Abstract
Background and Aims
Clonal growth is a common feature in flowering plants. As clone size increases, the selfing rate in self-compatible species is likely to increase due to more frequent geitono-pollination events (i.e. pollination among flowers within the same genet). This study investigated the breeding system of the marsh cinquefoil (Comarum palustre) and assessed spatial distribution of clones, clone size and architecture, and their effects on realized outcrossing rates. In addition, pollen dispersal was investigated in two patchy populations.
Methods
The species' breeding system was investigated under controlled conditions through hand pollinations (self- vs. cross-pollination). Using microsatellite markers, an assessment was made of the realized outcrossing rates and the genetic diversity in four natural populations, the clonal structure in two populations within five 15 × 15 m sampling plots following 0·5 × 0·5 m grids, and the pollen dispersal through paternity assignment tests in those two populations.
Key Results Comarum palustre
is a self-compatible species but only presents a low rate of spontaneous self-pollination. The occurrence of inbreeding depression was not detected at the seed set stage (δSS = 0·04). Clones were spatially clumped (AC = 0·60–0·80), with intermediate to no intermingling of the ramets (DC = 0·40–1·00). Genet size ranged from one to 171 ramets. Patchy populations had low outcrossing rates (tm = 0·33–0·46). Large clones showed lower outcrossing rates than small clones. Pollen dispersal mainly occurred within patches as only 1–7 % of the pollination events occurred between patches of >25 m separation. Seedling recruitment events were detected.
Conclusions
Genet size together with distances between patches, through increasing geitono-pollination events, appeared to be important factors influencing realized outcrossing rates. The study also revealed seed flow allowing seedling recruitment, which may contribute to increasing the number of new patches, and potentially further enhance gene flow within populations.
Keywords: Breeding system, clonal growth, gene flow, genet size, genetic diversity, microsatellites, marsh cinquefoil, Comarum palustre, outcrossing rate
INTRODUCTION
A majority of flowering plant species propagate by both vegetative clonal multiplication and sexual reproduction (Widén et al., 1994; Klimeš et al., 1997; Rautiainen et al., 2004). Clonal growth through the development of vegetative structures such as rhizomes increases the size of individuals and leads to spatial clusters of genetically identical, yet morphologically independent flowering shoots (ramets) originating from a single individual (genet; Klimeš et al., 1997; Charpentier, 2001; Vallejo-Marín et al., 2010). Clonal growth influences the spatial arrangement of ramets within a genet, which is referred to as clonal architecture (Klimeš et al., 1997). It is often classified into two extreme propagation strategies. On the one hand, the ‘phalanx type’ is characterized by short internodes and densely clumped ramets, leading to large monoclonal patches. On the other hand, the ‘guerilla type’ presents long internodes and widely dispersed ramets leading to an intermingling of genets (Lovett-Doust, 1981).
Clonal growth confers several advantages to a genet, including attractiveness to pollinators by aggregated floral display, increased persistence when sexual recruitment fails and the ability to explore for resources in heterogeneous environments (Wilcock and Neiland, 2002; Honnay and Bossuyt, 2005; Louâpre et al., 2012). However, when pollen is dispersed over short distances, clonal growth and clonal architecture can negatively affect mating opportunities (Charpentier, 2001; Albert et al., 2008). Restricted pollen dispersal usually results from short-distance movements of pollinators, preferentially foraging among near-neighbour flowers (Williams, 2007; Fontaine et al., 2008; Karron et al., 2009). The flowering ramets of large, spatially clumped genets may therefore face increased levels of geitono-pollination, i.e. pollination among flowers within the same genet. In particular, ramets situated within large clumped clones face high levels of geitono-pollination (Albert et al., 2008; Honnay and Jacquemyn, 2008).
Reduced connectivity due to low pollen dispersal between spatially isolated patches of clones within populations with a subsequent low number of genotypes involved in seed production might further reduce the outcrossing rate (Schmucki and de Blois, 2009). Geitono-pollination coupled with low mate diversity might induce reproductive costs. In self-compatible plant species, geitono-pollination will increase the selfing rate (Barrett, 1998; Eckert, 2000). If the species suffers from inbreeding depression, geitono-pollination can lead to a reduction in reproductive success (e.g. seed set; Albert et al., 2008). In self-incompatible species, geitono-pollination might induce stigma clogging with self pollen, preventing deposition and germination of compatible outcross pollen (Wilcock and Neiland, 2002; Honnay and Bossuyt, 2005; Vandepitte et al., 2010).
In this study, we investigated the breeding system and estimated the mating system of the marsh cinquefoil Comarum palustre (Rosaceae). This pioneer species occurs in wet meadows and peat bogs across Europe (Macek and Lepš, 2007; Sarneel et al., 2011; Sarneel and Soons, 2012). These biotopes are particularly threatened, with a subsequent decline of plant species occurring therein (Frankard et al., 1998; Lamers et al., 2002). The species reproduces sexually but also propagates clonally via long lignifying rhizomes (Olesen and Warncke, 1992; Macek and Lepš, 2007). Plants grow sympodially with a terminal inflorescence. If not flowering, the annual increments are separated by short internodes and reduced leaves. The oldest parts of the rhizome may decay, resulting in clone splitting and subsequent independence of ramets (Macek and Lepš, 2007). The main pollinators of C. palustre are bumble bees and solitary bees (Olesen and Warncke, 1992; Guillén et al., 2005; Somme et al., unpubl. res.). The protandrous flowers last for about 5 d, where the female phase follows the male phase 2 d after anthesis (Olesen and Warncke, 1992).
Assessing asexual propagation and sexual reproductive success within and among the remaining threatened populations, as well as species' dispersal abilities by means of pollen and seed flow, is crucial to design appropriate management plans for long-term persistence of the species. Indeed, conservation strategies usually aim at enhancing connectivity among populations through increased pollen and/or seed flow (Tischendorf et al., 2003; Kindlmann and Burel, 2008), by restoring large biotope areas favouring plant dispersal and establishment, as well as plant pollinator movements. When it is not possible to restore large continuous plant populations, management of conspecific patches among populations might serve as guiding bridges for pollinators from one patch to another and thus act as stepping-stones (Beier and Noss, 1998; Van Rossum and Triest, 2012).
The breeding system of C. palustre was investigated by performing hand pollinations under controlled conditions to estimate the species self-fertility, self-compatibility and inbreeding depression at seed set stage. We also assessed clonal growth and clone architecture in two patchy populations using microsatellite markers. To estimate the mating system under natural conditions, the realized outcrossing rate was investigated in four natural populations. We linked the spatial distribution of clones within populations, clone architecture and size to the realized outcrossing rate and effective gene flow, inferred by paternity assignment tests. Specifically, the following questions were addressed. (1) Is C. palustre self-compatible and does it suffer from inbreeding depression? (2) What is the clonal structure of C. palustre in natural populations? (3) At the population level, does the distribution of clones influence the realized outcrossing rate? (4) At a fine scale, do clone architecture and size influence the realized outcrossing rate? (5) How far does pollen dispersal occur between patches in patchy populations? As some of the study populations are managed through extensive cattle grazing, we discuss our findings in the light of this management practice.
MATERIALS AND METHODS
Breeding system under controlled conditions
In May 2009, the breeding system was assessed on flowering shoots of 45 genets of Comarum palustre planted in 5 L pots (one plant per pot) filled with compost. The genets originated from two natural Belgian populations, Regné (50°14′32·57″N, 5°46′51·49″E) and Fouches (49°41′14·59″N, 5°43′6·75″E), separated from each other by 65 km. The genets were kept in an open greenhouse until being transferred just before flowering under controlled conditions in growth chambers at the Université catholique de Louvain (Louvain-la-Neuve, 50°39′58″N, 4°37′9″E). Light was supplied by Philips HPIT 400 W lamps (Philips Lighting, Brussels). Temperature was kept at 20/17 ± 3 °C (day/night) and relative humidity at 65 ± 12 %, with a day/night cycle of 16 h/8 h. Plants were watered with rainwater three times a week. Four flowers at the female stage were selected per flowering ramet (45 ramets per treatment) and randomly assigned to the following treatments: (1) hand cross-pollination (CP), with pollen transfer from a male-phase flower to a female-phase flower from another individual; (2) hand geitono-pollination (GP), with pollen transfer from a male-phase flower to a female-phase flower of a same individual; (3) hand self-pollination (SP), with pollen transfer from the stamens of a flower at male-phase to the stigma of the same flower at female-phase; and (4) spontaneous self-pollination (SSP) with unmanipulated flowers.
Fruits, consisting of a dry aggregate of achenes on an accrescent spongious receptacle (Olesen and Warncke, 1992), were harvested 3 weeks later and stored in individual envelopes at room temperature before seed counting. Seeds were scanned at 600 dpi and counted using a macro created with ImageJ (National Institute of Mental Health, Bethesda, MD, USA). They were automatically sorted into three categories according to size and shape: plump seeds, partly filled seeds and unfertilized ovules. The morphological screening was confirmed by testing seed viability (Kearns and Inouye, 1993). Seeds were longitudinally cut and soaked in a 1 % 2,3,5-triphenyltetrazolium chloride solution for 24 h at 35 °C. The colorimetric test showed that 90 % of the plump seeds were viable and 97 % of the partly filled seeds were aborted. The fertilization rate was calculated as the ratio between the number of viable plus aborted seeds and total number of ovules. Seed set was calculated as the ratio between the number of viable seeds and total number of ovules.
Seed set data from the hand pollination treatments were used to estimate the self-fertility index (SFI) and inbreeding depression at seed set stage (δSS). The SFI gives an estimate of the capacity of a plant to produce fruits and seeds in the absence of pollen vectors. It is calculated as the seed set of spontaneous self-pollination relative to that of hand cross-pollination (Lloyd and Schoen, 1992). Inbreeding depression (δSS), the reduced fitness of progeny derived from inbreeding relative to outbreeding, was estimated at the seed set stage as: δSS = 1 – ws/wo, where ws and wo are, respectively, the mean seed set following geitono- and cross-pollination (Husband and Schemske, 1995).
Study sites for genetic analyses
The realized outcrossing rate was assessed in four natural populations of C. palustre in South Belgium (Table 1). The study site located in Bihain (50°14′25·89″N, 5°48′22·60″E) consisted of a wet meadow used as an extensive pasture and displayed a continuous population of C. palustre covering 2700 m2. The population located in Sainte-Marie (49°40′3·31″N, 5°32′56·81″E) consisted of floating mats of C. palustre on 250 m2, intermingling with willow thickets. The study site located in Fosse (50°15′45·77″N, 5°38′37·44″E) displayed six patches of C. palustre separated from each other by 25–210 m, summing up to a total population size of 600 m2. This study site was managed through extensive cattle grazing. The study site located in Bra (50°19′5·51″N, 5°44′35·54″E) displayed three patches of C. palustre separated from each other by 52–100 m, summing up to a total population size of 1450 m2. This study site was also managed through extensive cattle grazing. The two latter sites (i.e. Fosse and Bra) were studied for fine-scale clonal structure and pollen dispersal inferred by paternity assignment tests.
Table 1.
Characteristics of four Belgian study populations of Comarum palustre: geographic co-ordinates, population area (i.e. area covered by C. palustre; m2), flower density (number of flowers m−2), number of sampled ramets (N), number of multilocus genotypes (G), clonal richness [R = (G – 1)/(N – 1)], number of sampled maternal ramets (Nm) and number of seedlings used to estimate the realized outcrossing rate (NS)
| Population | Co-ordinates: latitude (N), longitude (E) | Population area (m2) | Flower density (flowers m−2) | N | G | R | Nm | NS |
|---|---|---|---|---|---|---|---|---|
| Bihain | 50°14′25·89″, 5°48′22·60″ | 2500 | 18·5 ± 9·1 | 16 | 13 | 0·80 | 16 | 128 |
| Sainte-Marie | 49°40′3·31″, 5°32′56·81″ | 250 | 23·0 ± 10·8 | 11 | 9 | 0·80 | 11 | 88 |
| Bra | 50°19′5·51″, 5°44′35·54″ | 1450 | 55·8 ± 35·7 | 737 | 21 | 0·03 | 14 | 112 |
| Fosse | 50°15′45·77″, 5°38′37·44″ | 600 | 59·9 ± 25·5 | 480 | 22 | 0·04 | 17 | 136 |
Flower density, defined as the number of open flowers recorded on 15 plots of 1 m2 randomly chosen per population, was significantly different among the four populations, with Bra and Fosse displaying higher flower density (56–60 flowers m−2) than Bihain and Sainte-Marie (19–23 flowers m−2; χ2 = 29·76, d.f. = 3, P < 0·001; Table 1).
Sampling for genetic analyses
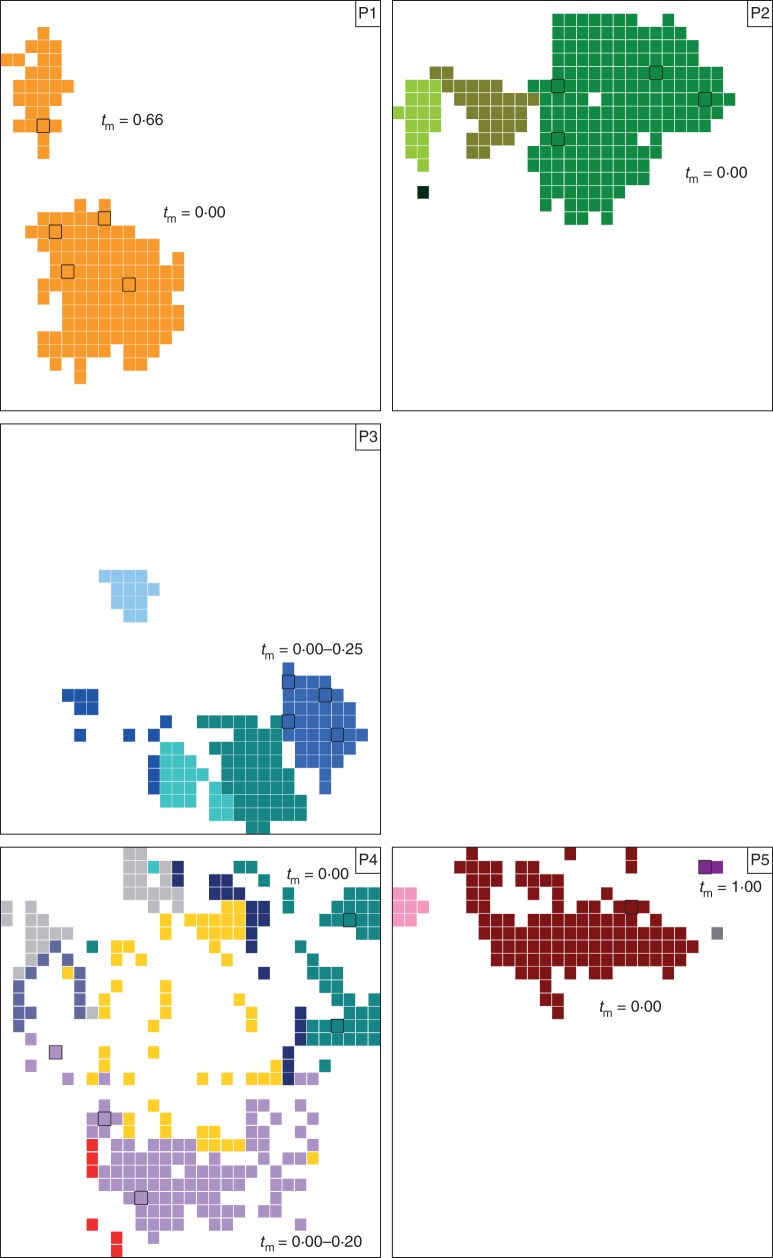
In July 2011, we randomly collected one young leaf and one fruit from 25 ramets per population in the four populations to estimate the effect of spatial distribution of ramets on the realized outcrossing rate at the population level. In order to investigate the effect of clonal structure and clone size on fruit realized outcrossing rate, we collected one fresh leaf per ramet at each node of a 0·5 × 0·5 m grid within two and three 15 × 15 m plots in the patchy populations of Fosse and Bra, respectively (Fig. 2; Arnaud-Haond et al., 2007).
Fig. 2.
Spatial distribution of the clones in five sampling plots of Comarum palustre: P1, P2, P3 from the population of Bra, and P4, P5 from the population of Fosse. In each plot, samples were collected in a 15 × 15 m quadrat at each point of intersection of a 0·5 × 0·5 m grid. Samples belonging to the same genet are represented by the same colour. The absence of C. palustre individuals is indicated by blank squares. Bold squares indicate sampled fruits for outcrossing rate estimates within clone patches.
In order to be confident that full sampling of the potential paternal plants was achieved for the paternity assignment tests in the two patchy populations, the sampling was extended to the entire population. It resulted in a sampling of 480 and 737 ramets in Fosse and Bra, respectively.
A total of 50 plump seeds were selected per fruit for germination. The germination procedure was adapted from Whittington et al. (1988), Baskin and Baskin (1990) and Lyanguzova (2011). Seeds were scarified with sand paper (3M 255P) and soaked in GA3 (3·5 mg L−1) for 2 h before sterilization in 500 mL of BRAVO fungicide (0·01 %, Syngenta, Basel, Switzerland) for about 30 s. Batches of ten seeds per fruit were sown on moist filter paper in five Petri dishes (a total of 500 Petri dishes) and placed in germination chambers (Snijders, The Netherlands; 12 h daylength and 12/12 h alternating temperature regimes of 15/25 °C). After 10 weeks, each seedling weighing approx. 25 mg was individually stored at –80 °C until DNA isolation.
DNA extraction and microsatellite genotyping
DNA was extracted from 75 mg of leaf tissue and 25 mg of seedlings using the cetyltrimethyl ammonium bromide (CTAB) method (Doyle and Doyle, 1990) and standardized DNA extracts to a concentration of 5 ng μL−1. Among the sampled fruits, we selected those that provided a minimum of eight seedlings, which allowed for a sufficient number of maternal ramets for a correct estimate of the realized outcrossing rate at the population level (Table 1). We used 16 of the 20 polymorphic microsatellite markers previously developed (Somme et al., 2012). The laboratory protocols were the same as those described in Somme et al. (2012). Subsequently, the PCR products were electrophoresed on an ABI 3100 capillary sequencer with GS400HD size standard (Applied Biosystems, Foster City, CA, USA) and the electropherograms were analysed using GeneMapper version 3·7 (Applied Biosystems).
Genetic diversity and clonal structure
To discriminate between ramets that might represent identical genets resulting either from sexual reproduction events or from clonal propagation, the probability of the first re-encounter (n = 1) of identical genets via sexual reproduction Psex (Parks and Werth, 1993) for each genet was calculated among the four populations using GENCLONE 2·0. (Arnaud-Haond and Belkhir, 2007). To ascertain the uniqueness of genets with missing data (i.e. unamplified loci), these genets were examined separately after removing the missing loci from the entire data set. It further allowed estimate of genet size (NR) among populations (i.e. the number of ramets per genet).
After clone identification, clonal diversity was assessed among the four populations by the genotypic richness R, computed as R = (G – 1)/(N – 1), where G is the number of identified genets, and N is the number of sampled ramets within each population (Dorken and Eckert, 2001).
For the assessment of population genetic measures, multiple ramets with identical genets were reduced into a single genet (Tables 1, 2). For the four populations, the average number of alleles per locus (NA), the expected and observed heterozygosity (HE and HO, respectively) and the Weir and Cockerham (1984) inbreeding coefficient (FIS) were calculated separately for adult plants and offspring using GENETIX 4·05·2 (Montpellier University, France). The HE values were corrected for small sampling size for adult plants, by applying the 2N/(2N – 1) correction. The significance of the F-statistics was tested at the 95 % confidence interval (CI) with 1000 permutations, and the mean F-statistics were considered significant when CIs did not overlap zero.
Spatial clone architecture and clonal structure were estimated among the five 15 × 15 m sampling plots in the two patchy populations of Fosse and Bra by calculation of the aggregation index Ac at the patch level and of the clonal dominance index Dc for each genet. The aggregation index Ac was calculated using GENCLONE 2·0. (Arnaud-Haond and Belkhir, 2007) as Ac = (psg – psp)/psg, where psg is the average probability of clonal identity of all sample unit pairs, and psp the average probability of clonal identity among pairwise nearest neighbours. The significance of Ac was tested with 10 000 permutations. To determine the level of intermingling between genets, the clonal dominance index Dc was calculated for each genet as Dc = (NR – 1)/(NT – 1), where NR is the genet size (i.e. the number of ramets per genet) and NT the total number of ramets present within the genet range (i.e. within the minimal polygon containing all ramets of the given genet; Ohsako, 2010). A clonal dominance index of 1 indicates that the spatial range of a genet is occupied exclusively by ramets of the same genet. Only genets representing more than two ramets were subjected to calculation of clonal dominance.
Outcrossing rates
To estimate the realized outcrossing rate, i.e. the outcrossing rate that produces viable seeds, in the study populations, the genotypes of the maternal ramets and their offspring were analysed with MLTR 3·3 (Ritland, 2002). Default initial values of the parameters were used (i.e. t = 0·9, rt = 0·1, rp = 0·1, F = 0·1). We used 1000 bootstraps to derive CIs with resampling performed over families. On the basis of maximum likelihood procedures, the program estimates the single- and multilocus outcrossing rates (ts and tm, respectively), biparental inbreeding (tm – ts), single locus maternal inbreeding coefficient (F), correlated selfing rate (rs) and the multilocus paternity correlation (rp; Ritland, 2002). The multilocus paternity correlation is the proportion of full-sibs among outcrossed sibs and indicates whether offspring are the result of single or multiple paternities. The reciprocal of rp (Nep = 1/rp) gives the effective number of pollen donors (Ritland, 2002).
Paternity analysis
A paternity analysis was carried out for the populations of Fosse and Bra. Paternity was assigned on a total of 112 and 136 seedlings from 17 and 14 fruits (i.e. eight seedlings per fruit), respectively, using CERVUS 3·0 (Marshall et al., 1998; Kalinowski and Taper, 2006). All potential parent genets within populations (22 and 21, respectively) were included in the analysis. CERVUS implements a likelihood-based assignment method using the allelic frequencies from the population, the number of candidate paternal plants, the proportion of paternal plants that are sampled, as well as possible errors in the genetic data (Marshall et al., 1998). For the simulations, we used the allelic frequencies computed on the parental population and applied the following simulation parameters to find the confidence level (CL) of paternity analysis assignment: 10 000 simulated mating events; 22 and 21 candidate paternal plants, respectively; 16 as the minimum number of loci; 0·90 as the proportion of candidate parents sampled; 0·05 as the genotyping error rate; all adult plants treated as candidate paternal plants. The relaxed and strict CLs, which corresponded to the statistical confidence of the assignments, were set to 80 and 95 %, respectively. We only considered fully genotyped offspring (at 16 loci) that presented no more than one mismatch to the maternal plant. A total of seven paternity assignments with more than two mismatches in the final trio mother/father/offspring were discarded.
Statistical analyses
Kruskal–Wallis and Mann–Whitney-U tests were used to compare the fertilization rate (i.e. ratio between the number of viable plus aborted seeds and total number of ovules) and seed set (i.e. ratio between the number of viable seeds and total number of ovules) per fruit from the different pollination treatments performed under controlled conditions.
To assess any effect of spatial distribution of clones (continuous or patchy) on the outcrossing rate, we performed Mann–Whitney-U tests with the multilocus outcrossing rate from the four populations. Within the two patchy populations, intercorrelation between the multilocus outcrossing rate within the 15 × 15 m sampling plots and genet size (NR) was tested using Kendall correlation analyses. We used Kruskal–Wallis tests to compare the expected and observed heterozygosities, and the Weir and Cockerham (1984) inbreeding coefficient of the offspring and the adults. If not indicated otherwise, data are presented as the mean ± s.d. All analyses were performed with R 2·13·0 (R Development Core Team, 2012).
RESULTS
Breeding system
Hand pollinations performed under controlled conditions led to significant differences in fertilization rate and seed set (χ2 = 62·56–76·23, d.f. = 3, P < 0·001; Fig. 1). Hand cross-, self- and geitono-pollination led to a higher fertilization rate and seed set than spontaneous self-pollination (Z = 407–621, P < 0·001). Consequently, C. palustre showed a low SFI (0·15), as flowers left to self-pollinate spontaneously led to a low mean seed set (5·5 ± 4·1) compared with hand cross-pollinated flowers (37·6 ± 3·5; Z = 504, P < 0·001; Fig, 1).
Fig. 1.
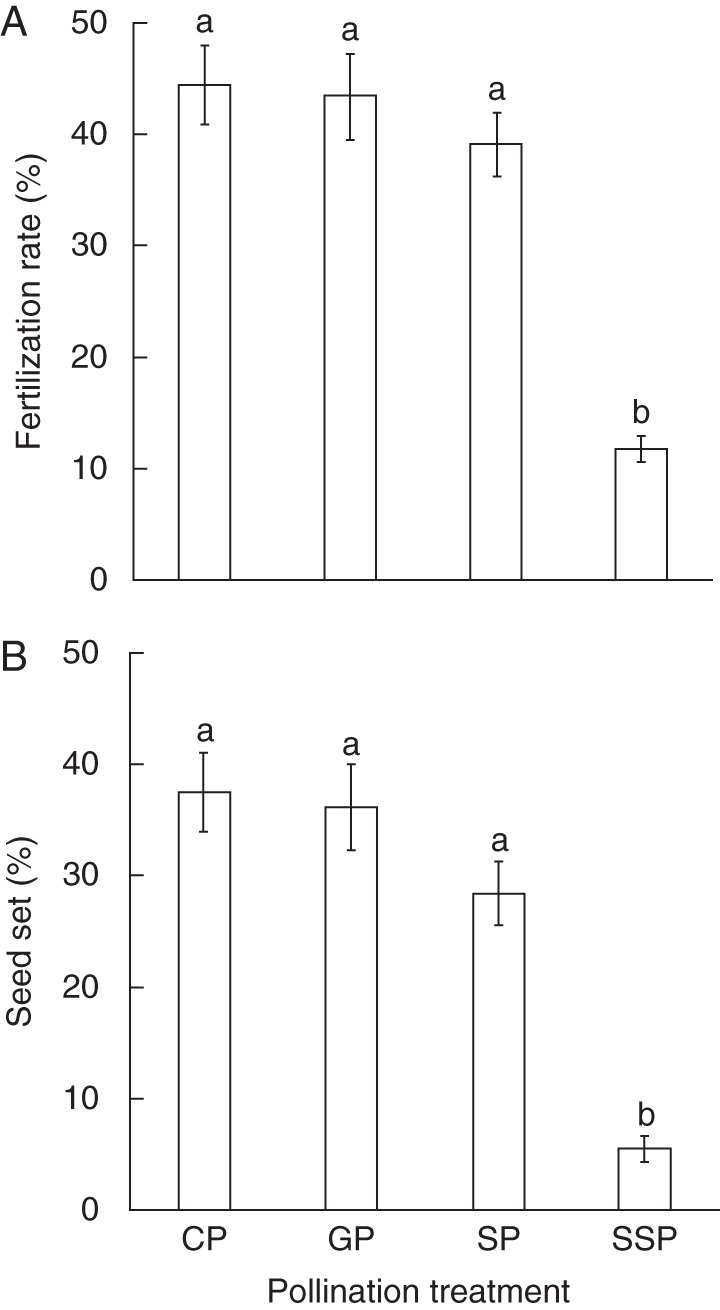
(A) Mean fertilization rate and (B) seed set (% ± s.e.) for the pollination treatments performed under controlled conditions: hand cross-pollination (CP), hand geitono-pollination (GP), hand self-pollination (SP) and spontaneous self-pollination (SSP). Group means that were significantly different (P < 0·05) are indicated with different letters.
Pollen origin, i.e. cross- vs. self-pollen, had no significant effect on the seed set, which did not differ significantly among the different hand pollination treatments (Z = 986–1392, P > 0·05; Fig. 1). The inbreeding depression estimate at the seed set stage was low, with δSS = 0·04.
Genetic diversity and clonal structure
Among the four populations, the number of genets ranged from nine in Sainte-Marie to 22 in Fosse (Table 1). All ramets within genets in the four populations were associated with a Psex value <10−9. The genotypic richness R ranged from 0·03 in Bra to 0·80 in Sainte-Marie and Bihain (Table 1).
Potential parental plants genotyped at 16 loci showed moderate to high expected and observed heterozygosity (HE = 0·65–0·74; HO = 0·59–0·75; Z = 83–114, P > 0·05) and low inbreeding coefficient values (FIS = –0·03–0·05; Table 2), except for the population of Fosse, which showed significant deviation from Hardy–Weinberg equilibrium (FIS = 0·09; Table 2). In the patchy populations of Bra and Fosse, the offspring showed high expected heterozygosity (HE = 0·52–0·60) but low observed heterozygosity (HO = 0·33–0·39; Z = 28–58, P < 0·01) and high inbreeding coefficient values (FIS = 0·37–0·38; Table 2). In the non-patchy populations of Bihain and Sainte-Marie, the offspring showed both high expected and observed heterozygosity (HE = 0·70–0·72; HO = 0·61–0·66; Z = 60–101, P < 0·05) and low inbreeding coefficient values (FIS = 0·07–0·14; Table 2). The HO and FIS values were significantly lower for the offspring than for the adults in the two patchy populations (Z = 22–28, P < 0·001; Z = 15–27, P < 0·001, respectively).
Table 2.
Genetic diversity measures for Comarum palustre in the four study populations
| NA | HE | HO | FIS | ||
|---|---|---|---|---|---|
| Bihain | Adults | 6·8 (0·6) | 0·74 (0·15) | 0·75 (0·22) | –0·01 (–0·10, 0·05) |
| Offspring | 8·9 (0·7) | 0·72 (0·14) | 0·61 (0·13) | 0·14 (0·10, 0·21) | |
| Sainte–Marie | Adults | 5·2 (0·5) | 0·72 (0·13) | 0·72 (0·21) | –0·03 (–0·17, 0·11) |
| Offspring | 6·8 (0·5) | 0·70 (0·10) | 0·66 (0·11) | 0·07 (0·02, 0·12) | |
| Bra | Adults | 6·6 (0·7) | 0·69 (0·04) | 0·64 (0·04) | 0·05 (–0·05, 0·11) |
| Offspring | 5·1 (0·4) | 0·52 (0·05) | 0·33 (0·04) | 0·38 (0·31, 0·44) | |
| Fosse | Adults | 6·0 (0·5) | 0·65 (0·03) | 0·59 (0·04) | 0·09 (0·01, 0·17) |
| Offspring | 6·0 (0·4) | 0·60 (0·04) | 0·39 (0·03) | 0·37 (0·32, 040) |
For each population, the mean number of alleles (NA), expected heterozygosity (HE), observed heterozygosity (HO) and Weir and Cockerham's inbreeding coefficient (FIS) are shown for adult plants and offspring, with standard error in parentheses. The 95 % confidence intervals are given in parentheses for FIS. Significant deviations from Hardy–Weinberg equilibrium of FIS values are in italics.
For the analyses, multiple ramets with identical genets were reduced into a single genet.
Analysis of the clonal structure in the patchy populations of Bra and Fosse revealed a number of genets within the five 15 × 15 m sampling plots ranging from two in patch P1 to eight in patch P4 (Fig. 2; Table 3). Among these sampling plots, the estimated aggregation index (Ac) ranged from 0·60 in patch P3 to 0·80 in patch P2 (P < 0·001), indicating significant spatial clustering of clonal ramets. The mean clonal dominance index 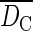 ranged from 0·76 in patch P4 (DC = 0·54–0·92) to 1·00 in the four other patches (Table 3; Fig. 2), indicating that the spatial range of a genet was almost exclusively occupied by ramets belonging to that genet. Genet size (NR) within the sampling plots ranged from one to 171 ramets.
ranged from 0·76 in patch P4 (DC = 0·54–0·92) to 1·00 in the four other patches (Table 3; Fig. 2), indicating that the spatial range of a genet was almost exclusively occupied by ramets belonging to that genet. Genet size (NR) within the sampling plots ranged from one to 171 ramets.
Table 3.
Clonal structure of the five 15 × 15 m sampling plots in the populations of Bra and Fosse: number of genotypes (G), clonal richness [R = (G –1)/(N – 1)], aggregation index (AC), mean clonal dominance index (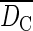 ), clone identity of the sampled maternal ramets, clone size (m2), genet size (NR), clonal dominance index (DC ), single- and multilocus outcrossing rates (ts and tm) and biparental inbreeding (tm – ts)
), clone identity of the sampled maternal ramets, clone size (m2), genet size (NR), clonal dominance index (DC ), single- and multilocus outcrossing rates (ts and tm) and biparental inbreeding (tm – ts)
| Sampling plot | Population | G | R | AC | 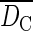 |
Clone identity | Clone size (m2) | NR | DC | ts | tm | tm – ts |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | Bra | 2 | 0·01 | 0·67 | 1·00 | B1 | 7·0 | 28 | 1·00 | 0·55 | 0·66 | 0·11 |
| B2 | 28·8 | 115 | 1·00 | 0·00 | 0·00 | 0·00 | ||||||
| 0·00 | 0·00 | 0·00 | ||||||||||
| 0·00 | 0·00 | 0·00 | ||||||||||
| 0·00 | 0·00 | 0·00 | ||||||||||
| P2 | Bra | 4 | 0·02 | 0·80 | 1·00 | B5 | 42·8 | 171 | 0·99 | 0·00 | 0·00 | 0·00 |
| 0·00 | 0·00 | 0·00 | ||||||||||
| 0·00 | 0·00 | 0·00 | ||||||||||
| 0·00 | 0·00 | 0·00 | ||||||||||
| P3 | Bra | 5 | 0·02 | 0·60 | 1·00 | B10 | 10·0 | 44 | 1·00 | 0·11 | 0·25 | 0·14 |
| 0·00 | 0·00 | 0·00 | ||||||||||
| 0·00 | 0·00 | 0·00 | ||||||||||
| 0·00 | 0·00 | 0·00 | ||||||||||
| P4 | Fosse | 8 | 0·04 | 0·62 | 0·79 | F8 | 26·8 | 97 | 0·74 | 0·05 | 0·20 | 0·15 |
| 0·05 | 0·20 | 0·15 | ||||||||||
| 0·00 | 0·00 | 0·00 | ||||||||||
| 0·04 | 0·13 | 0·09 | ||||||||||
| F10 | 11·3 | 46 | 0·40 | 0·00 | 0·00 | 0·00 | ||||||
| 0·00 | 0·00 | 0·00 | ||||||||||
| P5 | Fosse | 4 | 0·02 | 0·64 | 1·00 | F2 | 25·0 | 104 | 1·00 | 0·00 | 0·00 | 0·00 |
| 0·00 | 0·00 | 0·00 | ||||||||||
| F3 | 0·5 | 2 | 1·00 | 0·91 | 1·00 | 0·09 |
Each row represents a fruit of the same genet.
Outcrossing rates
At the population level, single- and multilocus outcrossing rates were highly variable and ranged from 0·14 to 0·79 and from 0·33 to 0·84, respectively (Table 4). We found a significant effect of spatial distribution of clones on outcrossing rates, with higher outcrossing rates in the non-patchy populations of Bihain and Sainte-Marie compared with the patchy populations of Bra and Fosse (Z = 33, P < 0·001). Within plots, the multilocus outcrossing rate was significantly correlated to genet size (Kendall rank correlation, τ = –0·54, P < 0·01) and ranged from 0·00 to 1·00 (Fig. 3; Table 3).
Table 4.
Mating system variables (± s.d.) of four Belgian study populations of Comarum palustre: single- and multilocus outcrossing rates (tm and ts), biparental inbreeding (tm – ts), maternal inbreeding (F), correlated selfing rate (rs), multilocus paternity correlation (rp) and the number of effective pollen donors (Nep = 1/rp)
| Population | ts | tm | tm – ts | F | rs | rp | Nep |
|---|---|---|---|---|---|---|---|
| Bihain | 0·75 ± 0·06 | 0·84 ± 0·09 | 0·09 ± 0·06 | 0·02 ± 0·03 | 0·42 ± 0·42 | 0·26 ± 0·14 | 3·86 |
| Sainte-Marie | 0·79 ± 0·07 | 0·83 ± 0·10 | 0·04 ± 0·08 | –0·16 ± 0·06 | 0·32 ± 0·23 | 0·49 ± 0·07 | 2·05 |
| Bra | 0·14 ± 0·06 | 0·33 ± 0·07 | 0·19 ± 0·06 | –0·20 ± 0·02 | 0·14 ± 0·09 | 0·88 ± 0·33 | 1·14 |
| Fosse | 0·34 ± 0·05 | 0·46 ± 0·06 | 0·12 ± 0·04 | 0·06 ± 0·03 | 0·09 ± 0·09 | 0·36 ± 0·10 | 2·79 |
Fig. 3.
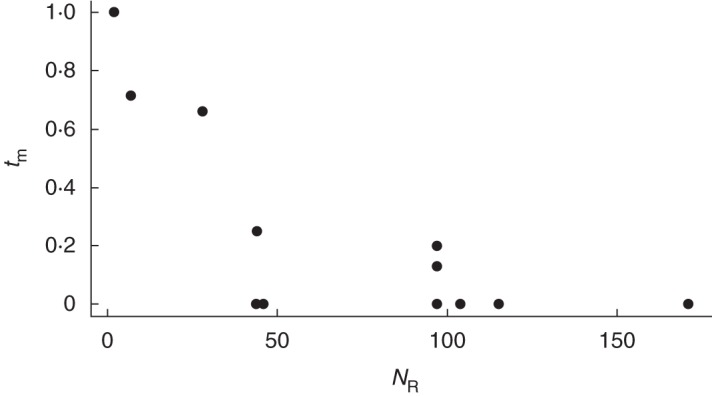
Relationship between genet size (NR) and multilocus outcrossing rate (tm).
Between 4 and 19 % of matings occurred between genetically related genets (biparental inbreeding, tm – ts = 0·04–0·19; Table 4). Maternal inbreeding coefficients varied among populations (F = –0·20–0·06; Table 4), with negative values in the populations of Bra and Sainte-Marie. The multilocus paternity correlation within fruit was high and ranged from 0·26 to 0·88. Consequently, the effective number of pollen donors (Nep) per fruit ranged from 1·14 to 3·86 (Table 4).
Paternity analyses and gene flow
The assignment rate reached 96 % in Fosse and 98 % in Bra. While 97 and 88 offspring in Bra were assigned at the 80 % CL and 95 % CL, respectively, all of the 127 assigned offspring in Fosse were assigned at the 95 % CL.
Paternity assignments revealed that only 1 and 7 % of the analysed offspring were sired by pollen dispersed between patches in Bra and Fosse, respectively. Nevertheless, some crossing events occurred between patches at a distance of 90 m in Bra and patches at a distance of 25 m up to 170 m in Fosse. Paternity analyses performed on genets in Bra revealed that seeds originating from patch P1 established in a third patch.
DISCUSSION
Breeding system
Comarum palustre depends on external agents to transfer pollen from the anthers onto the stigmas as we observed negligible seed set resulting from spontaneous self-pollination under controlled conditions. It might be explained by the protandry of the flowers of C. palustre, allowing a temporal separation of male and female stages. Protandrous species usually directly depend on animal pollination to set seeds (Lloyd and Schoen, 1992; Routley and Husband, 2003).
In this study, we did not observe any significant effect of pollen source on seed set under controlled conditions: flowers receiving outcross pollen showed a similar seed set to those receiving self pollen. This implies that C. palustre is a self-compatible species. These results are consistent with a previous study performed in a natural population of C. palustre in Denmark (Olesen and Warncke, 1992). Besides, our results did not support the occurrence of inbreeding depression at seed set stage. However, inbreeding depression has been reported to occur in C. palustre (Olesen and Warncke, 1992), with higher germination rates of outcrossed than of selfed seeds (0·48 and 0·41, respectively). Moreover, in our study, FIS values were found to be higher at adult stage (see below), suggesting selective pressure related to inbreeding depression at early life stages. Further investigation on the effect of the origin of pollen over the entire development cycle of individuals of C. palustre under controlled conditions is certainly needed to determine whether this species suffers from inbreeding depression and how it can affect population dynamics.
Clonal structure
Clonal structure analyses by means of the aggregation index revealed that genets were spatially clumped in the patchy populations of Bra and Fosse. The clonal dominance index revealed that genets of C. palustre presented a phalanx-type spatial architecture with little or no intermingling of the ramets. Such a spatial distribution pattern has been reported to result from a slow radial spread of the ramets, inducing short internodes between ramets (Lovett-Doust, 1981). However, clones in one sampling plot showed higher intermingling than in the other plots. Such a pattern corresponds rather to a guerrilla-type spatial architecture. It might reflect the foraging or the escape strategy of clonal plant species, which aims to optimize resource acquisition by concentrating ramets in favourable environments (de Kroon and Hutchings, 1995). Genet size ranged from one to 171 ramets, which corresponded to a genet area of <0·5 m2 to 42·8 m2. Similar genet areas have been reported for the clonal pioneer species Menyanthes trifoliata, which occurs in similar biotopes to C. palustre (J. Raabová et al., unpubl. data). The extents of clone persistence and spreading have been reported to be genetically determined (Huber et al., 1999; Rautiainen et al., 2004), and influenced by biotic (e.g. herbivory) and abiotic (e.g. low temperatures, seasonal flooding, resource availability) factors (Stuefer and Huber, 1998; Huhta et al., 2000; Lenssen et al., 2004; Louâpre et al., 2012). In particular, Macek and Lepš (2007) demonstrated that growth traits of C. palustre (i.e. rhizome length and branching) were positively correlated with surrounding vegetation density and height. As the two patchy populations of Fosse and Bra have been extensively grazed, actual clone size might also have been influenced by cattle trampling probably leading to breaking of rhizomes. As a consequence, cattle trampling might further limit clone size (see, for example, Eriksson, 1993; Austin et al., 2007; Rosenthal and Lederbogen, 2008). Moreover, cattle trampling may increase the bare soil area which represents optimal conditions for seed germination and recruitment of pioneer species like C. palustre (Rosenthal et al., 2012; Sarneel and Soons, 2012).
Outcrossing rates
The wide range of realized outcrossing rates observed for all four populations further supports the self-compatibility of C. palustre. Our findings highlighted the influence of the spatial distribution of clones and genet size (i.e. the number of ramets) at the population level on the realized outcrossing rate: patchy populations and large clones showed low outcrossing rates. In addition, paternity assignment tests revealed that most of the gene flow occurred within patches. Patchy populations are commonly expected to present low levels of gene flow between patches with a subsequent low number of genotypes involved in seed production within patches (Ellstrand and Elam, 1993; Young et al., 1996; Sork et al., 1999). Indeed, a patchy distribution of ramets may influence insect foraging behaviour. In particular, bumble bees and solitary bees, which are known to be pollinators of C. palustre in the study regions (Somme et al., unpubl. res.), preferentially forage among near-neighbour flowers (Chittka et al., 1997; Karron et al., 2009) and tend to stay longer within patches, consequently reducing pollen dispersal distances (Kwak et al., 1998; Goverde et al., 2002). In self-compatible species, short-distance pollen dispersal may favour geitono-pollination, especially when genet size is large, inducing low outcrossing rates due to self pollen deposition and germination or stigma clogging, whereas small clones receive more immigrant pollen flow from large clones. This might reflect preferential pollen flow originating from large clones, as found for Convallaria majalis (Vandepitte et al., 2013). Moreover, the two patchy populations are characterized by high flower densities. As bumble bees are known preferentially to forage sequentially near-neighbour flowers, high flower density patches may induce self pollen deposition by pollinators, which further favours low outcrossing rates (Karron et al., 2003; Raspé et al., 2004).
A low outcrossing rate within patches resulted in higher proportions of homozygotes among the offspring (HO = 0·33–0·39) compared with the adults (HO = 0·59–0·64) and also in highly inbred progeny (FIS = 0·37–0·38) compared with the adults (FIS = 0·05–0·09). Selective pressure throughout life stages might be involved in this pattern as it may favour outcrossed offspring of C. palustre. Indeed, Olesen and Warncke (1992) reported inbreeding depression acting on seedling germination, while Sarneel and Soons (2012) reported negative selective pressure on seedlings at high density. Therefore, we may hypothesize that at recruitment and later life stages, offspring resulting from cross-pollination might be favoured over those resulting from self-pollination, especially under physiological stress or competition (see, for example, Price and Waser, 1979; Waller, 1984).
Pollen dispersal
We detected little realized gene flow among patches. Long-distance pollinator movements between patches may increase the likelihood of foraging on other plant species and thus induce pollen loss or increase deposition of heterospecific pollen (Kwak et al., 1998). However, these long-distance movements are essential to maintain genetic diversity through gene flow within but also between populations (Hardy et al., 2004; Buehler et al., 2012), and stepping-stone patches are usually considered to increase the population connectivity by guiding pollinators at low energetic cost from one patch to another, enhancing pollen dispersal (Kwak et al., 1998; Van Rossum and Triest, 2012). In the patchy population of Fosse, patches at distances from 25 to 70 m may act as stepping-stones and contribute to increase pollen transfer, therefore favouring dispersal distances up to 170 m. This population consequently showed pollen dispersal distances similar to those reported in other populations of C. palustre using fluorescent dye as a pollen analogue (Mayer et al., 2012). Although Mayer et al. (2012) already demonstrated that bumble bees flew through the entire population by using fluorescent dye to mimic pollen, we further showed that long-distance and between-patch pollinator movements might lead to effective pollination and pollen flow, also followed by seedling recruitment.
Implications for conservation
To ensure connectivity by pollen and seed flow within patchy populations in a context of habitat fragmentation, it is essential to characterize species response to habitat fragmentation in relation to life history traits such as the mating system and dispersal abilities (Charpentier, 2001; Aguilar et al., 2006). Our findings showed that (1) pollinators play an essential role in pollen transfer for the protandrous flowers of C. palustre; (2) the species is self-compatible and inbreeding depression at seed set stage was not detected, though negative selective pressure on selfed progeny is likely to occur; (3) the clonal structure (i.e. genet size, aggregation and distance between patches) plays a significant role in the quality of pollen transfer and in population genetic diversity; and (4) short-distance dispersal events are more frequent than long-distance dispersal events within patchy populations.
These factors need to be taken into account in management strategies for ensuring population persistence. First, ensuring pollen flow between clone patches within plant populations may contribute to increase the outcrossing rate and then favour high genetic diversity, whereas a high selfing rate may lead to low seedling survival and impoverished genetic diversity. As pollen flow between clone patches occurs by means of pollinators, especially bumble bees, management practices at the population and landscape level should further favour habitats suitable for these pollinators (Moroń et al., 2008; Somme et al., unpubl. res.). Secondly, clonal structure within populations may be a key feature in the restoration or maintenance of efficient pollen dispersal. Clonal structure might be improved by management practices, such as mowing, topsoil removal and extensive cattle grazing. They are usually applied to restore wet meadows, fens and bogs or maintain these habitats at early succession stages, and favour pioneer plant species such as C. palustre (Middleton et al., 2006; Metsoja et al., 2012), and thus deserve attention. For instance, favouring the occurrence of bare soil areas might increase potentially suitable conditions for seedling recruitment of C. palustre (Sarneel and Soons, 2012). Finally, beside pollen dispersal, seed dispersal between patches, and subsequent seedling establishment, might increase the abundance of stepping-stone patches and gene dispersal, and further increase genetic diversity.
ACKNOWLEDGEMENTS
The authors thank the Belgian Fund for Scientific Research (FNRS contract 2·4540·09) for funding this research. L.S. holds a FRIA fellowship (Funds for training in Industry and Agriculture Research). The study was conducted in accordance with current Belgian laws. We thank the ‘Département de la Nature et des Forêts’ (DNF, Région Wallonne, Belgium) for the derogation concerning the sampling of plant individuals in nature reserves, and the DNF and Natagora for granting access to their properties. We thank Christel Buyens, Marc Migon, Marie-Christine Eloy and Marie-Christine Flamand for technical assistance, Xavier Draye for development of the macro for ImageJ, and Fabienne Van Rossum and anonymous referees for improving the manuscript.
LITERATURE CITED
- Aguilar R, Ashworth L, Galetto L, Aizen MA. Plant reproductive susceptibility to habitat fragmentation: review and synthesis through a meta-analysis. Ecology Letters. 2006;9:968–980. doi: 10.1111/j.1461-0248.2006.00927.x. [DOI] [PubMed] [Google Scholar]
- Albert T, Raspé O, Jacquemart A-L. Influence of clonal growth on selfing rate in Vaccinium myrtillus L. Plant Biology (Stuttgart, Germany) 2008;10:643–649. doi: 10.1111/j.1438-8677.2008.00067.x. [DOI] [PubMed] [Google Scholar]
- Arnaud-Haond S, Belkhir K. Genclone: a computer program to analyse genotypic data, test for clonality and describe spatial clonal organization. Molecular Ecology Notes. 2007;7:15–17. [Google Scholar]
- Arnaud-Haond S, Duarte CM, Alberto F, Serrão EA. Standardizing methods to address clonality in population studies. Molecular Ecology. 2007;16:5115–5139. doi: 10.1111/j.1365-294X.2007.03535.x. [DOI] [PubMed] [Google Scholar]
- Austin JE, Keough JR, Pyle WH. Effects of habitat management treatments on plant community composition and biomass in a Montane Wetland. Wetlands. 2007;27:570–587. [Google Scholar]
- Barrett SCH. The evolution of mating strategies in flowering plants. Trends in Plant Science. 1998;3:335–341. [Google Scholar]
- Baskin JM, Baskin CC. Role of temperature and light in the germination ecology of buried seeds of Potentilla recta. Annals of Applied Biology. 1990;117:611–616. [Google Scholar]
- Beier P, Noss RF. Do habitat corridors provide connectivity? Conservation Biology. 1998;12:1241–1252. [Google Scholar]
- Buehler D, Graf R, Holderegger R, Gugerli F. Contemporary gene flow and mating system of Arabis alpina in a Central European alpine landscape. Annals of Botany. 2012;109:1359–1367. doi: 10.1093/aob/mcs066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier A. Consequences of clonal growth for plant mating. Evolutionary Ecology. 2001;15:521–530. [Google Scholar]
- Chittka L, Gumbert A, Kunze J. Foraging dynamics of bumble bees: correlates of movements within and between plant species. Behavioral Ecology. 1997;8:239–249. [Google Scholar]
- Dorken ME, Eckert CG. Severely reduced sexual reproduction in Northern populations of a clonal plant, Decodon verticillatus (Lythraceae) Journal of Ecology. 2001;89:339–350. [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- Eckert CG. Contributions of autogamy and geitonogamy to self-fertilization in a mass-flowering, clonal plant. Ecology. 2000;81:532–542. [Google Scholar]
- Ellstrand NC, Elam DR. Population genetic consequences of small population size: implications for plant conservation. Annual Review of Ecology and Systematics. 1993;24:217–242. [Google Scholar]
- Eriksson O. Dynamics of genets in clonal plants. Trends in Ecology and Evolution. 1993;8:313–316. doi: 10.1016/0169-5347(93)90237-J. [DOI] [PubMed] [Google Scholar]
- Fontaine C, Collin CL, Dajoz I. Generalist foraging of pollinators: diet expansion at high density. Journal of Ecology. 2008;96:1002–1010. [Google Scholar]
- Frankard P, Ghiette P, Hindryckx M-N, Schumacker R, Wastiaux C. Peatlands of Wallony (S-Belgium) Suoseura–Finnish Peatland Society. 1998;49:33–47. [Google Scholar]
- Goverde M, Schweizer K, Baur B, Erhardt A. Small-scale habitat fragmentation effects on pollinator behaviour: experimental evidence from the bumblebee Bombus veteranus on calcareous grasslands. Biological Conservation. 2002;104:293–299. [Google Scholar]
- Guillén A, Rico E, Castroviejo S. Reproductive biology of the Iberian species of Potentilla L. (Rosaceae) Reproductive Biology. 2005;62:9–21. [Google Scholar]
- Hardy OJ, González-Martínez SC, Colas B, Fréville H, Mignot A, Olivieri I. Fine-scale genetic structure and gene dispersal in Centaurea corymbosa (Asteraceae). II. Correlated paternity within and among sibships. Genetics. 2004;168:1601–1614. doi: 10.1534/genetics.104.027714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnay O, Bossuyt B. Prolonged clonal growth: escape route or route to extinction? Oikos. 2005;108:427–432. [Google Scholar]
- Honnay O, Jacquemyn H. A meta-analysis of the relation between mating system, growth form and genotypic diversity in clonal plant species. Evolutionary Ecology. 2008;22:299–312. [Google Scholar]
- Huber H, Lukács S, Watson MA. Spatial structure of stoloniferous herbs: an interplay between structural blue-print, ontogeny and phenotypic plasticity. Plant Ecology. 1999;141:107–115. [Google Scholar]
- Huhta A-P, Lennartsson T, Tuomi J, Rautio P, Laine K. Tolerance of Gentianella campestris in relation to damage intensity: an interplay between apical dominance and herbivory. Evolutionary Ecology. 2000;14:373–392. [Google Scholar]
- Husband BC, Schemske DW. Magnitude and timing of inbreeding depression in a diploid population of Epilobium angustifolium (Onagraceae) Heredity. 1995;75:206–215. [Google Scholar]
- Kalinowski ST, Taper ML. Maximum likelihood estimation of the frequency of null alleles at microsatellite loci. Conservation Genetics. 2006;7:991–995. [Google Scholar]
- Karron JD, Mitchell RJ, Holmquist KG, Bell JM, Funk B. The influence of floral display size on selfing rates in Mimulus ringens. Heredity. 2003;92:242–248. doi: 10.1038/sj.hdy.6800402. [DOI] [PubMed] [Google Scholar]
- Karron JD, Holmquist KG, Flanagan RJ, Mitchell RJ. Pollinator visitation patterns strongly influence among-flower variation in selfing rate. Annals of Botany. 2009;103:1379–1383. doi: 10.1093/aob/mcp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns CA, Inouye DW. Techniques for pollination biologists. Niwot, CO: University Press of Colorado; 1993. [Google Scholar]
- Kindlmann P, Burel F. Connectivity measures: a review. Landscape Ecology. 2008;23:879–890. [Google Scholar]
- Klimeš L, Klimešová J, Hendriks R, van Groenendael J. Clonal plant architecture: a comparative analysis of form and function. In: de Kroon H, van Groenendael J, editors. The ecology and evolution of clonal plants. Leiden, The Netherlands: Bakhyus Publishers; 1997. pp. 1–29. [Google Scholar]
- de Kroon H, Hutchings MJ. Morphological plasticity in clonal plants: the foraging concept reconsidered. Journal of Ecology. 1995;83:143–152. [Google Scholar]
- Kwak MM, Velterop O, van Andel J. Pollen and gene flow in fragmented habitats. Applied Vegetation Science. 1998;1:37–54. [Google Scholar]
- Lamers LPM, Smolders AJP, Roelofs JGM. The restoration of fens in the Netherlands. Hydrobiologia. 2002;478:107–130. [Google Scholar]
- Lenssen JPM, Van Kleunen M, Fischer M, De Kroon H. Local adaptation of the clonal plant Ranunculus reptans to flooding along a small-scale gradient. Journal of Ecology. 2004;92:696–706. [Google Scholar]
- Lloyd DG, Schoen DJ. Self- and cross-fertilization in plants. I. Functional dimensions. International Journal of Plant Sciences. 1992;153:358–369. [Google Scholar]
- Louâpre P, Bittebière A-K, Clément B, Pierre J-S, Mony C. How past and present influence the foraging of clonal plants? PLoS One. 2012;7 doi: 10.1371/journal.pone.0038288. e38288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett-Doust L. Population dynamics and local specialization in a clonal perennial (Ranunculus repens): I. The dynamics of ramets in contrasting habitats. Journal of Ecology. 1981;69:743–755. [Google Scholar]
- Lyanguzova IV. Effect of industrial air pollution on wild plant seed germination and seedling growth. Russian Journal of Plant Physiology. 2011;58:991–998. [Google Scholar]
- Macek P, Lepš J. Environmental correlates of growth traits of the stoloniferous plant Potentilla palustris. Evolutionary Ecology. 2007;22:419–435. [Google Scholar]
- Marshall TC, Slate J, Kruuk LEB, Pemberton JM. Statistical confidence for likelihood-based paternity inference in natural populations. Molecular Ecology. 1998;7:639–655. doi: 10.1046/j.1365-294x.1998.00374.x. [DOI] [PubMed] [Google Scholar]
- Mayer C, Van Rossum F, Jacquemart A-L. Evaluating pollen flow indicators for an insect-pollinated plant species. Basic and Applied Ecology. 2012;13:690–697. [Google Scholar]
- Metsoja J-A, Neuenkamp L, Pihu S, Vellak K, Kalwij JM, Zobel M. Restoration of flooded meadows in Estonia – vegetation changes and management indicators. Applied Vegetation Science. 2012;15:231–244. [Google Scholar]
- Middleton BA, Holsten B, van Diggelen R. Biodiversity management of fens and fen meadows by grazing, cutting and burning. Applied Vegetation Science. 2006;9:307–316. [Google Scholar]
- Moroń D, Szentgyörgyi H, Wantuch M, et al. Diversity of wild bees in wet meadows: implications for conservation. Wetlands. 2008;28:975–983. [Google Scholar]
- Ohsako T. Clonal and spatial genetic structure within populations of a coastal plant, Carex kobomugi (Cyperaceae) American Journal of Botany. 2010;97:458–470. doi: 10.3732/ajb.0900262. [DOI] [PubMed] [Google Scholar]
- Olesen I, Warncke E. Breeding system and seasonal variation in seed set in a population of Potentilla palustris. Nordic Journal of Botany. 1992;12:373–380. [Google Scholar]
- Parks JC, Werth CR. A study of spatial features of clones in a population of bracken fern, Pteridium aquilinum (Dennstaedtiaceae) American Journal of Botany. 1993;80:537–544. doi: 10.1002/j.1537-2197.1993.tb13837.x. [DOI] [PubMed] [Google Scholar]
- Price MV, Waser NM. Pollen dispersal and optimal outcrossing in Delphinium nelsoni. Nature. 1979;277:294–297. [Google Scholar]
- Raspé O, Guillaume P, Jacquemart A-L. Inbreeding depression and biased paternity after mixed-pollination in Vaccinium myrtillus L. (Ericaceae) International Journal of Plant Sciences. 2004;165:765–771. [Google Scholar]
- Rautiainen P, Koivula K, Hyvärinen M. The effect of within-genet and between-genet competition on sexual reproduction and vegetative spread in Potentilla anserina ssp. egedii. Journal of Ecology. 2004;92:505–511. [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2012. [Google Scholar]
- Ritland K. Extensions of models for the estimation of mating systems using n independent loci. Heredity. 2002;88:221–228. doi: 10.1038/sj.hdy.6800029. [DOI] [PubMed] [Google Scholar]
- Rosenthal G, Lederbogen D. Response of the clonal plant Apium repens (Jacq.) Lag. to extensive grazing. Flora - Morphology, Distribution, Functional Ecology of Plants. 2008;203:141–151. [Google Scholar]
- Rosenthal G, Schrautzer J, Eichberg C. Low-intensity grazing with domestic herbivores: a tool for maintaining and restoring plant diversity in temperate Europe. Tuexenia. 2012;32:167–205. [Google Scholar]
- Routley MB, Husband BC. The effect of protandry on siring success in Chamerion angustifolium (Onagraceae) with different inflorescence sizes. Evolution. 2003;57:240–248. doi: 10.1111/j.0014-3820.2003.tb00259.x. [DOI] [PubMed] [Google Scholar]
- Sarneel JM, Soons MB. Post-dispersal probability of germination and establishment on the shorelines of slow-flowing or stagnant water bodies. Journal of Vegetation Science. 2012;23:517–525. [Google Scholar]
- Sarneel JM, Soons MB, Geurts JJM, Beltman B, Verhoeven JTA. Multiple effects of land-use changes impede the colonization of open water in fen ponds. Journal of Vegetation Science. 2011;22:551–563. [Google Scholar]
- Schmucki R, de Blois S. Pollination and reproduction of a self-incompatible forest herb in hedgerow corridors and forest patches. Oecologia. 2009;160:721–733. doi: 10.1007/s00442-009-1347-4. [DOI] [PubMed] [Google Scholar]
- Somme L, Raabová J, Jacquemart A-L, Raspé O. Development and multiplexing of microsatellite markers using pyrosequencing in the clonal plant Comarum palustre (Rosaceae) Molecular Ecology Resources. 2012;12:91–97. doi: 10.1111/j.1755-0998.2011.03072.x. [DOI] [PubMed] [Google Scholar]
- Sork VL, Nason J, Campbell DR, Fernandez JF. Landscape approaches to historical and contemporary gene flow in plants. Trends in Ecology and Evolution. 1999;14:219–224. doi: 10.1016/s0169-5347(98)01585-7. [DOI] [PubMed] [Google Scholar]
- Stuefer JF, Huber H. Differential effects of light quantity and spectral light quality on growth, morphology and development of two stoloniferous Potentilla species. Oecologia. 1998;117:1–8. doi: 10.1007/s004420050624. [DOI] [PubMed] [Google Scholar]
- Tischendorf L, Bender DJ, Fahrig L. Evaluation of patch isolation metrics in mosaic landscapes for specialist vs. generalist dispersers. Landscape Ecology. 2003;18:41–50. [Google Scholar]
- Vallejo-Marín M, Dorken ME, Barrett SCH. The ecological and evolutionary consequences of clonality for plant mating. Annual Review of Ecology, Evolution, and Systematics. 2010;41:193–213. [Google Scholar]
- Vandepitte K, Honnay O, Jacquemyn H, Roldán-Ruiz I. Effects of outcrossing in fragmented populations of the primarily selfing forest herb Geum urbanum. Evolutionary Ecology. 2010;24:1353–1364. [Google Scholar]
- Vandepitte K, Meyer TD, Jacquemyn H, Roldán-Ruiz I, Honnay O. The impact of extensive clonal growth on fine-scale mating patterns: a full paternity analysis of a lily-of-the-valley population (Convallaria majalis) Annals of Botany. 2013;111:623–628. doi: 10.1093/aob/mct024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rossum F, Triest L. Stepping-stone populations in linear landscape elements increase pollen dispersal between urban forest fragments. Plant Ecology and Evolution. 2012;145:332–340. [Google Scholar]
- Waller DM. Differences in fitness between seedlings derived from cleistogamous and chasmogamous flowers in Impatiens capensis. Evolution. 1984;38:427–440. doi: 10.1111/j.1558-5646.1984.tb00301.x. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Whittington WJ, Wilson GB, Humphries RN. The germination characteristics of seeds from Lychnis viscaria L. (Viscaria vulgaris Bernh.), Potentilla rupestris L. and Veronica spicata L. New Phytologist. 1988;109:505–514. [Google Scholar]
- Widén B, Cronberg N, Widén M. Genotypic diversity, molecular markers and spatial distribution of genets in clonal plants, a literature survey. 1994;29:245–263. [Google Scholar]
- Wilcock C, Neiland R. Pollination failure in plants: why it happens and when it matters. Trends in Plant Science. 2002;7:270–277. doi: 10.1016/s1360-1385(02)02258-6. [DOI] [PubMed] [Google Scholar]
- Williams CF. Effects of floral display size and biparental inbreeding on outcrossing rates in Delphinium barbeyi (Ranunculaceae) American Journal of Botany. 2007;94:1696–1705. doi: 10.3732/ajb.94.10.1696. [DOI] [PubMed] [Google Scholar]
- Young A, Boyle T, Brown T. The population genetic consequences of habitat fragmentation for plants. Trends in Ecology and Evolution. 1996;11:413–418. doi: 10.1016/0169-5347(96)10045-8. [DOI] [PubMed] [Google Scholar]