Abstract
Background
There is no method routinely used to predict response to anthracycline and cyclophosphamide–based chemotherapy in the clinic; therefore patients often receive treatment for breast cancer with no benefit. Loss of the Fanconi anemia/BRCA (FA/BRCA) DNA damage response (DDR) pathway occurs in approximately 25% of breast cancer patients through several mechanisms and results in sensitization to DNA-damaging agents. The aim of this study was to develop an assay to detect DDR-deficient tumors associated with loss of the FA/BRCA pathway, for the purpose of treatment selection.
Methods
DNA microarray data from 21 FA patients and 11 control subjects were analyzed to identify genetic processes associated with a deficiency in DDR. Unsupervised hierarchical clustering was then performed using 60 BRCA1/2 mutant and 47 sporadic tumor samples, and a molecular subgroup was identified that was defined by the molecular processes represented within FA patients. A 44-gene microarray-based assay (the DDR deficiency assay) was developed to prospectively identify this subgroup from formalin-fixed, paraffin-embedded samples. All statistical tests were two-sided.
Results
In a publicly available independent cohort of 203 patients, the assay predicted complete pathologic response vs residual disease after neoadjuvant DNA-damaging chemotherapy (5-fluorouracil, anthracycline, and cyclophosphamide) with an odds ratio of 3.96 (95% confidence interval [Cl] =1.67 to 9.41; P = .002). In a new independent cohort of 191 breast cancer patients treated with adjuvant 5-fluorouracil, epirubicin, and cyclophosphamide, a positive assay result predicted 5-year relapse-free survival with a hazard ratio of 0.37 (95% Cl = 0.15 to 0.88; P = .03) compared with the assay negative population.
Conclusions
A formalin-fixed, paraffin-embedded tissue-based assay has been developed and independently validated as a predictor of response and prognosis after anthracycline/cyclophosphamide–based chemotherapy in the neoadjuvant and adjuvant settings. These findings warrant further validation in a prospective clinical study.
Most chemotherapy regimens used for breast cancer in the adjuvant, neoadjuvant, or advanced settings contain agents that directly damage DNA, such as anthracyclines (epirubicin or doxorubicin) and alkylating agents (cyclophosphamide). Approximately 20% to 40% of early breast cancer patients have a complete clinical response and 10% have a complete pathological response (pCR) to these regimens (1–3), most likely because of a deficiency in normal DNA damage response (DDR) pathways (4,5). Many patients, however, do not respond and may not gain any benefit from this type of chemotherapy. In spite of this, there is no reliable method for predicting DDR deficiency from diagnostic material for the purpose of patient treatment selection.
One of the major DDR pathways disrupted in breast cancer is the Fanconi anemia (FA)/BRCA pathway (6). This pathway was first described as lost in a rare autosomal recessive condition characterized by extreme sensitivity to DNA-damaging agents (7). The FA/BRCA pathway coordinates the repair of stalled DNA replication after DNA damage and is therefore important for cancer cell survival after therapeutic DNA-damaging agents such as anthracyclines and cyclophosphamide (5,8). It is estimated to be deficient in approximately 25% of breast cancer patients through mutation or epigenetic silencing of several key components, including the BRCA1 and BRCA2 genes (9).
Although identification of FA/BRCA pathway–deficient, and therefore DDR-deficient, tumors could allow the selection of patients for anthracycline/cyclophosphamide–based chemotherapy treatment, the multiple mechanisms through which the pathway can be lost has made it difficult to develop assays suitable for clinical use. In this study, it was hypothesized that although the FA/BRCA pathway can be compromised by multiple mutational or epigenetic events, the resultant accumulation of DNA damage might activate common genetic processes, thereby defining a distinct molecular subgroup. Furthermore, an assay that identified this subgroup could predict which patients would benefit from chemotherapy. Taking this approach, a novel DDR deficiency (DDRD) assay that can be applied prospectively to patient samples has been developed and independently validated.
Methods
Patient samples
A public microarray dataset (Supplementary Table 1, available online) generated from bone marrow of 21 FA patients and 11 healthy control subjects (10) was used to define the molecular processes associated with FA/BRCA pathway dysfunction.
For subgroup identification, a dataset of 107 formalin-fixed, paraffin-embedded (FFPE) breast cancer samples enriched with 60 BRCA1/2 mutant tumors were collected in the Mayo Clinic, Rochester, Minnesota, after ethical approval from the Mayo Institutional Review Board. Sporadic control samples were matched to BRCA mutant samples based upon patient age at diagnosis, estrogen receptor (ER) and progesterone receptor (PR) status, FFPE block age, and diagnosis.The clinical parameters for the BRCA1/2 mutant and sporadic control sample set are provided in Supplementary Table 2 (available online).
The assay’s ability to predict pCR was assessed in the neoadjuvant setting using microarray data from 203 patients available in 3 public datasets. The first (11) and second (12) datasets were comprised of 86 and 51 ER-positive and ER-negative primary breast tumor samples, respectively, collected before treatment with fluorouracil, doxorubicin, and cyclophosphamide (FAC) (Supplementary Tables 3 and 4, available online). The third (13) was comprised of 66 ER-negative primary breast tumor samples collected before 5-fluorouracil, epirubicin, and cyclophosphamide (FEC) treatment (Supplementary Table 5, available online).
The assay’s ability to predict recurrence-free survival at 5 years after surgery was assessed in the adjuvant setting using an independent dataset of 191 N0-N1 ER-positive and ER-negative FFPE patient samples with a median follow-up of 4.93 years (Supplementary Table 6, available online). Historically, N0 and N1 patients were treated with adjuvant FEC; ER-positive patients also received hormone therapy. Among HER2-positive patients, 83.3% received 1 year of adjuvant trastuzumab. ER and PR status was determined by immunohistochemistry using the Quick Score method. T stage was determined according to the American Joint Committee on cancer TNM staging system (14). Tumor grade was defined according to the Nottingham grading system (15). HER2 status was assessed by immunohistochemistry: a score of 0 or 1 was negative; score of 3 was positive; and a score of 2 led to further assessment by chromogenic in situ hybridisation and was considered positive if HER2 amplification was detected. Lymphocytic infiltrate was determined by lymphocyte count of hematoxylin and eosin–stained full face sections.
The prognostic utility of the assay to predict recurrence-free survival was assessed using three public datasets (16–18) totalling 664 ER-positive and ER-negative patients that did not receive DNA-damaging chemotherapy with a median follow-up of 7.3 years (Supplementary Tables 7– 9, available online). Of these patients, 64 received tamoxifen.
Supplementary Tables 1– 9 provide the sample names and clinical information required to reproduce the results in this article for each dataset used in this study. The distribution of the clinical parameters within the training and validation datasets used within this study are provided within Supplementary Table 10 (available online). The publicly sourced FA dataset and neoadjuvant and prognostic validation datasets can be accessed using the GEO accession numbers provided within the Supplementary Materials (available online). Subgroup analysis based upon major ethnic groups was not performed because these data were not available for the tumor datasets. Flow diagrams of the discovery and validation of the DDRD assay is provided in Supplementary Figures 1 and 2 (available online).
Written informed consent was obtained for all tissue samples used.
Gene Expression Profiling
Total RNA was extracted from macrodissected FFPE tumor samples using the Roche High Pure RNA Paraffin Kit (Roche Diagnostics, Burgess Hill, UK) as described previously (19). Total RNA was amplified using the NuGEN WT-Ovation FFPE System (NuGEN, San Carlos, CA) and hybridized to the Almac Breast Cancer DSA (Affymetrix, Santa Clara, CA) as described previously (19,20).
Statistical Analysis Methods
Differentially expressed probesets between 21 FA patients and 11 healthy control subjects were identified using a fold-change threshold of absolute fold-change greater than 3 and a statistically significant t test P value threshold adjusted for false discovery rate of less than .001 (Supplementary Table 11, available online) (21). Statistically significantly enriched functional classes with a P value adjusted for false discovery rate of less than .05 [derived using the hypergeometric distribution test (22)] corresponding to differentially expressed genes were determined using the experiment analysis workflow from Metacore (GeneGo; Carlsbad, CA) (Supplementary Table 12, available online).
BRCA1/2 mutant and sporadic control training samples were split into 2 datasets based on the transcript levels of ESR1 (estrogen receptor 1); details are provided in the Supplementary Materials (available online). A combined background and variance filter was applied to each dataset (ER-negative and ER-positive) to identify the most variable probesets (Supplementary Tables 13 and 14, available online). Hierarchical clustering (Pearson correlation distance and Ward’s linkage) was applied to probesets and samples from each dataset separately. The number of subclusters was determined using the gap statistic (23). The sample clusters from the ER-positive and ER-negative datasets representing molecular processes identified in FA patient samples were selected for classification and labeled as DDRD-positive; the remaining samples were labeled as DDRD-negative.
Signature generation was performed using the partial least squares method (24) with selection of features based on recursive feature elimination during 10 repeats of fivefold cross-validation (Supplementary Figure 3, available online). Model development was nested within cross-validation, including an initial filter to remove 75% of probesets with the lowest variance and intensity, reference-based robust multichip averaging preprocessing, and recursive feature elimination discarding the least important 10% of probesets at each iteration. The total number of features in the final model was determined by the feature length maximizing the average area under the receiver operating characteristics curve under cross-validation. To facilitate validation of the signature across multiple microarray platforms, the selected probeset signature was regenerated at the gene level (Supplementary Tables 15– 17, available online). The threshold for dichotomization of the predictions was selected to maximize the sum of sensitivity and specificity from cross-validated training data (Supplementary Figure 3D, available online).
To calculate negative and positive predictive value within the training and neoadjuvant validation datasets, the prevalence of each endpoint (BRCA status/pCR) was estimated using the proportions of each class in the corresponding dataset. Supplementary Table 18 (available online) lists the priors for the training and each individual neoadjuvant dataset.
A logistic regression model was used to estimate the odds ratio (OR) comparing the odds of pCR in DDRD-positive vs DDRD-negative patients. Cox proportional hazards regression was used to investigate the prognostic effect of the DDRD signature on recurrence-free survival at 5 years after surgery. The estimated effect of the signature was adjusted for tumor stage, ER status, HER2 status, age, nodal status, and tumor grade by fitting a multivariable (logistic regression or Cox proportional hazards regression) model. The proportional hazard assumption was verified for the Cox model using a formal statistical test based on the Schoenfeld residuals (25). The P value for the test was calculated for original, log-transformed, and rank time scales. Samples with unknown clinical factors were excluded in this assessment.
All tests of statistical significance were two-sided and performed at a 5% alpha level.
Further details are available in the Supplementary Methods (available online).
Results
Identification of a DDRD Molecular Subgroup in Breast Cancer
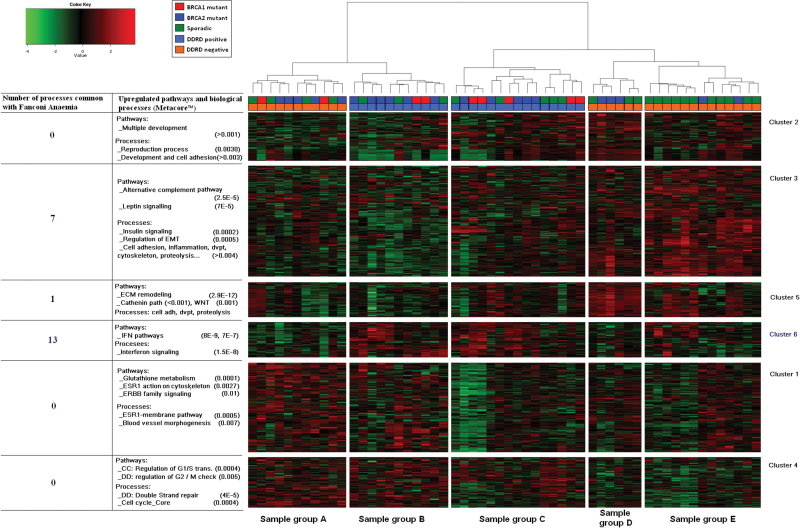
The biological processes modulated in response to loss of components of the FA/BRCA pathway were identified after analysis of a publicly available dataset from FA patients and healthy control subjects (10) and were found to be predominantly immune related (Supplementary Table 12, available online). To identify molecular subgroups in breast cancer defined by the same processes as those identified in FA patients, a DNA-microarray platform optimized for FFPE tissue (20) was used to analyze a BRCA1/2 mutant–enriched cohort of FFPE breast cancer samples. Molecular subgroups were identified using an unsupervised hierarchical clustering approach. ER-positive and ER-negative cohorts were considered separately because the dominant effect of the ER on hierarchical clustering (26) could prevent the identification of an ER-independent subgroup. Probeset lists displaying the most variation across the tumors were selected (Supplementary Tables 13 and 14, available online). A total of 6 probeset clusters and 3 sample groups were identified within the ER-negative dataset (Figure 1A), and 6 probeset clusters and 5 sample groups were identified within the ER-positive dataset (Figure 1B). Pathway analysis and molecular process characterization (Supplementary Tables 19 and 20, available online) identified probeset cluster 3 within the ER-negative cohort and probeset cluster 6 within the ER-positive cohort as representing the molecular processes (predominantly immune related) identified in FA patient samples, sharing 14 (Figure 1C; Supplementary Table 21, available online) and 13 (Figure 1D; Supplementary Table 22, available online) statistically significantly enriched processes, respectively.
Figure 1.
Hierarchical analysis of BRCA1/2 mutant and sporadic wild-type control breast cancer samples. Hierarchical analysis of estrogen receptor (ER)–negative (A) and ER-positive (B) BRCA1/2 mutant and sporadic wild-type control breast cancer samples. Probeset cluster groups are annotated on the right side. Pathway analysis of each probeset cluster group and the number of biological processes each probeset cluster group has in common with Fanconi anemia (FA) patients are annotated on the left side of each image. Molecular subgroups are labeled A–C for ER-negative samples and A–E for ER-positive samples. The legend for each image indicates the BRCA1/2 mutational status and the DNA damage response deficiency (DDRD) assay group each sample was assigned to for assay generation. C and D) Demonstration of the statistically significant common molecular processes between FA patient samples and cluster 3 of ER-negative samples and cluster 6 of ER-positive samples respectively. The bars represent the –log P value of statistical significance for each process in both the FA patient and breast cancer samples. P values are derived from the two-sided hypergeometric distribution test.
Using these data, a putative DDRD subgroup was generated by combining samples defined by the FA/BRCA–deficient biology: sample group B as defined by probeset cluster 3 in ER-negative samples (Figure 1A) and sample groups B and C as defined by probeset cluster 6 in ER-positive samples (Figure 1B). Pathway analysis of the genes differentially expressed between the tumors contained within the DDRD subgroup and the remaining tumors within the training set confirmed that immune signaling was the predominant biology associated with this subgroup (Supplementary Table 23, available online).
Development of an Assay to Identify the DDRD Subgroup
The putative DDRD subgroup was class-labeled DDRD-positive, whereas the remaining samples were class-labeled DDRD-negative. Computational classification resulted in a 44-gene assay that could be used to prospectively identify clinical samples within the DDRD molecular subgroup, referred to herein as the DDRD assay (Supplementary Table 17 and Supplementary Figure 3, available online). In the training set, the assay predicted BRCA1/2 mutational status with a specificity of 0.79 (95% confidence interval (Cl) = 0.64 to 0.86) and sensitivity of 0.58 (95% Cl = 0.48 to 0.65) (Table 1; Supplementary Figure 4A, available online), confirming an association between DDRD assay positivity and dysfunction of the FA/BRCA pathway.
Table 1.
Performance metrics of the DNA damage response deficiency (DDRD) assay within the training dataset*
| Prediction of BRCA mutation status using the DDRD assay | ||||||||
|---|---|---|---|---|---|---|---|---|
| Dataset | Sample No. | Clinical outcome | AUC (95% CI) | ACC (95% CI) | SENS (95% CI) | SPEC (95% CI) | PPV (95% CI) | NPV (95% CI) |
| Training | 107 | BRCA mutant V wildtype | 0.68 (0.56 to 0.78) | 0.70 (0.57 to 0.76) | 0.58 (0.48 to 0.65) | 0.79 (0.64 to 0.86) | 0.78 (0.63 to 0.85) | 0.60 (0.49 to 0.65) |
* The 95% confidence intervals (CIs) are from ±2 standard deviations from cross-validation. ACC = accuracy; AUC = area under the receiver operating characteristics curve; NPV = negative predictive value; PPV = positive predictive value; SENS = sensitivity; SPEC = specificity;.
Validation of the DDRD Assay in Predicting Response to Neoadjuvant Anthracycline-Based Chemotherapy
To assess the ability of the DDRD assay to predict response to DNA-damaging chemotherapy, it was applied to 3 microarray datasets (11–13). In each study, patients were treated with neoadjuvant 5-fluorouracil, anthracycline, and cyclophosphamide–based regimens (Supplementary Tables 3– 5, available online), and the endpoints were pCR or residual disease.
Multivariable analysis in the combined neoadjuvant dataset demonstrated that the DDRD assay predicted response in the DDRD-positive sample cohort relative to the DDRD-negative sample cohort independent of and superior to ER status, T stage, and grade (OR = 3.96; 95% Cl =1.67 to 9.41; P = .002) (Table 2; Supplementary Figure 4B, available online). The performance metrics of the DDRD assay within each individual neoadjuvant dataset are provided in Supplementary Table 24 and Supplementary Figure 4, C–E (available online).
Table 2.
Multivariable analysis of the predictive value of the DNA damage response deficiency (DDRD) assay for pathological complete response within the neoadjuvant fluorouracil, doxorubicin, and cyclophosphamide/5-fluorouracil, epirubicin, and cyclophosphamide validation dataset*
| Parameter | Multivariable analysis OR (95% CI) | P |
|---|---|---|
| DDRD-positive | 3.96 (1.67 to 9.41) | .002 |
| ER-negative | 2.93 (0.94 to 9.08) | .06 |
| Grade (3 vs 2) | 2.19 (0.95 to 5.07) | .07 |
| T stage | .32 | |
| T1 | 1.00 (referent) | |
| T2 | 0.60 (0.08 to 4.44) | |
| T3 | 0.45 (0.06 to 3.50) | |
| T4 | 0.09 (0.01 to 1.67) |
* Logistic regression was used to estimate the odds ratio (OR) and its 95% confidence interval (CI). DDRD-negative, ER-positive, grade level 2, and T stage 1 were the baseline (referent) or comparative levels. P values are derived from the two-sided Wald test. ER = estrogen receptor.
Clinical risk groups were defined by a response classifier based upon a logistic regression model of ER status, age, stage, and grade. Within each risk group, pCR rates were stratified by DDRD status further confirming the independent association of DDRD-positivity with pCR (Supplementary Table 25, available online). HER2 status was not assessed in this analysis as there was an insufficient number of samples with HER2 status information available.
Validation of the DDRD Assay in Predicting Clinical Outcome After Adjuvant FEC Chemotherapy
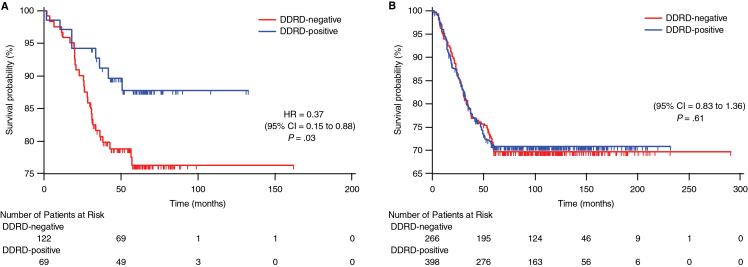
The assay was applied to FFPE samples from an independent validation cohort of breast cancer (N0–N1) patients who received adjuvant FEC chemotherapy. Overall, DDRD-positive patients had improved relapse-free survival relative to DDRD-negative patients with a 5-year hazard ratio (HR) of 0.37 (95% Cl = 0.15 to 0.88; P = .03) on multivariable analysis (Figure 2A; Table 3). Importantly, the assay predicted outcome better than and independent of ER status, HER2 status, T stage, grade, nodal status, and age (Table 3).
Figure 2.
Kaplan–Meier time to recurrence curves stratified by the DNA damage response deficiency (DDRD) assay. Kaplan–Meier time to recurrence curves censored at 5 years for 191 breast cancer patients treated with adjuvant 5-fluorouracil, epirubicin, and cyclophosphamide (FEC) (A) and 664 early breast cancer patients from 3 studies (14–16) in which patients did not receive DNA-damaging chemotherapy (B). P values are derived from the two-sided Wald test. The number of events/patients at risk at various time intervals are noted in the tables below the curves. CI = confidence interval; HR = hazard ratio.
Table 3.
Multivariable analysis of the predictive value of the DNA damage response deficiency (DDRD) assay for relapse-free survival 5 years after fluorouracil, epirubicin, and cyclophosphamide chemotherapy*
| Parameter | Multivariable analysis HR (95% CI) | P |
|---|---|---|
| DDRD-positive | 0.37 (0.15 to 0.88) | .03 |
| ER-negative | 0.46 (0.20 to 1.06) | .07 |
| HER2-positive | 1.72 (0.79 to 3.73) | .17 |
| Age | 0.59 (0.32 to 1.08) | .09 |
| Grade (3 vs 1 and 2)† | 1.14 (0.48 to 2.71) | .77 |
| T stage | .40 | |
| T1 | 1.00 (referent) | |
| T2 | 0.63 (0.29 to 1.40) | |
| T3 | 1.13 (0.35 to 3.65) | |
| Nodal status | .97 | |
| N0 | 1.00 (referent) | |
| N1 | 1.02 (0.47 to 2.21) |
* Cox proportional hazard model used to calculate hazard ratio (HR) and 95% confidence interval (CI). DDRD-negative, ER-negative, grade level 1 and 2, T stage 1, HER2-negative, and nodal status N0 were the baseline (referent) or comparative levels. Age was continuous with parameter estimate for a 16-year difference in age (representing ratio of hazard between patients at the 1st and 3rd quartiles of age distribution). P values are derived from the two-sided Wald test. ER = estrogen receptor.
† Grade 1 and 2 were combined because of small number of grade 1 patients.
The precision of the DDRD assay was also evaluated following Clinical and Laboratory Standards Institute guidelines (27). This demonstrated that the total standard deviation estimates of samples spanning the DDRD assay’s score range were acceptable for clinical application because they are less than 5% of the DDRD assay’s reportable range (see Supplementary Material, available online).
To determine whether the DDRD assay was prognostic independent of treatment, it was applied to 3 publicly available early breast cancer datasets where patients had no cytotoxic chemotherapy (16–18) (Supplementary Tables 7– 9, available online). No difference in survival was noted between the DDRD-positive and DDRD-negative populations (HR = 1.07; 95% Cl = 0.83 to 1.36; P = .61) (Figure 2B), confirming that the assay predicts patient outcome in breast cancer only in the context of FEC chemotherapy.
Assessment of the Association Between ER and HER2 Status With DDRD Assay Result
Because ER-negative tumors are more frequently sensitive to DNA-damaging chemotherapy than ER-positive tumors, the proportion of each population classified as DDRD-positive was assessed (Table 4). Consistent with what has been observed in the clinic, more ER-negative patients than ER-positive patients were DDRD-positive (52.0% vs 25.0%, P < .001; 67.9% vs 24.3%, P < .001 in the adjuvant dataset and neoadjuvant datasets, respectively). There was no statistically significant increase in DDRD positivity in HER2-positive patients (41.3% of HER2-positive vs 32.0% of HER2-negative; P = .26) in the adjuvant dataset.
Table 4.
Association of the DNA damage response deficiency (DDRD) assay with estrogen receptor (ER) status, HER2 status, and lymphocytic infiltration within validation datasets
| Adjuvant dataset | Total No. | DDRD- positive, % | P* |
|---|---|---|---|
| Adjuvant | |||
| ER-positive | 112 | 25.0 | <.001 |
| ER-negative | 77 | 52.0 | |
| HER2-positive | 46 | 41.3 | .26 |
| HER2-negative | 128 | 32.0 | |
| Triple negative | 44 | 54.6 | .001 |
| Non–triple negative | 135 | 28.2 | |
| Lymphocytic infiltrate positive | 35 | 74.3 | <.001 |
| Lymphocytic infiltrate negative | 155 | 27.7 | |
| Neoadjuvant | |||
| ER-positive | 70 | 24.3 | <.001 |
| ER-negative | 134 | 67.9 | |
* A two-sided χ2 test was used to assess the statistical significance of the univariate association between the DDRD assay and each clinical factor.
Assessment of the Association Between Immune Infiltration and DDRD Assay Result
It has previously been observed that lymphocytic infiltration (LI) associates with an increased sensitivity to chemotherapy (28). Because the DDRD molecular subgroup is largely defined by immune signaling, it was important to confirm that the assay signal was arising from tumor cells and not from lymphocytes. The association between the assay score and LI was therefore assessed in the adjuvant dataset. A statistically significant association between DDRD positivity and LI was observed (P < .001); however, 43 case patients out of 155 with no LI were classified as DDRD-positive and 9 case patients out of 35 with LI were classified as DDRD-negative. These data suggest that the DDRD assay does not directly represent LI.
To further assess whether the upregulation of immune signaling measured by the assay originated from DDRD tumor cells, the assay score of laboratory cell-line models representative of DDRD positivity (BRCA1 mutant HCC1937 empty vector control breast cancer cell line [29] as well as RKO-FANCC knockout and RKO-FANCG knockout colon cancer cell lines) vs DDRD-negative controls (HCC1937 cells in which BRCA1 was corrected and the RKO parental cell line) was assessed. In each case, the DDRD-positive cell line displayed a statistically significantly higher assay score when compared with the control cell lines (Supplementary Figure 5 and Supplementary Table 26, available online).
Together these data support the model that the immune signaling defining the DDRD subgroup and assay arises directly from tumor cells and this can be associated with LI.
Discussion
This study describes a 44-transcript assay that prospectively identifies a molecular subgroup of breast cancers deficient in the FA/BRCA DDR pathway and sensitive to anthracycline/cyclophosphamide–based chemotherapy. In the neoadjuvant setting, the DDRD assay was found to statistically significantly predict pCR to FAC/FEC. Stratification of pCR rates by ER status or a logistic regression model developed using standard clinical risk factors demonstrated the DDRD assay positively associated with increased response within each stratified group. In the adjuvant setting, the assay was prognostic in the context of FEC chemotherapy and was not prognostic in the absence of chemotherapy. Importantly, the assay provided additional information when assessed by multivariable analysis against other prognostic factors commonly assessed in the clinic.
To improve the ability to detect a FA/BRCA pathway–deficient subgroup, the training cohort of primary breast tumors was enriched with known BRCA1/2 mutant tumors. Interestingly, not all of the BRCA1/2 mutant samples fell within the DDRD subgroup, and 32.5% of DDRD-positive tumors were BRCA1/2 wild-type. This is consistent with what is observed in the clinic, as not all BRCA1/2 mutant tumors are sensitive to DNA-damaging agents. This may be because not all reported mutations affect DNA repair (30) or could be because of alternative mechanisms compensating for the DDR deficiency. Also not all BRCA1/2 wild-type tumors possess a normal DDR because epigenetic silencing of BRCA1/2 as well as loss of other components of the FA/BRCA pathway can result in DNA damage sensitivity (9).
Cell-line models representative of loss of the FA/BRCA pathway demonstrated a high DDRD score, indicating that the immune signal arises directly from tumor cells. Interestingly, LI has been reported in BRCA1 mutant cancers (31), and it has been reported that LI, as assessed on standard histopathology, associates with chemotherapy response (28), although there is only moderate interobserver agreement with this measurement (kappa = 0.6) (32). Although there was an association between the DDRD result and LI in this study, this was not absolute, indicating that one was not interchangeable for the other. The immune infiltrate may, however, be in response to the immune signal originating from DDRD tumor cells. In support of this, there is increasing evidence that damaged DNA, as occurs spontaneously in FA/BRCA–deficient tumors, can activate immune pathways similar to viral infection (33). Indeed, it has recently been reported that expression of a collection of immune genes associates with response to anthracycline-based chemotherapy in publically available datasets (34). Interestingly seven of these genes—CXCL10, MX1, IDO1, IFI44L, CD2, ETV7 and RSAD2—are also present in the DDRD assay. CXCL10 has recently been reported to be upregulated in BRCA1-deficient breast cancers, further supporting a link between a specific immune response and loss of the FA/BRCA pathway (35). It is therefore hypothesized that the DDRD assay identifies tumors with endogenous DNA damage secondary to an abnormal DDR, with resultant upregulation of immune signaling, failed senescence, and possible lymphocyte activation.
Triple negativity (estrogen, progesterone, and HER2 receptor negativity) has been suggested as a biomarker for abnormal DDR and therefore response to DNA-damaging therapies (36). Upon multivariable analysis, however, a positive DDRD result was found to predict outcome in the neoadjuvant and adjuvant settings independent of the ER status, indicating that triple negativity is distinct from a positive DDRD result.
There are conflicting data regarding the use of HER2 or the related TOP2A amplification as predictive markers of response to anthracyclines. In this study, we did not observe a statistically significant association between a positive result for DDRD and HER2 positivity (P = .26). Interestingly a large meta-analysis of 3452 breast cancer patients treated with adjuvant chemotherapy advised against the routine use of HER2 or TOP2A amplification as a selection biomarker for anthracycline-based regimens because women with non-HER2– amplified tumors still benefited from treatment (37). In the adjuvant dataset, we observed that 32.0% of HER2-negative patients were DDRD-positive and would therefore be predicted to have an increased probability of benefiting from DNA-damaging chemotherapy. Consistent with this, the DDRD assay provided prognostic information independent from HER2 status in the multivariable analysis of known clinical factors in the adjuvant dataset. Unfortunately the HER2 status was not available for a sufficient number of samples in the neoadjuvant chemotherapy dataset to further assess the interaction between HER2, DDRD, and anthracycline response in this setting. A recent publication may explain some of the inconsistencies around HER2 and TOP2A as predictive biomarkers. Chromosomal instability associates with a benefit from anthracyclines in breast cancer and can result in the amplification of chromosome 17 on which the TOP2A and HER2 genes are located. This study suggests that the mechanism of sensitivity to anthracyclines is not the level of HER2 or TOP2A expression but rather the underlying chromosomal instability (38). Interestingly loss of the FA/BRCA pathway is associated with chromosomal instability (39–41), which provides further evidence for the link between the DDRD molecular subgroup and DNA-damaging chemotherapy response.
The approach to developing a biomarker of benefit from DNA-damaging chemotherapy presented in this study is different to that adopted by others. The DDRD assay is based upon a molecular subgroup discovered by unsupervised hierarchical clustering and is applicable to FFPE tumor sections routinely taken before the administration of chemotherapy. Others have demonstrated molecular subtypes that associate with chemosensitivity but, unlike this study, have not developed these into assays that could be used to analyze FFPE tissue in a prospective manner (34,42).
A potential limitation of this study is that the validation of the assay in the adjuvant treatment setting was performed in a historical cohort of N0 and N1 patients to allow adequate follow-up for recurrence. In modern practice, there has been a move toward offering chemotherapy to lower-risk patients who may be underrepresented in this dataset. Also in modern practice, N1 patients commonly receive a taxane in the adjuvant setting. Additionally, although the neoadjuvant dataset supports the predictive utility of the assay and the non-chemotherapy-treated dataset suggests the assay is not prognostic for untreated patients, we acknowledge that the best study design to demonstrate its utility as a predictive assay in the adjuvant setting would be a prospective biomarker–treatment interaction design (43).
In summary, we have discovered a DDRD molecular subgroup in breast cancer that is predominantly defined by upregulation of immune signaling and can be identified by a 44-transcript DDRD assay. This assay has been demonstrated to predict increased tumor response and decreased recurrence after FEC/FAC chemotherapy. We hypothesize that patients classified as DDRD-negative may not respond to the same extent as DDRD-positive patients from anthracycline/cyclophosphamide–based treatment and may therefore benefit from alternative approaches such as taxane-based regimens. This could be investigated further using samples from ongoing adjuvant treatment studies that are comparing anthracycline-based vs taxane-based chemotherapy such as the NSABP-B49 clinical trial.
Funding
This work was supported by the the European Regional Development Fund; Invest Northern Ireland; an Experimental Cancer Medicine Centre grant (CR-UK and HSC R&D Northern Ireland); and by a National Institutes of Health grant (CA116167) and a National Institutes of Health Specialized Program of Research Excellence in Breast Cancer awarded to the Mayo Clinic (CA116201).
J. M. Mulligan, L. A. Hill, and S. Deharo contributed equally to this article. J. M. Mulligan, L. A. Hill, S. Deharo, and R. D. Kennedy were responsible for conception and experimental design; collection and assembly of data; implentation of experimental design, data analysis, and interpretation; manuscript writing, and final approval of the manuscript. G. Irwin was responsible for collection and assembly of data; implentation of experimental design, data analysis, and interpretation; manuscript writing, and final approval of the manuscript. D. Boyle, J. E. Quinn, P. B. Mullan, C. R. James, S. M. Walker, P. Kerr, V. Proutski, M. Salto-Tellez, and P. G. Johnston were responsible for collection and assembly of data; data analysis and interpretation; manuscript writing, and final approval of the manuscript. K. E. Keating, O. Y. Raji, F. A. McDyer, E. O’Brien, M. Bylesjo, T. S. Davison, and D. Paul Harkin were responsible for experimental design; collection and assembly of data; implentation of experimental design, data analysis, and interpretation; manuscript writing, and final approval of the manuscript. N. M. Lindor, J. James, and F. J. Couch were responsible for collection and assembly of data; data analysis and interpretation; manuscript writing, final approval of the manuscript, and provision of study material or patients.
The authors had full responsibility for the design of the study; the collection, the analysis, and interpretation of the data; the decision to submit the article for publication, and the writing of the article. We thank the patients involved in this study for their participation.
J. M. Mulligan, L. A. Hill, K. S. Deharo, E. Keating, O. Y. Raji, F. A. McDyer, E. O’Brien, M. Bylesjo, S. M. Walker, P. Kerr, T. S. Davison, V. Proutski, D. P. Harkin, and R. D. Kennedy are employees of Almac Diagnostics. M. Salto-Tellez is a consultant to Almac Diagnostics. P. G. Johnston is a board member of Almac Diagnostics and a consultant to Sanofi Aventis. F. J. Couch receives royalties from Myriad Genetics Laboratories. All other authors declare no conflicts of interest.
Part of this work has been presented at a poster discussion session at ASCO 2011 (abstract No. 10511) and a poster session at ASCO 2013 (abstract No. 109814).
References
- 1. Fisher B, Brown A, Mamounas E, et al. Effect of preoperative chemotherapy on local-regional disease in women with operable breast cancer: findings from National Surgical Adjuvant Breast and Bowel Project B-18. J Clin Oncol. 1997;15(7):2483–2493 [DOI] [PubMed] [Google Scholar]
- 2. Petit T, Wilt M, Velten M, et al. Comparative value of tumour grade, hormonal receptors, Ki-67, HER-2 and topoisomerase II alpha status as predictive markers in breast cancer patients treated with neoadjuvant anthracycline-based chemotherapy. Eur J Cancer. 2004;40(2):205–211 [DOI] [PubMed] [Google Scholar]
- 3. Arun BK, Dhinghra K, Valero V, et al. Phase III randomized trial of dose intensive neoadjuvant chemotherapy with or without G-CSF in locally advanced breast cancer: long-term results. Oncologist. 2011;16(11):1527–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kriege M, Seynaeve C, Meijers-Heijboer H, et al. Sensitivity to first-line chemotherapy for metastatic breast cancer in BRCA1 and BRCA2 mutation carriers. J Clin Oncol. 2009;27(23):3764–3771 [DOI] [PubMed] [Google Scholar]
- 5. Kennedy RD, D’, Andrea AD. DNA repair pathways in clinical practice: lessons from pediatric cancer susceptibility syndromes. J Clin Oncol. 2006;24(23):3799–3808 [DOI] [PubMed] [Google Scholar]
- 6. D’Andrea AD. Susceptibility pathways in Fanconi’s anemia and breast cancer. N Engl J Med. 2010;362(20):1909–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grompe M, D’, Andrea A. Fanconi anemia and DNA repair. Hum Mol Genet. 2001;10(20):2253–9 [DOI] [PubMed] [Google Scholar]
- 8. Kennedy RD, D’, Andrea AD. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev. 2005;19(24):2925–2940 [DOI] [PubMed] [Google Scholar]
- 9. Turner N, Tutt A, Ashworth A. Hallmarks of “BRCAness” in sporadic cancers. Nat Rev Cancer. 2004;4(10):814–819 [DOI] [PubMed] [Google Scholar]
- 10. Vanderwerf SM, Svahn J, Olson S, et al. TLR8-dependent TNF-(alpha) overexpression in Fanconi anemia group C cells. Blood. 2009;114(26):5290–5298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tabchy A, Valero V, Vidaurre T, et al. Evaluation of a 30-gene paclitaxel, fluorouracil, doxorubicin, and cyclophosphamide chemotherapy response predictor in a multicenter randomized trial in breast cancer. Clin Cancer Res. 2010;16(21):5351–5361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Iwamoto T, Bianchini G, Booser D, et al. Gene pathways associated with prognosis and chemotherapy sensitivity in molecular subtypes of breast cancer. J Natl Cancer Inst. 2011;103(3):264–272 [DOI] [PubMed] [Google Scholar]
- 13. Bonnefoi H, Potti A, Delorenzi M, et al. Validation of gene signatures that predict the response of breast cancer to neoadjuvant chemotherapy: a substudy of the EORTC 10994/BIG 00-01 clinical trial. Lancet Oncol. 2007;8(12):1071–1078 [DOI] [PubMed] [Google Scholar]
- 14. Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17(6):1471–1474 [DOI] [PubMed] [Google Scholar]
- 15. Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19(5):403–410 [DOI] [PubMed] [Google Scholar]
- 16. Desmedt C, Piette F, Loi S, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13(11):3207–3214 [DOI] [PubMed] [Google Scholar]
- 17. Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–679 [DOI] [PubMed] [Google Scholar]
- 18. Sotiriou C, Wirapati P, Loi S, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98(4):262–272 [DOI] [PubMed] [Google Scholar]
- 19. Kennedy RD, Bylesjo M, Kerr P, et al. Development and independent validation of a prognostic assay for stage II colon cancer using formalin-fixed paraffin-embedded tissue. J Clin Oncol. 2011;29(35):4620–4626 [DOI] [PubMed] [Google Scholar]
- 20. Tanney A, Oliver GR, Farztdinov V, et al. Generation of a non-small cell lung cancer transcriptome microarray. BMC Med Genomics. 2008;1:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Assoc. 1994;57(1):289–300 [Google Scholar]
- 22. Cho RJ, Huang M, Campbell MJ, et al. Transcriptional regulation and function during the human cell cycle. Nat Genet. 2001;27(1):48–54 [DOI] [PubMed] [Google Scholar]
- 23. Yan M, Ye K. Determining the number of clusters using the weighted gap statistic. Biometrics. 2007;63(4):1031–1037 [DOI] [PubMed] [Google Scholar]
- 24. DeJong S. SIMPLS: An alternative approach to partial least squares regression. Chemometrics Intell Lab Syst. 1993;18(3):251–263 [Google Scholar]
- 25. Schoenfeld DA. Sample-size formula for the proportional-hazards regression model. Biometrics. 1983;39(2):499–503 [PubMed] [Google Scholar]
- 26. Sorlie T, Perou CM, Fan C, et al. Gene expression profiles do not consistently predict the clinical treatment response in locally advanced breast cancer. Mol Cancer Ther. 2006;5(11):2914–2918 [DOI] [PubMed] [Google Scholar]
- 27. Clinical and Laboratory Standards Institute Defining, Establishing and Verifying Reference Intervals in the Clinical Laboratory; Approved Guideline. 2008, Published by Clinical and Laboratory Standards Institute, Philadelphia, PA [Google Scholar]
- 28. Denkert C, Loibl S, Noske A, et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol. 2010;28(1):105–113 [DOI] [PubMed] [Google Scholar]
- 29. Quinn JE, Kennedy RD, Mullan PB, et al. BRCA1 functions as a differential modulator of chemotherapy-induced apoptosis. Cancer Res. 2003;63(19):6221–6228 [PubMed] [Google Scholar]
- 30. Linger RJ, Kruk PA. BRCA1 16 years later: risk-associated BRCA1 mutations and their functional implications. FEBS J. 2010;277(15):3086–3096 [DOI] [PubMed] [Google Scholar]
- 31. Lakhani SR, Jacquemier J, Sloane JP, et al. Multifactorial analysis of differences between sporadic breast cancers and cancers involving BRCA1 and BRCA2 mutations. J Natl Cancer Inst. 1998;90(15):1138–1145 [DOI] [PubMed] [Google Scholar]
- 32. Longacre TA, Ennis M, Quenneville LA, et al. Interobserver agreement and reproducibility in classification of invasive breast carcinoma: an NCI breast cancer family registry study. Mod Pathol. 2006;19(2):195–207 [DOI] [PubMed] [Google Scholar]
- 33. Sabatel H, Pirlot C, Piette J, et al. Importance of PIKKs in NF-kappaB activation by genotoxic stress. Biochem Pharmacol. 2011;82(10):1371–1383 [DOI] [PubMed] [Google Scholar]
- 34. Ignatiadis M, Singhal SK, Desmedt C, et al. Gene modules and response to neoadjuvant chemotherapy in breast cancer subtypes: a pooled analysis. J Clin Oncol. 2012;30(16):1996–2004 [DOI] [PubMed] [Google Scholar]
- 35. Mulligan AM, Raitman I, Feeley L, et al. Tumoral lymphocytic infiltration and expression of the chemokine CXCL10 in breast cancers from the Ontario Familial Breast Cancer Registry. Clin Cancer Res. 2013;19(2):336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363(20):1938–1948 [DOI] [PubMed] [Google Scholar]
- 37. Di Leo A, Desmedt C, Bartlett JM, et al. HER2 and TOP2A as predictive markers for anthracycline-containing chemotherapy regimens as adjuvant treatment of breast cancer: a meta-analysis of individual patient data. Lancet Oncol. 2011;12(12):1134–1142 [DOI] [PubMed] [Google Scholar]
- 38. Munro AF, Twelves C, Thomas JS, et al. Chromosome instability and benefit from adjuvant anthracyclines in breast cancer. Br J Cancer. 2012;107(1):71–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Popova T, Manie E, Rieunier G, et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 2012;72(21):5454–5462 [DOI] [PubMed] [Google Scholar]
- 40. Daniels MJ, Wang Y, Lee M, et al. Abnormal cytokinesis in cells deficient in the breast cancer susceptibility protein BRCA2. Science. 2004;306(5697):876–879 [DOI] [PubMed] [Google Scholar]
- 41. Naim V, Rosselli F. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat Cell Biol. 2009;11(6):761–768 [DOI] [PubMed] [Google Scholar]
- 42. Rodriguez AA, Makris A, Wu MF, et al. DNA repair signature is associated with anthracycline response in triple negative breast cancer patients. Breast Cancer Res Treat. 2010;123(1):189–196 [DOI] [PubMed] [Google Scholar]
- 43. Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst. 2009;101(21):1446–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]