Abstract
Background and Aims
Shining a laser onto biological material produces light speckles termed biospeckles. Patterns of biospeckle activity reflect changes in cell biochemistry, developmental processes and responses to the environment. The aim of this work was to develop methods to investigate the biospeckle activity in roots and to characterize the distribution of its intensity and response to thigmostimuli.
Methods
Biospeckle activity in roots of Zea mays, and also Jatropha curcas and Citrus limonia, was imaged live and in situ using a portable laser and a digital microscope with a spatial resolution of 10 μm per pixel and the ability to capture images every 0·080 s. A procedure incorporating a Fujii algorithm, image restoration using median and Gaussian filters, image segmentation using maximum-entropy threshold methods and the extraction of features using a tracing algorithm followed by spline fitting were developed to obtain quantitative information from images of biospeckle activity. A wavelet transform algorithm was used for spectral decomposition of biospeckle activity and generalized additive models were used to attribute statistical significance to changes in patterns of biospeckle activity.
Key Results
The intensity of biospeckle activity was greatest close to the root apex. Higher frequencies (3–6 Hz) contributed most to the total intensity of biospeckle activity. When a root encountered an obstacle, the intensity of biospeckle activity decreased abruptly throughout the root system. The response became attenuated with repeated thigmostimuli.
Conclusions
The data suggest that at least one component of root biospeckle activity resulted from a biological process, which is located in the zone of cell division and responds to thigmostimuli. However, neither individual cell division events nor root elongation is likely to be responsible for the patterns of biospeckle activity.
Keywords: Biospeckle imaging, Citrus limonia, image analysis, Jatropha curcas, model, root, maize, Zea mays, thigmostimulus, touch
INTRODUCTION
Speckle is an optical phenomenon that occurs when a heterogeneous material is illuminated by a laser (Hetch, 2002). Lasers are coherent light sources that produce parallel, single-frequency and in-phase electromagnetic waves. When a beam of laser light passes through a material, heterogeneities in the material modify the path and phase of the light rays, and the resulting interactions between light rays create brighter and dimmer spots. The distribution of these spots is termed an interference pattern. Interference patterns are therefore a measure of the heterogeneity of a material. Since heterogeneities in biological material vary with time, for example as cells divide, elongate and differentiate, interference patterns from biological materials are dynamic. The dynamic interference patterns observed on biological materials are termed patterns of biospeckle activity.
The patterns of biospeckle activity can be used to extract information about changes in the heterogeneity of biological materials. This technique has been applied to several biological phenomena. It has been used (1) to quantify the motion of fluids or cells, for example when describing blood flow in a tissue (Briers, 1975), the growth of bacteria and motility of semen (Carvalho et al., 2009; Murialdo et al., 2012) or the rate of root deformation (Rathnayake et al., 2008), (2) to assess differences between individuals or treatments, for example when analysing the maturity of tissues (Rabelo et al., 2005), determining the efficacy of drugs on parasites (Pomarico et al., 2004; Amaral et al., 2013), or detecting fungi in bean seeds (Braga et al., 2005) or diagnosing cancer (Ul'yanov et al., 2012) and (3) to quantify spatial or temporal changes in specific tissues, for example from the patterns of biospeckle activity of roots grown in tissue culture (Braga et al., 2009). The origins of the patterns of biospeckle activity are often unknown. However, it has been speculated that they can arise from biological processes, including tissue maturation, cell division, movement of organelles, cytoplasmic streaming and biochemical reactions (Rabelo et al., 2005).
The plant root exhibits indeterminate growth and provides an excellent material to investigate the application of biospeckle imaging to biology. The root has a defined longitudinal pattern of cell division, elongation and differentiation (Gregory, 2008), and responds rapidly to environmental perturbations (Robinson, 1996; White et al., 2005; Monshausen and Gilroy, 2009a; Hodge, 2009). Thus, one can investigate whether patterns of biospeckle activity differ along a root and associate these with defined developmental processes, or investigate whether patterns of biospeckle activity are influenced by defined environmental challenges, such as thigmostimuli (Richter et al., 2009). This paper describes improvements to the spatial resolution, temporal acquisition and processing of biospeckle images that have allowed the location of biospeckle activity in roots to be identified live and in situ, and its response to thigmostimuli to be investigated.
MATERIAL AND METHODS
Plant cultivation
In this study, most experiments were performed on maize (Zea mays) seedlings. Experiments on roots of other plant species were also carried out to analyse similarities in the patterns of biospeckle activity across species. Two seedlings of jatropha (Jatropha curcas) were observed at hourly intervals for up to 14 h and one citrus root (Citrus limonia) was observed at hourly intervals for 10 h. Seeds were surface-sterilized using a solution of 50 % sodium hypochlorite and then rinsed with distilled water. Sterilized seeds were then agitated for 1 min in a 70 % alcohol solution. Plants were grown on soft agar gels (5·5 g L−1) containing 1·65 g L−1 NH4HO3, 1·90 g L−1 KNO3, 170 mg L−1 KH2PO4, 0·83 mg L−1 KCl, 0·25 g L−1 CaCl2, 0·370 g L−1 MgSO4.7H2O, 22·3 mg L−1 MnSO4.4H2O, 8·6 mg L−1 ZnSO4.7H2O, 27·8 mg L−1 FeSO4.7H2O, 6·2 mg L−1 H3BO3, 0·25 mg L−1 Na2MoO4.2H20, 0·025 mg L−1 CuSO4.5H2O, 0·025 mg L−1 CoCl2.6H20, 37·2 mg L−1 Na2EDTA, 100 mg L−1 myoinositol, 0·5 mg L−1 thiamine HCl, 0·5 mg L−1 pyridoxine HCl, 0·5 mg L−1 niacin, 2·0 mg L−1 glycine and 30·0 mg L−1 sucrose (Murashige and Skoog, 1962). The pH of the nutrient solution was adjusted to 5·5. Seeds were placed in transparent plastic tubes containing set agar. The tubes were placed in a controlled environment at 26 ± 2 °C, with 16 h of daylight (photosynthetic photon flux (area) density, PPFD = 42 µmol m−2 s−1) and 8 h of darkness.
Micro-biospeckle imaging
Upon germination, jatropha, maize and citrus seedlings produced a variable number of roots. The roots that emerged first from the seed were imaged to characterize biospeckle activity quantitatively at steady state. Other roots were also imaged for qualitative assessment. Roots were illuminated with a coherent HeNe laser light source (632 nm, 5 mW). The laser beam was filtered with a neutral filter to reduce the intensity of light and the beam was expanded using a concave lens to generate homogeneous illumination of samples (Fig. 1A). Since lenses introduce smooth variations in the path of light, illumination remains coherent and biospeckle patterns can be observed on a larger portion of the sample. The distance between the laser and the sample was 75 cm. Images were captured with a portable digital microscope (AM 413zt, Dino-Lite). The field of view consisted of a window of 0·96 × 1·26 cm. The working distance of the microscope was 7·3 cm and a 25× zoom was used. The angle between the laser beam and the microscope Z-axis was 67 °. Data at each sampling time consisted of 128 grey-scale images, termed participant images. Participant images had a 1280 × 1024 pixel resolution and were captured every 0·080 s (Fig. 1B). In order to obtain better image quality, roots were imaged when close to the surface of the tubes.
Fig. 1.
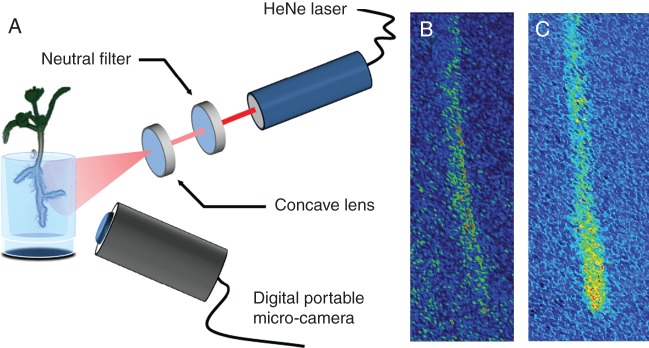
Continuous high-resolution micro-biospeckle imaging. (A) Root tissues are illuminated with a HeNe laser source whose intensity is attenuated using an adjustable neutral filter in order to avoid photo-damage. The beam is then expanded using a concave lens to obtain uniform illumination of the sample. The light emitted/scattered from root tissues is captured using a portable digital microscope. (B) Pseudo-coloured image obtained using this system (participant image) showing the random interference patterns (speckle) obtained in a root tissue. (C) The participant image is then processed to determine the intensity of biospeckle activity using the Fujii method.
Analysis of biospeckle intensity patterns and the effects of death and thigmostimulation
An initial set of experiments was performed to characterize root growth and root responses to laser light. The size of the elongation zone of maize plants was determined on four different seedlings grown as described above. Five days after sowing, seedlings were imaged with a portable digital microscope every 5 min for 20 min. Time-lapse images were then analysed to quantify the size of the root elongation zone. A separate experiment was performed to study the effects of exposure to laser light on root growth. Eleven seedlings were grown for 5 days in the controlled environment. Four of these seedlings were then put in the biospeckle imaging system but did not receive laser illumination. The remaining seven plants were placed in the biospeckle imaging system and received the light doses required to create biospeckle images for 3 h. Root length was measured on all 11 seedlings at 30-min intervals for 3 h. Measured root length was used to calculate elongation rate.
A second set of experiments was carried out to determine the biological processes linked to biospeckle activity. Five maize seedlings were grown in plastic tubes to test the hypothesis that the biospeckle patterns are created by biological processes. Seedlings were grown in agar for 5 days after sowing. Seedlings were then imaged using the biospeckle imaging system to determine biospeckle intensity in living roots. The seedlings were then killed by freeze–thawing in liquid nitrogen and again imaged using the biospeckle imaging system. Biospeckle intensity before and after freeze–thawing was compared. Another experiment was carried out to study the effect of thigmostimuli on the biospeckle activity in root tissue. Four seedlings were imaged using the biospeckle imaging system to establish the reference biospeckle intensity distribution along the root. Then, a thin metal blade was introduced through a slit in the plastic tube containing the seedling. The blade was placed perpendicularly to the root so that it touched the root apex directly. The slit was cut 1 day before imaging so that the sample did not receive mechanical stimulation during the 24 h that preceded the insertion of the blade. The blade was left in place to impose continuous touch stimulation and biospeckle images were recorded at 30-min intervals for 2 h. Contact with the blade was defined as a thigmostimulus. The effects of thigmostimuli on root biospeckle activity were also studied in an experiment in which the soft agar gel was layered over a harder agar gel (10 mg L−1) containing the same amendments. In this experiment, physical obstacles to root growth included the sides of the glass tube and the hard agar gel. Contact with these obstacles was defined as a thigmostimulus. In this situation, the time and place of the contact was less predictable, but it allowed the observation of systemic responses throughout a plant root system.
Analysis of participant images
The set of 128 participant images were processed to quantify the intensity of patterns of biospeckle activity across root tissues. Numerous methods have been developed to analyse temporal variations in speckle patterns, such as generalized difference (Arizaga et al., 2002) and laser speckle contrast analysis (LASCA) (Briers and Webster, 1996). Here, we used the method developed by Fujii et al. (1987) because it provides better results when samples have been underexposed (Braga et al., 2009). The Fujii method is based on the summation of the differences between pixels of two consecutive images with a weighting factor:
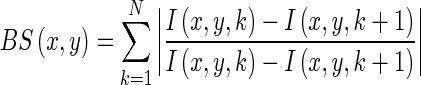 |
(1) |
where I(x, y, k) is the intensity of pixel at coordinates (x, y) in the kth image. The result is a new grey scale image, BS, in which intensity levels reflect the biospeckle activity in the root (Fig. 1C).
Analysis of image data
An image analysis pipeline was developed to automate the processing of images generated in this study. Bright-field images (Fig. 2A) were used to characterize the size of the elongation zone. The velocity of material points at the surface of the root were obtained using particle image velocimetry (Fig. 2B) (Bengough et al., 2006). The magnitude of the velocity of points on the surface of the root was then expressed as a function of the distance from the root tip. A flexible logistic growth curve (Morris and Silk, 1992) was then fitted to the data points (Fig. 2C):
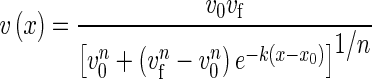 |
(2) |
where x is the distance from the root tip, vf is the velocity at the root tip and v0 is the velocity at the base of the root at position x0; k and n are the parameters to be adjusted to fit the curve to the data. Fitting was achieved by minimization of the mean square error using the Levenberg–Marquardt algorithm implemented in the Scipy library (Moré, 1978). The flexible logistic function was then used to determine the size of the elongation zone. The velocity of material points fitted to the data (eqn 2) was used to determine the strain rate (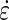 ):
):
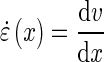 |
(3) |
Fig. 2.
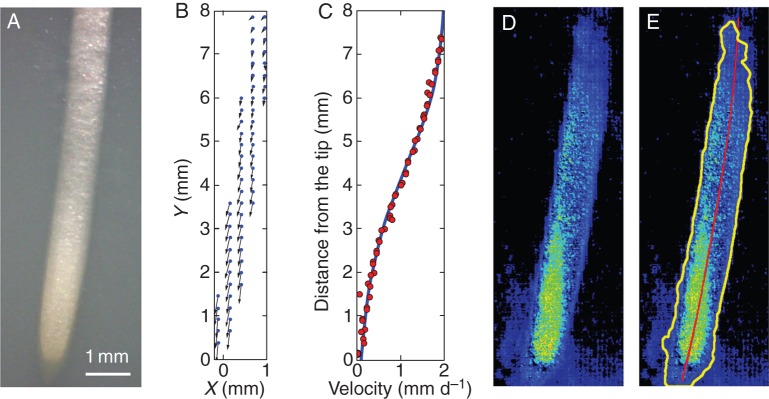
Analysis of root image data. (A) Bright-field image of a maize root used to analyse the root elongation zone. (B) Velocity of material points on the root surface was determined by particle image velocimetry. (C) Velocity of material points was expressed as a function of distance from the root tip. (D) A biospeckle intensity or activity map was obtained from participant images using the Fujii method. Image restoration filters were applied to remove image artefacts. Images were smoothed using a median filter followed by a Gaussian filter with a radius of 37 μm. Smoothing was followed by normalization to reduce variations in contrast at different times. Images were segmented using maximum-entropy thresholding. (E) Segmented binary data were then used to detect the root boundaries (black contour) to extract relevant pixel intensities from the initial activity maps. A cubic spline was fitted to the distribution of root pixels to determine the root centre line (red line) and to calculate the distance from the root tip for each pixel in the image.
The end of the cell division zone was assumed to be approximately 2 mm from the tip, as observed in previous experiments on maize (Sacks et al., 1997). The end of the elongation zone was defined as the point where the velocity of points on the root fell below 5 % of the maximum velocity of the tip.
The BS images (Fig. 2D) were transformed by an image restoration step. This corrected for stochasticity in the biospeckle data using a median filter (each pixel is replaced by the median value of pixels in a neighbourhood) and a Gaussian filter (convolution of the image by a Gaussian function). Both filters used a window radius of 37 μm. Image restoration was followed by normalization of the images to limit intensity variations across frames. The normalized image was a smoothed description of variations of biospeckle intensity across the image. Image restoration was followed by a segmentation step in which individual pixels were classified into root and non-root types. The main difficulty inherent in segmentation is the identification of a criterion for the discrimination of pixel types. An efficient way to achieve this objective is to use the entropy of the image histogram. Entropy is a measure of the uncertainty of a random variable, and it can be used to find the optimal threshold to separate histograms into two parts. The threshold value was defined as the pixel intensity that maximized the entropy of root and non-root histograms (Fleury et al., 1996). The binary image obtained by segmentation was then processed to extract the edges of the root using an edge-tracing algorithm. The edge contour was then used as a mask to collect pixel position and intensity from the initial activity maps (yellow contour in Fig. 2E). A cubic spline was fitted to the distribution of root pixels to determine the root centre line (red line in Fig. 2E) and to calculate the distance from the root tip for each pixel in the image. Results were then exported to text files for statistical analyses. Image analysis procedures were implemented as a plug-in in ImageJ and will be made available upon request.
Spectral analysis of root biospeckle data using a wavelet transform algorithm
Laser speckles are associated with material points in the root tissue, and biological activity induces changes in the tissue that alter the intensity of the speckles. To gain an understanding of the frequencies at which the intensity of the laser speckles change with time, it is useful to apply methods developed for the field of signal processing (Sendra et al., 2005). In particular, numerous mathematical techniques have been proposed to characterize the spectral composition of periodic signals, i.e. the set of wavelengths that constitute a signal. For example, the Fourier transform can be used to decompose a signal into the sum of sine and cosine (called basis functions) and provide information about the contributions of different frequencies in the global signal. However, sine and cosine functions expand their fluctuations infinitely, and for this reason Fourier analysis is mainly limited to the study of stationary signals. Here, we applied a wavelet transform algorithm (Torrence and Compo, 1998), which relies on local periodic functions, called the mother wavelet Ψ. The basis functions are then constructed incrementally by displacing the centre and stretching (scale parameter) the mother wavelet:
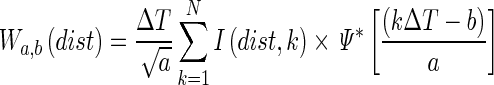 |
(4) |
where a is the scale parameter, b is the centre of the basis function, dist is the distance from the root apex, k is the time index and Ψ* represents the complex conjugate. By varying the scale (a) of the mother wavelet and translating its centre (b) along the time index (k), it is possible to determine how the position along the root (dist) and time (b) affect the amplitude (W) of each scale parameter of the biospeckle signal. Participant images were acquired every 0·080 s; the highest frequency observable according to the sampling theorem is 6·25 Hz. For statistical analyses, we divided the 0–6·25 Hz frequency window into 25 spectral bands of 0·25 Hz.
Statistical analysis of biospeckle data
Generalized additive models (Hastie and Tibshirani, 1995) were constructed to quantify the patterns of biospeckle activity across species (Fig. 3), along the root axis (Figs 3–6) or as a function of frequencies (Figs 4–6). The models used in this paper can be formalized as the sum of two terms:
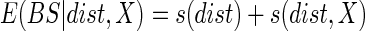 |
(5) |
where dist is the distance from the root tip and X denotes another predictor of the biospeckle intensity, such as frequency or plant species. The biospeckle intensity BS is the dependent variable and its expectation E(BS) is assumed to be the sum of two spline interpolators s. The first term accounts for the average variations of the biospeckle intensity along the root. The second term indicates interactions with other factors (X). When the interaction term is significantly different from zero, it is concluded that factor X has an effect on the patterns of biospeckle activity along the root. Generalized additive models were constructed in this work to study the effect of plant species, frequency and time after touch sensing. Analyses were carried out using the R software and the mgcv package (Wood, 2008). Data on the size of the meristem, the elongation rates of roots and the effects of light, freeze thawing and thigmostimuli are expressed as mean biospeckle intensity ± s.e.
Fig. 3.
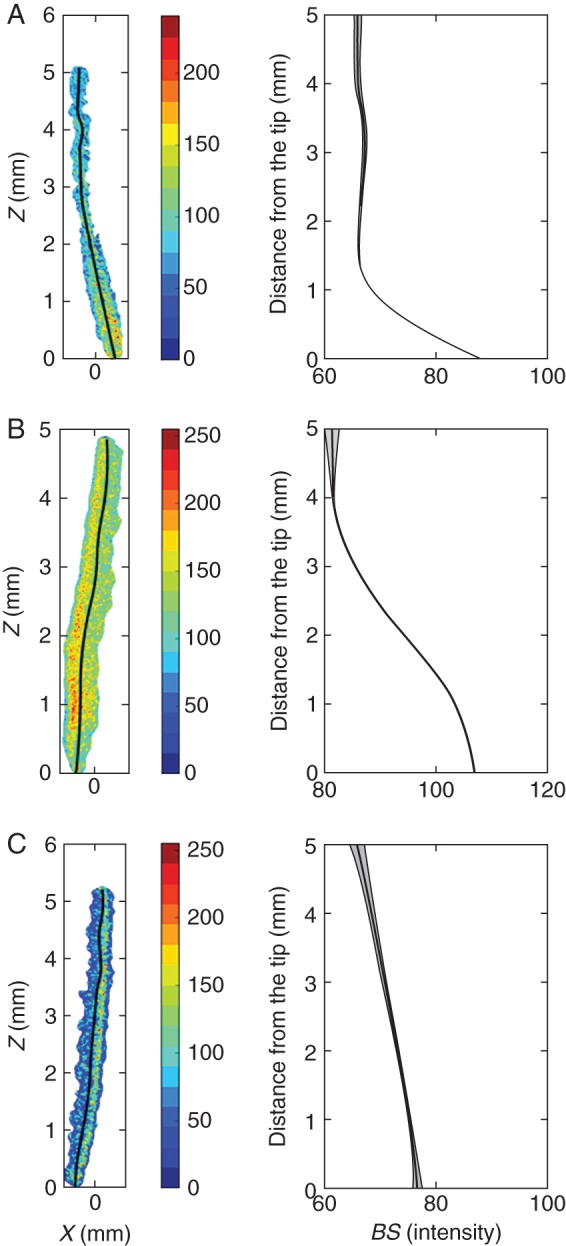
Plant species showed different root biospeckle activity maps. Images on the left show reconstructed activity maps using the Fujii method. Images on the right show the variations of biospeckle intensity (BS) along the root axis. (A) Jatropha roots had moderate overall biospeckle intensity, with a sharp decrease in biospeckle intensity in the first millimetre from the root tip. (B) Maize roots had high biospeckle intensity, and biospeckle intensity decreased over the first 3 mm from the root tip. (C) Citrus had the lowest root biospeckle intensity of the three species studied, and the decrease in biospeckle intensity with distance from the root tip was less marked in citrus than in jatropha or maize. Data represent means (lines) ± s.e. (grey shading) from 18 images of biospeckle activity in roots of jatropha, 72 images in roots of maize and 10 images in roots of citrus.
Fig. 4.
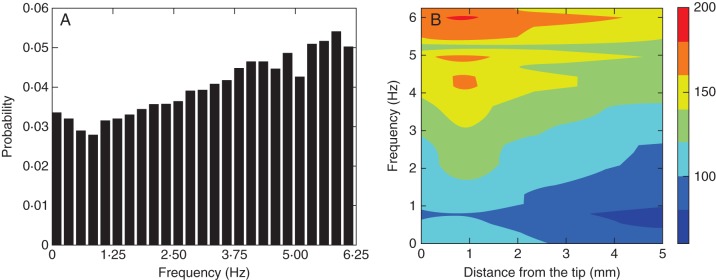
Spectral decomposition of the mean pattern of biospeckle activity from 50 measurements of maize roots growing without obstruction. (A) Biospeckle intensity across the 0–6·25 Hz window was decomposed into bands of 0·25 Hz. The X-axis represents the frequency of the biospeckle signal and the Y-axis represents the proportion of the total signal. (B) Space–frequency matrices were used to analyse the patterns of biospeckle activity along the root at different frequencies. These figures show that high-frequency variations contributed most to the patterns of biospeckle activity.
Fig. 5.
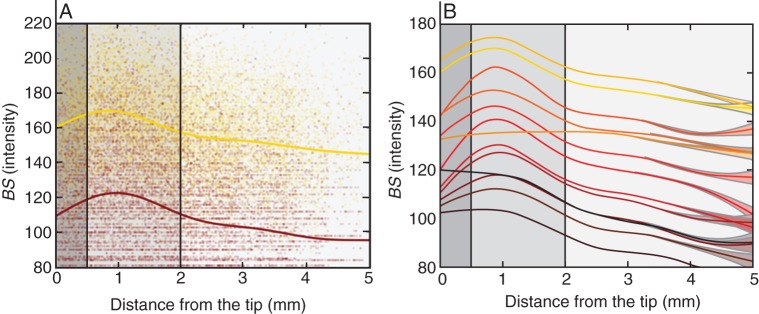
Statistical analysis of frequency data. (A) Biospeckle intensity data for high frequencies (bright yellow, 6·0–6·25 Hz) and low frequencies (dark brown, 1·5–1·75 Hz). (B) Model prediction of the change in biospeckle intensity as a function of distance from the tip and frequency 0–6·25 Hz (bright yellow to dark brown, respectively). Shades of grey indicate the root cap, cell division and cell elongation zones (dark grey, mild grey and light grey, respectively).
Fig. 6.
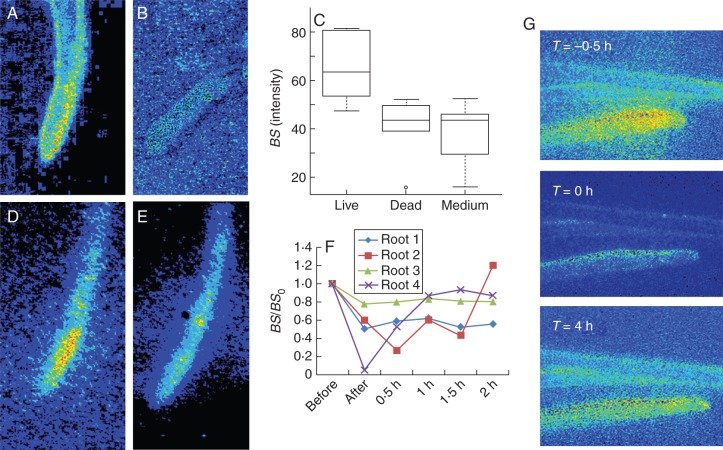
Thigmostimuli affect the pattern of biospeckle activity in roots. (A, D) Living roots maintained a high level of biospeckle intensity. (B) Dead roots exhibited strongly reduced biospeckle intensity. (C) Biospeckle intensity in living roots was significantly higher than in dead roots, and the biospeckle intensity of dead roots was not statistically different from the biospeckle intensity of the medium. (D, E) Thigmostimuli, effected using a thin metal blade, reduced biospeckle intensity. (F) Thigmostimulation initially reduced biospeckle intensity to a minimum value before it was gradually restored with time. (G) Time-lapse data showing a decrease in overall biospeckle intensity in maize roots within 30 min of touching a physical barrier (T = 0 h) and its restoration after a lag period (T = 4 h).
RESULTS AND DISCUSSION
High-resolution, live micro-biospeckle imaging of roots
Biospeckle images have previously been obtained from germinating beans (Braga et al., 2005), orange fruit (Rabelo et al., 2005) and roots of pine (Rathnayake et al., 2008), coffee and eucalyptus (Braga et al., 2009). The spatial resolution of previous studies investigating patterns of biospeckle activity associated with root developmental processes was low (typically >20 μm per pixel), and the biospeckle activities of different regions of the root could not be distinguished clearly (Braga et al., 2009). Using a portable digital microscope, the spatial resolution of images of biospeckle activity could be increased to approximately 10 μm per pixel, which allowed biospeckle activities associated with different root tissues or cell types to be determined and thinner roots to be studied (Fig. 1). Higher biospeckle activities were observed in tissues close to the root apex (Fig. 1). This observation is consistent with the low-resolution images of Braga et al. (2009). The temporal resolution of previous studies investigating patterns of biospeckle activity associated with root developmental processes was also low (Braga et al., 2009). Therefore, an experimental system was designed to collect higher-magnification images of biospeckle activity of roots continuously for an extended period, which allowed any changes in biospeckle activity in response to developmental or environmental stimuli to be monitored (Fig. 1).
Results of this study showed that maize plants grew healthily in the system for the duration of the experiment. Kinematic analysis of root growth was used to determine the size of the elongation zone. The end of the elongation zone was observed at 6·2 ± 0·76 mm from the apex (n = 4 roots). The thickness of the root cap at the root apex was 0·6 ± 0·05 mm (n = 4 roots). The light dose received by roots is limited since illumination is required for a maximum of 12 s every 30 min. There was no statistically significant difference in the root elongation rates of plants illuminated by laser light (1·4 ± 0·20 mm h−1, n = 4 roots) and plants that did not receive laser light (1·5 ± 0·18 mm h−1, n = 7 roots).
Spatial patterns of biospeckle activity in roots
To characterize differences in patterns of biospeckle activity associated with root development and environmental challenges, it was necessary to develop methods to quantify variation in their frequency and intensity. To extract quantitative information rapidly and objectively from images of biospeckle activity of growing roots, the following automated procedure was adopted (Fig. 2). A Fujii algorithm was implemented to generate biospeckle activity maps (Fig. 2D). Images were restored using median and Gaussian filters and segmented using maximum-entropy thresholding methods. Root features were extracted using a tracing algorithm followed by spline fitting (Fig. 2E). Intensity of biospeckle activity along the root was compared with the position of the cell elongation zone obtained from kinematic analysis (Fig. 3) and the cell division zone reported in previous studies (Sacks et al., 1997). A data file containing the location of root pixels and their intensity of biospeckle activity for each original image was produced using this procedure (for details see Materials and methods). This data file was used for the spectral decomposition of biospeckle activity using a wavelet transform algorithm. Generalized additive models were then used to attribute statistical significance to changes in patterns of biospeckle activity.
Biospeckle activities were examined in roots of jatropha, maize and citrus (Fig. 3). In general, the intensity of biospeckle activity was greatest close to the root apex and decreased to a minimal value at a distance of 1–5 mm from the apex, depending upon plant species. There were significant differences in the intensity of biospeckle activity between plant species, with maize exhibiting the highest intensity of biospeckle activity (Fig. 3B) and citrus the lowest intensity (Fig. 3C). The region of high intensity of biospeckle activity extended the shortest distance from the root tip in jatropha (Fig. 3A) and the longest distance in citrus (Fig. 3C). Similar spatial patterns of biospeckle activity were maintained during steady-state growth. The differences in patterns of biospeckle activity were unrelated to the growth rates of the roots of different plant species. These were 0·70 cm day−1 for jatropha, 0·92 cm day−1 for maize and 0·89 cm day−1 for citrus.
The total intensity of biospeckle activity can be decomposed into the intensities of biospeckle activity at different frequencies. Such spectral decomposition can be illustrated with data from maize, which exhibited the highest intensity of biospeckle activity of the three plant species studied (Fig. 4). The highest frequency investigated in the experiments reported here was 6·25 Hz, which was determined by the rate of acquisition of images of biospeckle activity. It was observed that higher frequencies contributed most to the total intensity of biospeckle activity (Fig. 4A) and that a decrease in the intensity of biospeckle activity at higher frequencies accounted for most of the decrease in the total intensity of biospeckle activity with distance from the root apex (Fig. 4B). Quantitative analysis of data from three roots imaged every 30 min for a period of 6 h supported these conclusions (Fig. 5). The intensity of biospeckle activity at higher frequencies (e.g. 5·5–5·7 Hz) decreased more sharply than the intensity of biospeckle activity at lower frequencies (e.g. 0·25–0·5 Hz) with distance from the root apex (Fig. 5A). Statistical models indicated that the relationships between the intensity of biospeckle activity and distance from the root apex differed between frequencies (P < 0·001). The models predicted the maximum intensity of biospeckle activity at all frequencies at a distance of 0·5–1·5 mm from the root apex (Fig. 5B). This region corresponds to the zone of cell division in the maize root (Sacks et al., 1997). The intensity of biospeckle activity decreased most sharply in the zone of cell elongation, which in our experimental system began at about 2·7 mm from the tip.
Biological activity creates biospeckle activity
When roots were growing, high levels of biospeckle activity were observed in root tissues (Fig. 6A). However, biospeckle activity was reduced to a level similar to the background after freeze–thawing (Fig. 6B). The average biospeckle intensity of living roots was 65·2 ± 6·93 (n = 5), that of dead roots was 40·5 ± 5·45 (n = 6) and that of the medium was 37·6 ± 5·03 (n = 6). Differences in biospeckle intensity between living roots and dead roots was statistically significant (two sample t-test, P < 0·05). Differences in biospeckle intensity between the medium and dead roots were not statistically significant (Fig. 6C). These results indicate that biospeckle activity is created by biological activity, and are consistent with results obtained with apple fruit killed with DMSO (Kurenda et al., 2013).
It is likely that epidermal or cortical cells contribute most to the patterns of biospeckle activity. A fraction of the light passing through a cell is absorbed. Absorbed light is then emitted back but in random direction and phase. Emitted light is no longer coherent and therefore does not produce light speckles. Thus, as laser light loses coherence as a function of depth inside the root tissue due to absorption, biospeckle signals are dominated by cells at the surface of tissues.
In all experiments presented here, significant background biospeckle activity was also observed. This background biospeckle activity appeared to decrease when the biospeckle activity of the root decreased. This phenomenon can be explained because background biospeckle activity in the medium has two origins. First, the gel contains water, and microscopic water movement creates genuine background activity. Second, light originating from the root is scattered by the medium and creates background activity dependent on root biospeckle activity. The background activity is reduced, and therefore background biospeckle activity is reduced, when root biospeckle activity decreases.
Biospeckle activity in roots responds to thigmostimuli
When root growth was unobstructed, the intensity and spatial pattern of biospeckle activity in the roots remained high (Fig. 6D). However, when a root encountered a physical obstacle, a decrease in the intensity of biospeckle activity was observed (Fig. 6E).
The quotient between biospeckle intensity before and after thigmostimulation was significantly smaller than 1 (P = 0·06). After thigmostimulation, the biospeckle intensity was gradually restored to a greater value on average, although this increase was not statistically significant (Fig. 6E). The roots exhibiting the largest reduction in biospeckle intensity also showed recovery towards higher biospeckle intensity. A possible explanation for differences in recovery between roots might be related to the time or the angle at which they are in contact with the obstacle. The reduction in biospeckle intensity was observed throughout the root system, not just in the root that had touched the obstacle (Fig. 6G, data not shown n = 4 events). When root growth was unobstructed, biospeckle activity was greatest close to the root apex. However, when a root encountered an obstacle, and the intensity of biospeckle activity was reduced, spatial differences in the intensity of biospeckle activity were also reduced.
Conclusions and perspectives
In conclusion, in all three plant species studied the intensity of biospeckle activity was greatest close to the root apex (Figs 3, 5 and 6) and in maize roots this appeared to be associated with the zone of cell division. No biospeckle activity was generated on dead roots. When a maize root encountered an obstacle, the intensity of biospeckle activity decreased abruptly not only in the root that touched the obstacle but also throughout the root system (Fig. 6). These three observations suggest that at least one component of biospeckle activity results from a biological process and that this process is located in the zone of cell division and responds to thigmostimuli. Higher frequencies (3–6 Hz) contributed most to the total intensity of biospeckle activity (Figs 4 and 5). Individual cell division events are unlikely to determine the biospeckle activity at this frequency. The frequency of cell division is less than 3·3 × 10−5 Hz (0·12 cells pixel−1 h−1) in the cell division zone of a maize root (Sacks et al., 1997). Furthermore, although thigmostimuli can reduce rates of cell division in roots, in a response linked to cytosolic Ca2+ signals (Legue et al., 1997; White and Broadley, 2003; Monshausen and Gilroy, 2009b), this is unlikely to produce a systemic response. A recent study on apple fruit tissue has suggested biological mechanisms associated with biospeckle activity (Kurenda et al., 2013). This study employed a variety of pharmaceuticals and demonstrated that those affecting the (de)polymerization of actin microfilaments influenced biospeckle activity most. It is well known that the organization of actin microfilaments is modified by mechanical stimulation (Shi et al., 2011; Asnacios and Hamant, 2012) and it is possible that the changes in biospeckle activity observed upon thigmostimulation result from alterations in the polymerization of actin microfilaments. The intensity of biospeckle activity at lower frequencies is relatively unaffected by thigmostimuli. It is likely that biospeckle activity at lower frequencies is related to slowly changing macroscopic processes or homeostatic cellular processes. Although the exact biological nature of the biospeckle signal remains enigmatic, the technique can be used to screen large plant populations non-destructively for developmental variation or responses to environmental parameters. The equipment is relatively inexpensive and portable, the analysis can be automated, and the technique provides a rapid and sensitive functional assay.
ACKNOWLEDGEMENTS
We thank Vantuil Antônio Rodrigues, who helped prepare seedlings for analysis, and Dr Tim George and Dr Tracy Valentine, who provided comments on the original manuscript. The James Hutton Institute receives support from the Scottish Government Rural and Environment Science and Analytical Services Division (RESAS, Workpackage 3·3). The research at Universidade Federal de Lavras received support from CNPq, Fapemig, Capes and Finep.
LITERATURE CITED
- Amaral IC, Júnior RAB, Ramos EM, Ramos ALS, Roxael EAR. Application of biospeckle laser technique for determining biological phenomena related to beef aging. Journal of Food Engineering. 2013;119:135–139. [Google Scholar]
- Arizaga R, Cap NL, Rabal H, Trivi M. Display of local activity using dynamical speckle patterns. Optical Engineering. 2002;41:287–294. [Google Scholar]
- Asnacios A, Hamant O. The mechanics behind cell polarity. Trends in Cell Biology. 2012;22:584–591. doi: 10.1016/j.tcb.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Bengough AG, Bransby MF, Hans J, McKenna SJ, Roberts TJ, Valentine TA. Root responses to soil physical conditions; growth dynamics from field to cell. Journal of Experimental Botany. 2006;57:437–447. doi: 10.1093/jxb/erj003. [DOI] [PubMed] [Google Scholar]
- Braga RA, Dupuy LX, Pasqual M, Cardoso RR. Live biospeckle laser imaging of root tissues. European Biophysics Journal. 2009;38:679–686. doi: 10.1007/s00249-009-0426-0. [DOI] [PubMed] [Google Scholar]
- Braga RA, Rabelo GF, Granato LR, et al. Detection of fungi in beans by the laser biospeckle technique. Biosystems Engineering. 2005;91:465–469. [Google Scholar]
- Briers JD. Wavelength dependence of intensity fluctuations in laser speckle patterns from biological specimens. Optics Communications. 1975;13:324–326. [Google Scholar]
- Briers JD, Webster S. Laser speckle contrast analysis (LASCA): a nonscanning, full-field technique for monitoring capillary blood flow. Journal of Biomedical Optics. 1996;1:174–179. doi: 10.1117/12.231359. [DOI] [PubMed] [Google Scholar]
- Carvalho PHA, Barreto JB, Braga RA, Jr, Rabelo GF. Motility parameters assessment of bovine frozen semen by biospeckle laser (BSL) system. Biosystems Engineering. 2009;102:31–35. [Google Scholar]
- Fleury M, Hayat L, Clark AF. Parallel entropic auto-thresholding. Image and Vision Computing. 1996;14:247–263. [Google Scholar]
- Fujii H, Nohira K, Yamamoto Y, Ikawa H, Ohura T. Evaluation of blood flow by laser speckle image sensing. Part 1. Applied Optics. 1987;26:5321–5325. doi: 10.1364/AO.26.005321. [DOI] [PubMed] [Google Scholar]
- Gregory P. Plant roots: growth, activity and interaction with soils. Oxford: Blackwell Publishing; 2008. [Google Scholar]
- Hastie TJ, Tibshirani RJ. Generalized additive models. London: Chapman and Hall; 1995. [DOI] [PubMed] [Google Scholar]
- Hetch E. Optics. San Francisco: Addison-Wesley; 2002. [Google Scholar]
- Hodge A. Root decisions. Plant, Cell & Environment. 2009;32:628–640. doi: 10.1111/j.1365-3040.2008.01891.x. [DOI] [PubMed] [Google Scholar]
- Kurenda A, Pieczywek PM, Adamiak A, Zdunek A. Effect of cytochalasin B, lantrunculin B, colchicine, cycloheximid, dimethyl sulfoxide and ion channel inhibitors on biospeckle activity in apple tissue. Food Biophysics. 2013:1–7. doi: 10.1007/s11483-013-9302-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legue V, Blancaflor E, Wymer C, Perbal G, Fantin D, Gilroy S. Cytoplasmic free Ca2+ in arabidopsis roots changes in response to touch but not gravity. Plant Physiology. 1997;114:789–800. doi: 10.1104/pp.114.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monshausen GB, Gilroy S. The exploring root – root growth responses to local environmental conditions. Current Opinion in Plant Biology. 2009a;12:766–772. doi: 10.1016/j.pbi.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Monshausen GB, Gilroy S. Feeling green: mechanosensing in plants. Trends in Cell Biology. 2009b;19:228–235. doi: 10.1016/j.tcb.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Moré JJ. Lecture notes in mathematics 630. Watson GA: 1978. The Levenberg-Marquardt algorithm: implementation and theory. Berlin: Springer. [Google Scholar]
- Morris AK, Silk WK. Use of a flexible logistic function to describe axial growth of plants. Bulletin of Mathematical Biology. 1992;54:1069–1081. [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Murialdo SE, Passoni LI, Guzman MN, et al. Discrimination of motile bacteria from filamentous fungi using dynamic speckle. Journal of Biomedical Optics. 2012;17:0560111–0560115. doi: 10.1117/1.JBO.17.5.056011. [DOI] [PubMed] [Google Scholar]
- Pomarico JA, Di Rocco HO, Alvarez L, et al. Speckle interferometry applied to pharmacodynamic studies: evaluation of parasite motility. European Biophysics Journal. 2004;33:694–699. doi: 10.1007/s00249-004-0413-4. [DOI] [PubMed] [Google Scholar]
- Rabelo GF, Braga RA, Jr, Fabbro IMD, Arizaga R, Rabal H, Trivi MR. Laser speckle techniques applied to study quality of fruits. Revista Brasileira de Engenharia Agrícola e Ambiental. 2005;9:570–575. [Google Scholar]
- Rathnayake AP, Kadono H, Toyooka S, Miwa M. A novel optical interference technique to measure minute root elongations of Japanese red pine (Pinus densiflora Seibold & Zucc.) seedlings infected with ectomycorrhizal fungi. Environmental and Experimental Botany. 2008;64:314–321. [Google Scholar]
- Richter GL, Monshausen GB, Krol A, Gilroy S. Mechanical stimuli modulate lateral root organogenesis. Plant Physiology. 2009;151:1855–1866. doi: 10.1104/pp.109.142448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D. Resource capture by localized root proliferation: why do plants bother? Annals of Botany. 1996;77:179–186. [Google Scholar]
- Sacks MM, Silk WK, Burman P. Effect of water stress on cortical cell division rates within the apical meristem of primary roots of maize. Plant Physiology. 1997;114:519–527. doi: 10.1104/pp.114.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendra GH, Arizaga R, Rabal H, Trivi M. Decomposition of biospeckle images in temporary spectral bands. Optics Letters. 2005;30:1641–1643. doi: 10.1364/ol.30.001641. [DOI] [PubMed] [Google Scholar]
- Shi L, Wang B, Gong W, Zhang Y, Zhu L, Yang X. Actin filaments and microtubules of arabidopsis suspension cells show different responses to changing turgor pressure. Biochemical and Biophysical Research Communications. 2011;405:632–637. doi: 10.1016/j.bbrc.2011.01.081. [DOI] [PubMed] [Google Scholar]
- Torrence C, Compo GP. A practical guide to wavelet analysis. Bulletin of the American Meteorological Society. 1998;79:61–78. [Google Scholar]
- Ul'yanov S, Laskavyi V, Glova AB, et al. Adaptation of LASCA method for diagnostics of malignant tumours in laboratory animals. Quantum Electronics. 2012;42:399–404. [Google Scholar]
- White PJ, Broadley MR. Calcium in plants. Annals of Botany. 2003;92:487–511. doi: 10.1093/aob/mcg164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PJ, Broadley MR, Greenwood DJ, Hammond JP. Genetic modifications to improve phosphorus acquisition by roots. York: International Fertiliser Society.; 2005. [Google Scholar]
- Wood SN. Fast stable direct fitting and smoothness selection for generalized additive models. Journal of the Royal Statistical Society. 2008;70:495–518. [Google Scholar]