5322584 [Google Scholar]
- 39.Baer CF, Miyamoto MM, Denver DR. 2007. Mutation rate variation in multicellular eukaryotes: causes and consequences. Nat. Rev. Genet. 8, 619–631 (doi:10.1038/nrg2158) [DOI] [PubMed] [Google Scholar]
- 40.Flibotte S, et al. 2010. Whole-genome profiling of mutagenesis in Caenorhabditis elegans. Genetics 185, 431–441 (doi:10.1534/genetics.110.116616) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manoel D, Carvalho S, Phillips PC, Teotónio H. 2007. Selection against males in Caenorhabditis elegans under two mutational treatments. Proc. R. Soc. B 274, 417–424 (doi:10.1098/rspb.2006.3739) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morran LT, Parmenter MD, Phillips PC. 2009. Mutation load and rapid adaptation favour outcrossing over self-fertilization. Nature 462, 350–352 (doi:10.1038/nature08496) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morran LT, Ohdera AH, Phillips PC. 2010. Purging deleterious mutations under self fertilization: paradoxical recovery in fitness with increasing mutation rate in Caenorhabditis elegans. PLoS ONE 5, e14473 (doi:10.1371/journal.pone.0014473) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cutter AD. 2005. Mutation and the experimental evolution of outcrossing in Caenorhabditis elegans. J. Evol. Biol. 18, 27–34 (doi:10.1111/j.1420-9101.2004.00804.x) [DOI] [PubMed] [Google Scholar]
- 45.Estes S, Phillips PC, Denver DR, Thomas WK, Lynch M. 2004. Mutation accumulation in populations of varying size: the distribution of mutational effects for fitness correlates in Caenorhabditis elegans. Genetics 166, 1269–1279 (doi:10.1534/genetics.166.3.1269) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pothof J, van Haaften G, Thijssen K, Kamath RS, Fraser AG, Ahringer J, Plasterk RHA, Tijsterman M. 2003. Identification of genes that protect the C. elegans genome against mutations by genome-wide RNAi. Gene Dev. 17, 443–448 (doi:10.1101/gad.1060703) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schulenburg H, Ewbank JJ. 2004. Diversity and specificity in the interaction between Caenorhabditis elegans and the pathogen Serratia marcescens. BMC Evol. Biol. 4, 49 (doi:10.1186/1471-2148-4-49) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schulte RD, Hasert B, Makus C, Michiels NK, Schulenburg H. 2012. Increased responsiveness in feeding behaviour of Caenorhabditis elegans after experimental coevolution with its microparasite Bacillus thuringiensis. Biol. Lett. 8, 234–236 (doi:10.1098/rsbl.2011.0684) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schulte RD, Makus C, Hasert B, Michiels NK, Schulenburg H. 2011. Host-parasite local adaptation after experimental coevolution of Caenorhabditis elegans and its microparasite Bacillus thuringiensis. Proc. R. Soc. B 278, 2832–2839 (doi:10.1098/rspb.2011.0019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schulte RD, Makus C, Schulenburg H. 2013. Host-parasite coevolution favours parasite genetic diversity and horizontal gene transfer. J. Evol. Biol. 26, 1836–1840 (doi:10.1111/jeb.12174) [DOI] [PubMed] [Google Scholar]
- 51.Masri L, Schulte RD, Timmermeyer N, Thanisch S, Crummenerl LL, Jansen G, Michiels NK, Schulenburg H. 2013. Sex differences in host defence interfere with parasite-mediated selection for outcrossing during host–parasite coevolution. Ecol. Lett. 16, 461–468 (doi:10.1111/ele.12068) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morran LT, Schmidt OG, Gelarden IA, Parrish RC, Lively CM. 2011. Running with the red queen: host–parasite coevolution selects for biparental sex. Science 333, 216–218 (doi:10.1126/science.1206360) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morran LT, Parrish RC, Gelarden IA, Lively CM. 2013. Temporal dynamics of outcrossing and host mortality rates in host–pathogen experimental coevolution. Evolution 67, 1860–1868 (doi:10.1111/evo.12007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Félix M-A, et al. 2011. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 9, e1000586 (doi:10.1371/journal.pbio.1000586) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dennehy JJ, Friedenberg NA, Yang YW, Turner PE. 2006. Bacteriophage migration via nematode vectors: host–parasite–consumer interactions in laboratory microcosms. Appl. Environ. Microbiol. 72, 1974–1979 (doi:10.1128/AEM.72.3.1974-1979.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Friedenberg NA. 2003. Determinism in a transient assemblage: the roles of dispersal and local competition. Am. Nat. 162, 586–596 (doi:10.1086/378782) [DOI] [PubMed] [Google Scholar]
- 57.Friedenberg NA. 2003. Experimental evolution of dispersal in spatiotemporally variable microcosms. Ecol. Lett. 6, 953–959 (doi:10.1046/j.1461-0248.2003.00524.x) [Google Scholar]
- 58.Lockery SR, et al. 2008. Artificial dirt: microfluidic substrates for nematode neurobiology and behavior. J. Neurophysiol. 99, 3136–3143 (doi:10.1152/jn.91327.2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keightley PD, Caballero A. 1997. Genomic mutation rates for lifetime reproductive output and lifespan in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 94, 3823–3827 (doi:10.1073/pnas.94.8.3823) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sicard M, Hering S, Schulte R, Gaudriault S, Schulenburg H. 2006. The effect of Photorhabdus luminescens (Enterobacteriaceae) on the survival, development, reproduction and behaviour of Caenorhabditis elegans (Nematoda: Rhabditidae). Environ. Microbiol. 9, 12–25 (doi:10.1111/j.1462-2920.2006.01099.x) [DOI] [PubMed] [Google Scholar]
- 61.Ajie BC, Estes S, Lynch M, Phillips PC. 2005. Behavioral degradation under mutation accumulation in Caenorhabditis elegans. Genetics 170, 655–660 (doi:10.1534/genetics.104.040014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Azevedo RBR, Keightley PD, Laurén-Määttä C, Vassilieva LL, Lynch M, Leroi AM. 2002. Spontaneous mutational variation for body size in Caenorhabditis elegans. Genetics 162, 755–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Estes S, Ajie BC, Lynch M, Phillips PC. 2005. Spontaneous mutational correlations for life-history, morphological and behavioral characters in Caenorhabditis elegans. Genetics 170, 645–653 (doi:10.1534/genetics.104.040022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vassilieva LL, Lynch M. 1999. The rate of spontaneous mutation for life-history traits in Caenorhabditis elegans. Genetics 151, 119–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Joyner-Matos J, Hicks KA, Cousins D, Keller M, Denver DR, Baer CF, Estes S. 2013. Evolution of a higher intracellular oxidizing environment in Caenorhabditis elegans under relaxed selection. PLoS ONE 8, e65604 (doi:10.1371/journal.pone.0065604) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Estes S, Lynch M. 2003. Rapid fitness recovery in mutationally degraded lines of Caenorhabditis elegans. Evolution 57, 1022–1030 [DOI] [PubMed] [Google Scholar]
- 67.Denver DR, Morris K, Lynch M, Thomas WK. 2004. High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature 430, 679–682 (doi:10.1038/nature02697) [DOI] [PubMed] [Google Scholar]
- 68.Baer CF, Joyner-Matos J, Ostrow D, Grigaltchik V, Salomon MP, Upadhyay A. 2010. Rapid decline in fitness of mutation accumulation lines of gonochoristic (outcrossing) Caenorhabditis nematodes. Evolution 64, 3242–3253 (doi:10.1111/j.1558-5646.2010.01061.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baer CF, et al. 2005. Comparative evolutionary genetics of spontaneous mutations affecting fitness in rhabditid nematodes. Proc. Natl Acad. Sci. USA 102, 5785–5790 (doi:10.1073/pnas.0406056102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baer CF, Phillips N, Ostrow D, Avalos A, Blanton D, Boggs A, Keller T, Levy L, Mezerhane E. 2006. Cumulative effects of spontaneous mutations for fitness in Caenorhabditis: role of genotype, environment and stress. Genetics 174, 1387–1395 (doi:10.1534/genetics.106.061200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matsuba C, Ostrow DG, Salomon MP, Tolani A, Baer CF. 2013. Temperature, stress and spontaneous mutation in Caenorhabditis briggsae and Caenorhabditis elegans. Biol. Lett. 9, 20120334 (doi:10.1098/rsbl.2012.0334) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chasnov JR, Chow KL. 2002. Why are there males in the hermaphroditic species Caenorhabditis elegans? Genetics 160, 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wegewitz V, Schulenburg H, Streit A. 2008. Experimental insight into the proximate causes of male persistence variation among two strains of the androdioecious Caenorhabditis elegans (Nematoda). BMC Ecol. 8, 12 (doi:10.1186/1472-6785-8-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lopes PC, Sucena É, Santos ME, Magalhães S. 2008. Rapid experimental evolution of pesticide resistance in C. elegans entails no costs and affects the mating system. PLoS ONE 3, e3741 (doi:10.1371/journal.pone.0003741) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dolgin ES, Charlesworth B, Baird SE, Cutter AD. 2007. Inbreeding and outbreeding depression in Caenorhabditis nematodes. Evolution 61, 1339–1352 (doi:10.1111/j.1558-5646.2007.00118.x) [DOI] [PubMed] [Google Scholar]
- 76.LaMunyon CW, Ward S. 1999. Evolution of sperm size in nematodes: sperm competition favours larger sperm. Proc. R. Soc. Lond. B 266, 263–267 (doi:10.1098/rspb.1999.0631) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chandler CH, Chadderdon GE, Phillips PC, Dworkin I, Janzen FJ. 2012. Experimental evolution of the Caenorhabditis elegans sex determination pathway. Evolution 66, 82–93 (doi:10.1111/j.1558-5646.2011.01420.x) [DOI] [PubMed] [Google Scholar]
- 78.Walker DW, McColl G, Jenkins NL, Harris J, Lithgow GJ. 2000. Natural selection: evolution of lifespan in C. elegans . Nature 405, 296–297 (doi:10.1038/35012693) [DOI] [PubMed] [Google Scholar]
- 79.Anderson JL, Reynolds RM, Morran LT, Tolman-Thompson J, Phillips PC. 2011. Experimental evolution reveals antagonistic pleiotropy in reproductive timing but not life span in Caenorhabditis elegans. J. Gerontol. Ser. A, Biol. Sci. Med. Sci. 66, 1300–1308 (doi:10.1093/gerona/glr143) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Denver DR, Howe DK, Wilhelm LJ, Palmer CA, Anderson JL, Stein KC, Phillips PC, Estes S. 2010. Selective sweeps and parallel mutation in the adaptive recovery from deleterious mutation in Caenorhabditis elegans. Genome Res. 20, 1663–1671 (doi:10.1101/gr.108191.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hodgkin J. 2002. One lucky XX male: isolation of the first Caenorhabditis elegans sex-determination mutants. Genetics 162, 1501–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Janzen FJ, Phillips PC. 2006. Exploring the evolution of environmental sex determination, especially in reptiles. J. Evol. Biol. 19, 1775–1784 (doi:10.1111/j.1420-9101.2006.01138.x) [DOI] [PubMed] [Google Scholar]
- 83.Bell G. 1982. The masterpiece of nature: the evolution and genetics of sexuality. London, UK: Croom Helm [Google Scholar]
- 84.Goddard MR. 2007. Why bother with sex? Answers from experiments with yeast and other organisms. In Sex in fungi: molecular determination and evolutionary implications (ed. Heitman JKJ, JW Kronstad, JW Taylor, LA Casselton), pp. 489–506. Washington, DC: ASM Press [Google Scholar]
- 85.Barrière A, Félix M-A. 2005. High local genetic diversity and low outcrossing rate in Caenorhabditis elegans natural populations. Curr. Biol. 15, 1176–1184 (doi:10.1016/j.cub.2005.06.022) [DOI] [PubMed] [Google Scholar]
- 86.Barrière A, Félix M-A. 2007. Temporal dynamics and linkage disequilibrium in natural Caenorhabditis elegans populations. Genetics 176, 999–1011 (doi:10.1534/genetics.106.067223) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cutter AD, Avilés L, Ward S. 2003. The proximate determinants of sex ratio in C. elegans populations. Genet. Res. 81, 91–102 (doi:10.1017/S001667230300613X) [DOI] [PubMed] [Google Scholar]
- 88.Stewart AD, Phillips PC. 2002. Selection and maintenance of androdioecy in Caenorhabditis elegans. Genetics 160, 975–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Garcia LR, LeBoeuf B, Koo P. 2007. Diversity in mating behavior of hermaphroditic and male–female Caenorhabditis nematodes. Genetics 175, 1761–1771 (doi:10.1534/genetics.106.068304) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Teotónio H, Manoel D, Phillips PC. 2006. Genetic variation for outcrossing among Caenorhabditis elegans isolates. Evolution 60, 1300–1305 [PubMed] [Google Scholar]
- 91.Agrawal AF. 2006. Evolution of sex: why do organisms shuffle their genotypes? Curr. Biol. 16, R696–R704 (doi:10.1016/j.cub.2006.07.063) [DOI] [PubMed] [Google Scholar]
- 92.Morran LT, Cappy BJ, Anderson JL, Phillips PC. 2009. Sexual partners for the stressed: facultative outcrossing in the self-fertilizing nematode Caenorhabditis elegans. Evolution 63, 1473–1482 (doi:10.1111/j.1558-5646.2009.00652.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gimond C, Jovelin R, Han S, Ferrari C, Cutter AD, Braendle C. 2013. Outbreeding depression with low genetic variation in selfing Caenorhabditis nematodes. Evolution 67, 3087–3101 (doi:10.1111/evo.12203) [DOI] [PubMed] [Google Scholar]
- 94.LaMunyon CW, Bouban O, Cutter AD. 2007. Postcopulatory sexual selection reduces genetic diversity in experimental populations of Caenorhabditis elegans. J. Hered. 98, 67–72 (doi:10.1093/jhered/esl052) [DOI] [PubMed] [Google Scholar]
- 95.Ellis RE. 2008. Sex determination in the Caenorhabditis elegans germ line. Curr. Top. Dev. Biol. 83, 41–64 (doi:10.1016/S0070-2153(08)00402-X) [DOI] [PubMed] [Google Scholar]
- 96.Antebi A. 2007. Genetics of aging in Caenorhabditis elegans. PLoS Genet. 3, e129 (doi:10.1371/journal.pgen.0030129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Williams GC. 1957. Pleiotropy, natural selection, and the evolution of senescence. Evolution 11, 398–411 (doi:10.2307/2406060) [Google Scholar]
- 98.Williams PD, Day T, Fletcher Q, Rowe L. 2006. The shaping of senescence in the wild. Trends Ecol. Evol. 21, 458–463 (doi:10.1016/j.tree.2006.05.008) [DOI] [PubMed] [Google Scholar]
- 99.Cutter AD. 2004. Sperm-limited fecundity in nematodes: how many sperm are enough? Evolution 58, 651–655 [PubMed] [Google Scholar]
- 100.Woodruff GC, Eke O, Baird SE, Félix M-A, Haag ES. 2010. Insights into species divergence and the evolution of hermaphroditism from fertile interspecies hybrids of Caenorhabditis nematodes. Genetics 186, 997–1012 (doi:10.1534/genetics.110.120550) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kozlowska JL, Ahmad AR, Jahesh E, Cutter AD. 2011. Genetic variation for postzygotic reproductive isolation between Caenorhabditis briggsae and Caenorhabditis sp. 9. Evolution 66, 1180–1195 (doi:10.1111/j.1558-5646.2011.01514.x) [DOI] [PubMed] [Google Scholar]
- 102.Yan C, Bi Y, Yin D, Zhao Z. 2012. A method for rapid and simultaneous mapping of genetic loci and introgression sizes in nematode species. PLoS ONE 7, e43770 (doi:10.1371/journal.pone.0043770) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dey A, Jeon Y, Wang G-X, Cutter AD. 2012. Global population genetic structure of Caenorhabditis remanei reveals incipient speciation. Genetics 191, 1257–1269 (doi:10.1534/genetics.112.140418) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Herron MD, Doebeli M. 2013. Parallel evolutionary dynamics of adaptive diversification in Escherichia coli. PLoS Biol. 11, e1001490 (doi:10.1371/journal.pbio.1001490) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jarrell TA, Wang Y, Bloniarz AE, Brittin CA, Xu M, Thomson JN, Albertson DG, Hall DH, Emmons SW. 2012. The connectome of a decision-making neural network. Science 337, 437–444 (doi:10.1126/science.1221762) [DOI] [PubMed] [Google Scholar]
- 106.Bendesky A, Bargmann CI. 2011. Genetic contributions to behavioural diversity at the gene–environment interface. Nat. Rev. Genet. 12, 809–820 (doi:10.1038/nrg3065) [DOI] [PubMed] [Google Scholar]
- 107.Smith JR, Stanfield GM. 2011. TRY-5 is a sperm-activating protease in Caenorhabditis elegans seminal fluid. PLoS Genet. 7, e1002375 (doi:10.1371/journal.pgen.1002375) [DOI] [PMC free article] [PubMed] [Google Scholar]