Abstract
Background
Loss of 18q22.3 is a prognostic marker in pancreatic ductal adenocarcinoma (PDAC). This study investigated genes encoded by this cytoband.
Methods
We studied mRNA/protein expression in radically resected (n = 130) and metastatic patients (n = 50). The role of CYB5A was tested in 11 PDAC cell lines and five primary cultures through retrovirus-mediated upregulation and small interfering RNA using wound-healing, invasion, annexin-V, electron microscopy, and autophagic assays, as well as autophagy genes and kinases arrays. CYB5A+ orthotopic models (n = 6 mice/group) were monitored by Firefly and Gaussia-luciferase bioluminescence, magnetic resonance imaging, and high-frequency ultrasound. Data were analyzed by t test, Fisher exact-test, log-rank test and Cox proportional hazards models. All statistical tests were two-sided.
Results
Both resected and metastatic patients with low mRNA or protein expression of CYB5A had statistically significantly shorter survival (eg, median = 16.7 months, 95% confidence interval [CI] = 13.5 to 19.9; vs median = 24.8 months, 95% CI = 12.8 to 36.9; P = .02, two-sided log-rank test; n = 82 radically resected PDACs), and multivariable analyses confirmed prognostic relevance. Moreover, we characterized a novel function to CYB5A, autophagy induction, concomitant with reduced proliferation and migration/invasion of PDAC cells. Network analysis of proautophagic pathways suggested CYB5A interaction with TRAF6, which was confirmed by TRAF6 downregulation after CYB5A reconstitution (−69% in SU.86.86-CYB5A+; P = .005, two-sided t test). CYB5A silencing had opposite effects, restoring TRAF6 expression and wound healing. In vivo studies showed that CYB5A induced autophagy while inhibiting tumor growth/metastasis and increasing survival (median = 57 days, 95% CI = 52 to 61; vs median = 44 days, 95% CI = 21 to 57; P = .03, two-sided log-rank test).
Conclusions
These results define CYB5A as a novel prognostic factor for PDAC that exerts its tumor-suppressor function through autophagy induction and TRAF6 modulation.
Pancreatic ductal adenocarcinoma (PDAC) carries one of the worst prognoses of any major malignancy and exhibits profound chemoresistance (1–3). The inefficacy of currently available therapeutic strategies has been attributed to the dense desmoplastic reaction, which reduces drug penetration, and to the high rate of genetic alterations affecting multiple pathways (4,5). Genetic analyses uncovered mechanisms controlling pancreatic carcinogenesis (6) and studies to identify aberrancies associated with outcome are warranted.
We previously investigated genomic imbalances using array-comparative genomic hybridization in a cohort of 44 radically resected patients, the largest PDAC series ever investigated by array-comparative genomic hybridization (7). In this series, the median overall survivals (OS) for patients with and without loss of the cytoband 18q22.3 were 7.6 and 21.4 months, respectively (P = .02, two-sided log-rank test).
The cytoband 18q22.3 contains five known genes (FBXO15, c18orf55, CYB5A, c18orf51, and CPGL). In agreement with previous findings (8), CPGL reduced proliferation and inhibited migration in SU.86.86 cells carrying FLAG-tagged CPGL. However, knockdown of CPGL in the PANC-1 cells did not affect proliferation, cell cycle distribution, and wound healing (7).
The aim of this study was to evaluate whether the mRNAs and/or proteins coded by the genes in the 18q22.3 cytoband were associated with outcome in two cohorts of radically resected patients and one cohort of metastatic PDAC patients. Further, we aimed to characterize key factors affecting proliferative and invasive capacity, as well as autophagy induction, which may provide mechanistic insights on PDAC aggressive behavior and contribute to the rational development of new prognostic and therapeutic approaches.
Methods
Cell Lines
AsPc1, BxPc-3, Capan-2, CFPAC-1, HPAC, MIA PaCa-2, PANC-1, PL45, SU.86.86, SUIT2-007, SUIT2-028, and hTERT-HPNE were from American Type Culture Collection (Manassas, VA). Five primary cultures (PDAC-1/-2/-3/-4/-5) were isolated at Pisa University Hospital (9).
Patient Samples
The primary tumors (n = 130) of the two cohorts of PDAC patients were resected with pancreatico-duodenectomy or total/distal-pancreatectomy before adjuvant treatment, which consisted of gemcitabine-based combined modality (eg, gemcitabine 1000mg/m2/day on days 1, 8, and 15 every 28 days, followed by gemcitabine 300mg/m2 weekly plus concomitant radiation therapy to a total of 45 Gray). Clinicopathological characteristics of these patients are reported in Supplementary Table 1, A and B (available online).
Fresh-frozen samples from the first cohort (n = 48, stage IIb, pT3N1Mx according to American Joint Committee on Cancer - Tumor Node Metastasis staging system), which were collected from December 2001 to October 2004, were stored until laser microdissection. Similarly, 33 biopsies from metastatic tumors were collected before treatment (gemcitabine 1000mg/m2/day on days 1, 8, and 15). Patients were followed-up for 1.6 to 56.5 months (median = 18.2 months) until January 2010.
Patients were stratified according to the median expression of target genes. Disease-free survival was defined as the time from the diagnosis to the first relapse or last follow-up in radically resected patients. Progression-free survival was defined as the time from the diagnosis to the first progression or last follow-up in metastatic patients. OS was calculated from the diagnosis to the death or last follow-up.
Tissue microarrays (TMAs) were constructed using core tissue biopsies from formalin-fixed paraffin-embedded specimens of the second cohort (n = 82). Two additional TMAs were constructed with 50 samples from metastatic patients.
All specimens were obtained after patient’s written consent approved by the Pisa University Hospital Ethics Committee. Detailed methods for laser microdissection and construction of TMAs can be found in the Supplementary Methods (available online).
Quantitative Reverse-Transcription Polymerase Chain Reaction and Immunohistochemistry
Expression of FBXO15, C18orf55, CYB5A, CPGL, and CPGL-B was evaluated in 48 patients undergoing surgical resection and 33 metastatic patients (7,9), whereas CYB5A protein expression was evaluated in TMAs from a second cohort (n = 82) of radically resected (10) patients and 50 metastatic patients, as described in the Supplementary Methods (available online).
In Vitro Studies
PDAC-2 and SU.86.86 cells were transduced with PLNCX2/CYB5A-YFP constructs (Supplementary Figure 1, A and B, available online), or transfected with 25 nmol/L CYB5A small interfering RNA or negative control (s223365 and Silencer Negative Control #1; Ambion, Foster City, CA). CYB5A expression was compared with hTERT-HPNE using the 2-ΔΔCt method.
Ultrastructural analysis and CYB5A and LC-3 immunoblotting/fluorescence were performed as previously described (9,11).
Autophagy-related genes were assessed using the Autophagy-RT2-Profiler PCR-Array (Qiagen, Hilden, Germany). Kinase activity was evaluated by peptide substrate array (PamGene, ‘s-Hertogenbosch, the Netherlands), as described (12).
Functional and physical interactions of CYB5A were retrieved using STRING (13). Data were loaded in Cytoscape version 2.8.3, and networks were merged with embedded analysis tools. Clustering of the final network was obtained with ClusterOne plug-in version 1.1, whereas the BINGO plug-in provided ontologies for each cluster (14,15).
Further details on proliferation, migration, invasion, immunoblotting, DNA damage, apoptosis, and autophagy assays are in the Supplementary Methods (available online).
In Vivo Studies
PDAC-2 cells were transduced with lentiviral vectors containing Firefly luciferase (Fluc)/mCherry, and Gaussia luciferase(Gluc)/CFP, as described (16,17). Cells were then transduced with PLNCX2/CYB5A-YFP or control constructs and injected in the pancreas of 12 randomized mice: six PLNCX2/CYB5A-YFP (CYB5A+) and six PLNCX2/YFP (CTR group). Tumor growth was monitored by charge-coupled device camera imaging. Experiments were carried out according to a protocol approved by the VU University Amsterdam, the Netherlands. Further details are in the Supplementary Methods (available online).
Statistical Analyses
All experiments were performed in triplicate and repeated three times. Data were expressed as mean values ± standard errors and analyzed by Student t test or Mann–Whitney test followed by the Tukey multiple comparison. Associations between clinicopathologic features were tested by Fisher exact test. Kaplan–Meier and log-rank methods were used to compare disease-free survival/progression-free survival and OS curves using SPSS version 20 (IBM, Chicago, IL). Statistically significant variables in the univariable analysis were included in multivariable analysis using Cox proportional hazards model. The proportional hazards assumption was satisfied using the Schoenfeld residuals method. All statistical tests were two-sided. A P value of less than .05 was considered statistically significant.
Results
CYB5A mRNA and Protein Expression and Outcome
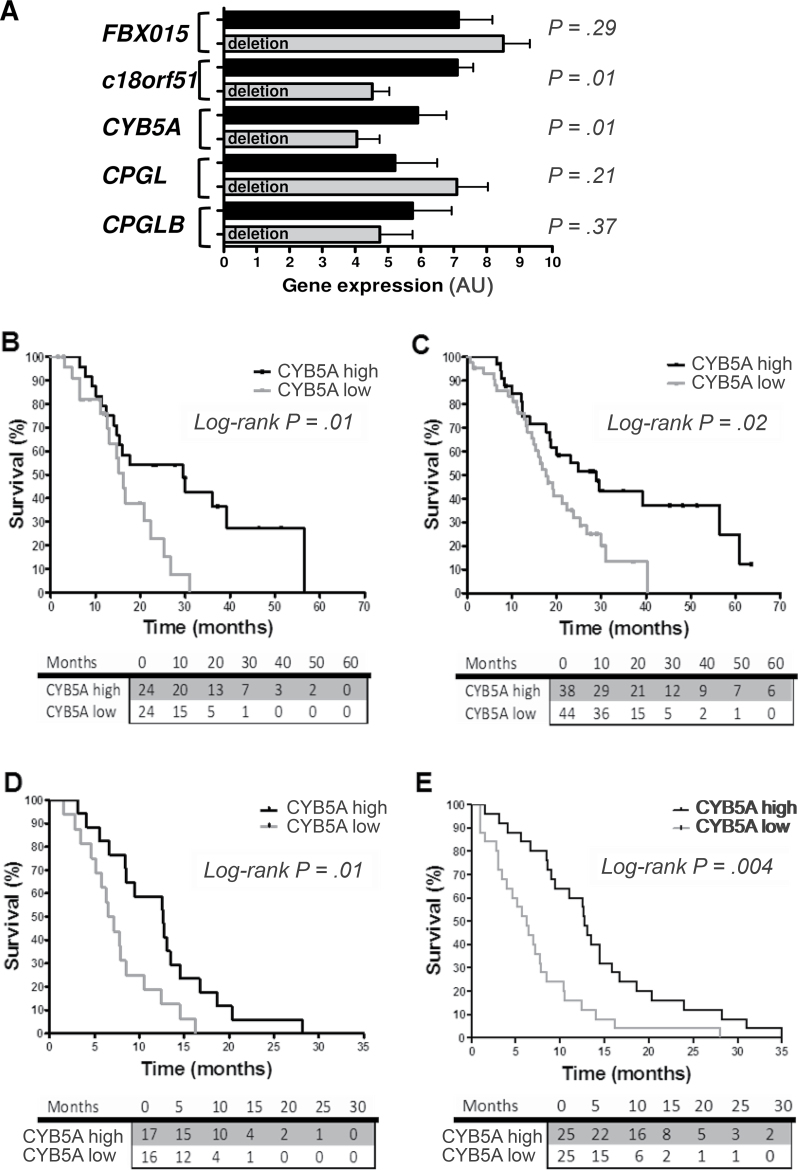
Figure 1A depicts FBXO15, c18orf55, CYB5A, CPGL, and CPGL-B transcripts variation according to CPGL deletion, which is used as an indicator of cytoband 18q22.3 deletion (7). Median expression values of CYB5A and c18orf55 were statistically significantly lower in deleted samples (P < .05, two-sided Mann–Whitney test). CPGL deletion was confirmed to have prognostic value (Table 1), as well as grading. However, considering the five genes studied, only patients with CYB5A expression below median had a statistically significantly shorter OS (median OS = 16.3, 95% confidence interval [CI] = 13.4 to 19.1; vs median OS = 29.5 months, 95% CI = 11.1 to 47.8; hazard ratio [HR] = 2.3, 95% CI = 1.2 to 6.0; P = .01, two-sided log-rank test) (Figure 1B). Similar results were observed for disease-free survival (Table 1), whereas there was no difference in CYB5A levels according to grade (Supplementary Table 1C, available online).
Figure 1.
Expression of CYB5A and clinical outcome. A) Polymerase chain reaction data of the five genes in the 48 radically resected pancreatic ductal adenocarcinoma patients (PDAC) patients of the first cohort, with and without CPGL deletion. Data are expressed as arbitrary units (AUs), as explained in the Supplementary Methods (available online). P values were calculated with two-sided Mann–Whitney test. C18orf51 was not included because it was not detectable in preliminary analyses in PDAC cells. B) Survival curves according to polymerase chain reaction data of CYB5A mRNA expression in the patients from the first cohort, grouped according to high/low vs median values. P values were calculated with two-sided log-rank test. C) Survival curves according to CYB5A protein expression in the 82 radically resected PDAC patients of the second cohort (high/low staining score values, as explained in the Supplementary Methods, available online). All patients received the same gemcitabine-based adjuvant chemotherapy, but 13 patients in this cohort had stage IIA PDACs (pT3N0Mx), and fewer tumors exhibited grade 3 histological differentiation in comparison with those in the first cohort (Supplementary Table 1, available online). P values were calculated with two-sided log-rank test. D and E) Survival curves according to CYB5A mRNA and protein expression in the metastatic patients (mRNA expression data were available for 33 patients, whereas protein expression data were available for 50 patients). P values were calculated with two-sided log-rank test. The number of patients at risk in each group is given in the tables below the graphs.
Table 1.
Outcome according to clinical characteristics and mRNA expression of the genes in the 18q22.3 cytoband in the first cohort of pancreatic ductal adenocarcinoma patients*
| Univariate analysis | No. (%) | OS months (95% CI) | P† | DFS month (95% CI) | P† |
|---|---|---|---|---|---|
| 48 | 16.7 (9.8 to 23.5) | 11.8 (7.7 to 15.8) | |||
| Age, y | .67 | .65 | |||
| ≤65 | 28 (58) | 15.9 (14.1 to 17.7) | 9.8 (6.4 to 13.2) | ||
| >65 | 20 (42) | 22.3 (12.8 to 31.7) | 14.5 (7.6 to 21.3) | ||
| Sex | .26 | .49 | |||
| Male | 21 (44) | 15.9 (9.2 to 19.5) | 11.4 (7.8 to 14.9) | ||
| Female | 27 (56) | 25.2 (12.4 to 41.3) | 13.0 (7.2 to 18.8) | ||
| Resection status | .49 | .78 | |||
| R0 | 38 (79) | 17.7 (7.5 to 27.8) | 10.0 (6.8 to 13.2) | ||
| R1 | 10 (21) | 16.2 (13.2 to 19.3) | 14.8 (10.2 to 19.3) | ||
| Vascular infiltration | .77 | .72 | |||
| No | 29 (60) | 15.9 (11.1 to 20.8) | 9.8 (5.2 to 14.3) | ||
| Yes | 19 (40) | 20.9 (12.7 to 29.0) | 15.7 (9.5 to 21.8) | ||
| Neural infiltration | .78 | .85 | |||
| No | 22 (46) | 16.7 (7.8 to 25.5) | 13.0 (5.3 to 20.7) | ||
| Yes | 26 (54) | 17.7 (8.7 to 26.6) | 11.8 (8.7 to 14.8) | ||
| PanINs | .63 | .64 | |||
| No | 23 (48) | 16.3 (14.1 to 18.4) | 9.3 (3.9 to 14.8) | ||
| Yes | 25 (52) | 22.3 (9.9 to 34.6) | 14.5 (9.2 to 19.8) | ||
| WHO grading | <.001 | .01 | |||
| 1–2 | 21 (44) | 31.0 (28.6 to 33.4) | 24.1 (15.2 to 33.0) | ||
| 3 | 27 (46) | 14.8 (11.6 to 17.9) | 9.3 (5.9 to 12.8) | ||
| CPGL deletion | .04 | .10 | |||
| No | 32 (67) | 15.2 (10.6 to 19.9) | 10.0 (6.5 to 13.5) | ||
| Yes | 12 (33) | 31.0 (21.7 to 40.3) | 14.7 (5.7 to 23.8) | ||
| FBXO15 expression | .20 | .17 | |||
| ≤median = low | 23 (49) | 16.3 (12.5 to 20.0) | 7.7 (2.4 to 13.0) | ||
| >median = high | 24 (51) | 25.2 (18.0 to 32.4) | 13.0 (9.1 to 16.9) | ||
| c18orf55 expression | .17 | .46 | |||
| ≤median = low | 21 (50) | 16.3 (14.3 to 18.3) | 10.0 (4.9 to 15.0) | ||
| >median = high | 21 (50) | 26.6 (17.6 to 35.7) | 14.8 (8.1 to 21.4) | ||
| CYB5A expression | .01 | .05 | |||
| ≤median = low | 24 (50) | 16.3 (13.4 to 19.1) | 7.3 (3.9 to 18.8) | ||
| >median = high | 24 (50) | 29.5 (11.1 to 47.8) | 11.9 (3.2 to 20.5) | ||
| CPGL expression | .68 | .09 | |||
| ≤median = low | 24 (50) | 16.3 (14.2 to 18.4) | 7.7 (5.1 to 10.3) | ||
| >median = high | 24 (50) | 22.3 (9.8 to 34.7) | 14.8 (10.3 to 19.2) | ||
| CPGL-B expression | .69 | .66 | |||
| ≤median = low | 23 (49) | 16.3 (13.8 to 18.7) | 10.0 (4.5 to 15.5) | ||
| >median = high | 24 (51) | 20.9 (7.3 to 34.5) | 11.8 (7.8 to 15.7) | ||
| Multivariable analysis | df | Risk of death HR (95% CI) | P‡ | Risk of relapse HR (95% CI) | P‡ |
| WHO grading | 1 | .001 | .03 | ||
| 1–2 | 0.3 (0.1 to 0.6) | 0.4 (0.2 to 0.8) | |||
| 3 | 1.0 (referent) | 1.0 (referent) | |||
| CYB5A expression | 1 | .02 | .05 | ||
| Low | 2.8 (1.2 to 6.7) | 1.8 (1.0 to 1.3) | |||
| High | 1.0 (referent) | 1.0 (referent) |
* CI = confidence Interval; df = degrees of freedom; DFS = disease-free survival; HR = hazard ratio; OS = overall survival; PanIN = pancreatic intraepithelial neoplasia; WHO = World Health Organization.
† Calculated using log-rank two-sided test.
‡ Calculated using Cox proportional hazards two-sided test.
The prognostic role of CYB5A was confirmed in an independent cohort of radically resected patients (Table 2), in whom protein expression was evaluated by immunohistochemistry (Supplementary Figure 2, available online). Patients with low CYB5A expression had median OS of 16.7 months (95% CI = 13.5 to 19.9), whereas remaining patients had median OS of 24.8 months (95% CI = 12.8 to 36.9; HR = 1.9, 95% CI = 1.2 to 3.6; P = .02, two-sided log-rank test) (Figure 1C). CYB5A was also associated with disease-free survival (Table 1), whereas there were no differences in CYB5A expression in relation to all the clinicopathological parameters (Supplementary Table 2, available online). Multivariable analyses demonstrated that CYB5A mRNA and protein expression and grading are independent prognostic factors (Tables 1 and 2). Similar results were obtained in 50 metastatic patients (Table 3; Figure 1, C and D). No differences were observed in analysis by sex (data not shown).
Table 2.
Outcome according to clinical characteristics and protein expression of CYB5A in the second cohort of radically resected pancreatic ductal adenocarcinoma patients*
| Univariate analysis | No. (%) | OS months (95% CI) | P† | DFS months (95% CI) | P† |
|---|---|---|---|---|---|
| 82 | 19.2 (15.4 to 23.0) | 12.0 (9.4 to 14.6) | |||
| Age, y | .30 | .68 | |||
| ≤65 | 43 (52) | 16.7 (6.0 to 27.4) | 12.0 (7.3 to 16.8) | ||
| >65 | 39 (48) | 19.2 (17.1 to 21.3) | 12.4 (8.3 to 16.5) | ||
| Sex | .86 | .80 | |||
| Male | 39 (48) | 18.4 (14.1 to 22.7) | 10.8 (8.6 to 19.5) | ||
| Female | 43 (52) | 21.1 (13.6 to 28.7) | 14.1 (8.6 to 19.5) | ||
| Resection status | .33 | .61 | |||
| R0 | 66 (80) | 19.2 (14.3 to 24.1) | 12.0 (8.9 to 15.1) | ||
| R1 | 16 (20) | 18.4 (16.3 to 20.5) | 10.8 (8.2 to 13.3) | ||
| Vascular infiltration | .35 | .67 | |||
| No | 38 (46) | 23.8 (12.6 to 35.0) | 12.0 (4.9 to 19.1) | ||
| Yes | 44 (54) | 18.4 (14.5 to 22.3) | 11.8 (9.4 to 14.1) | ||
| Neural infiltration | .47 | .86 | |||
| No | 23 (28) | 24.5 (8.1 to 40.9) | 12.0 (1.0 to 23.0) | ||
| Yes | 59 (72) | 18.8 (16.2 to 21.3) | 12.0 (9.6 to 14.4) | ||
| PanINs | .75 | 30 | |||
| No | 53 (64) | 18.4 (12.0 to 25.6) | 12.4 (9.7 to 15.0) | ||
| Yes | 29 (36) | 20.0 (14.3 to 25.7) | 10.8 (7.3 to 14.3) | ||
| WHO grading | .05 | .06 | |||
| 1–2 | 61 (74) | 20.5 (14.9 to 26.1) | 12.8 (8.4 to 17.3) | ||
| 3 | 21 (26) | 14.6 (7.3 to 21.8) | 7.5 (5.3 to 9.6) | ||
| AJCC stage | .16 | .24 | |||
| IIA | 13 (16) | 20.0 (18.5 to 21.9) | 12.0 (9.1 to 14.4) | ||
| IIB | 69 (84) | 18.4 (14.0 to 22.2) | 12.0 (9.0 to 15.0) | ||
| CYB5A protein expression | .02 | .02 | |||
| Low | 44 (54) | 16.7 (13.5 to 19.9) | 11.6 (8.3 to 14.9) | ||
| High | 38 (46) | 24.8 (12.8 to 36.9) | 16.4 (6.2 to 26.6) | ||
| Multivariable analysis | df | Risk of death HR (95% CI) | P‡ | Risk of relapse HR (95% CI) | P‡ |
| WHO grading | 1 | .05 | .14 | ||
| 1–2 | 0.6 (0.3 to 1.0) | 0.6 (0.4 to 1.2) | |||
| 3 | 1.0 (referent) | 1.0 (referent) | |||
| CYB5A expression | 1 | .02 | .03 | ||
| Low | 2.0 (1.1 to 3.6) | 1.8 (1.1 to 1.4) | |||
| High | 1.0 (referent) | 1.0 (referent) |
* AJCC American Joint Committee on Cancer; CI = confidence interval; df = degrees of freedom; DFS = disease-free survival; HR = hazard ratio; OS = overall survival; PanIN = pancreatic intraepithelial neoplasia; WHO = World Health Organization.
† Calculated using log-rank two-sided test.
‡ Calculated using Cox’s proportional hazards two-sided test.
Table 3.
Outcome according to clinical characteristics and protein expression of CYB5A in the cohort of metastatic patients*
| Univariate analysis | No. (%) | OS months (95% CI) | P† | PFS months (95% CI) | P‡ |
|---|---|---|---|---|---|
| 50 | 8.5 (6.5 to 10.4) | 5.8 (3.3 to 8.4) | |||
| Age, y | .20 | .14 | |||
| ≤65 | 28 (56) | 7.7 (5.7 to 9.7) | 12.0 (7.3 to 16.8) | ||
| >65 | 22 (44) | 9.0 (4.3 to 13.7) | 12.4 (8.3 to 16.5) | ||
| Sex | .12 | .10 | |||
| Male | 34 (68) | 6.7 (4.5 to 9.0) | 10.8 (8.6 to 19.5) | ||
| Female | 16 (32) | 12.5 (11.2 to 13.8) | 14.1 (8.6 to 19.5) | ||
| WHO grading | .007 | .05 | |||
| 1–2 | 23 (46) | 13.9 (9.9 to 15.4) | 12.8 (8.4 to 17.3) | ||
| 3 | 27 (54) | 7.8 (5.0 to 9.3) | 7.5 (5.3 to 9.6) | ||
| CYB5A protein expression | .004 | <.001 | |||
| Low | 25 (50) | 12.7 (9.8 to 15.5) | 11.6 (8.3 to 14.9) | ||
| High | 25 (50) | 6.5 (4.4 to 8.5) | 16.4 (6.2 to 26.6) | ||
| Multivariable analysis | df | Risk of death HR (95% CI) | P‡ | Risk of relapse HR (95% CI) | P‡ |
| WHO grading | 1 | .05 | .05 | ||
| 1–2 | 0.5 (0.3 to 0.9) | 0.7 (0.5 to 1.0) | |||
| 3 | 1.0 (referent) | 1.0 (referent) | |||
| CYB5A expression | 1 | .04 | <.001 | ||
| Low | 1.9 (1.2 to 3.0) | 2.1 (1.3 to 2.7) | |||
| High | 1.0 (referent) | 1.0 (referent) |
* CI = confidence interval; df = degrees of freedom; HR = hazard ratio; OS = overall survival; PFS = progression-free survival; WHO = World Health Organization.
† Calculated using log-rank two-sided test.
‡ Calculated using Cox’s proportional hazards two-sided test.
Effects of CYB5A on Cell Growth and Invasion
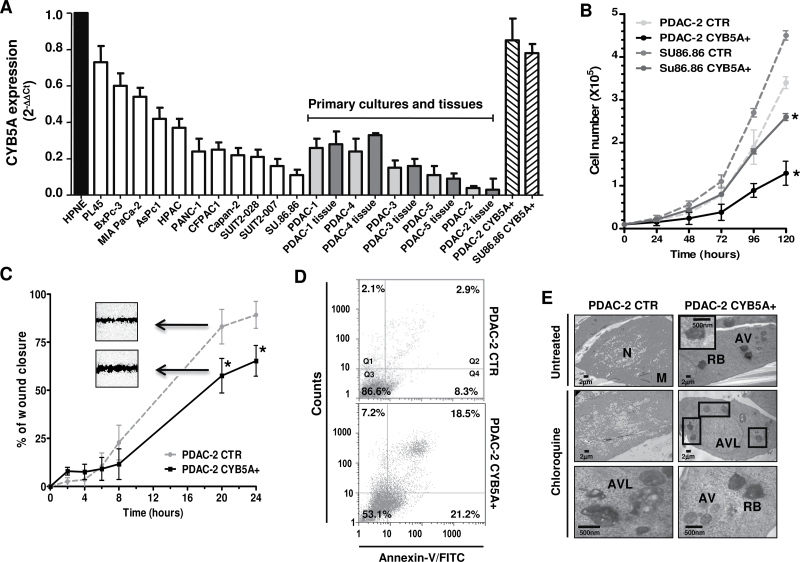
To study the biological role of CYB5A, we initially assessed its mRNA expression, demonstrating lower expression levels in all malignant cells compared with nontumorigenic cells (Figure 2A). Remarkably, CYB5A levels in primary cultures were correlated (R 2 = 0.901; P = .01, two-sided Spearman test) to the values detected in their tissues of origin, suggesting that these cultures are reliable models.
Figure 2.
Effects of CYB5A modulation on pancreatic cells. A) CYB5A mRNA expression in pancreatic cancer cells (vs normal pancreatic ductal cells HPNE), including the corresponding tissues for five primary cell cultures (gray bars) and cells transduced with the CYB5A vector (CYB5A+). B) Growth curves of SU.86.86 and PDAC-2 cells (CTR vs CYB5A+). C) In vitro migration of CTR vs CYB5A+ cells, showing statistically significant wound healing (representative pictures in the inserts). Statistically significant differences were already detected at 8-hour time points for SU.86.86 (−30% migration vs CTR, data not shown). D) Histograms of cytofluorometric analysis of apoptosis 24 hours after starvation, as detected by Annexin-V (Q2: late apoptosis; Q4: early apoptosis) in PDAC-2 cells. E) Electron microscopy phenotypes in PDAC-2 and SU.86.86 cells, showing autophagic vacuoles (AV), residual bodies (RB), and autophagolysosomes (AVL) near the nucleus (N) and mitochondria (M). Points indicate mean values from three independent experiments. Bars indicate standard deviation. All P values were calculated with two-sided Student t test. *P < .05.
SU.86.86 and PDAC-2 cells were selected for further investigation because they carried the 18q22.3 cytoband loss (Supplementary Figure 3, available online) and expressed the lowest levels of CYB5A mRNA. We successfully established CYB5A-overexpressing subclones and empty vectors with more than 90% efficiency in each cell type (Supplementary Figure 4, A and B, available online). Furthermore, quantitative reverse-transcription polymerase chain reaction demonstrated a 7-fold and 20-fold increase of CYB5A expression in SU.86.86-CYB5A+ and PDAC-2-CYB5A+, respectively (Figure 2A). These CYB5A+ subclones showed slower proliferation rates, as well as less wound healing (Figure 2, B and C) and invasion (Supplementary Figure 4C, available online), indicating that CYB5A expression leads to both growth inhibition and attenuated cell migration. CYB5A small interfering RNA transfection of PDAC-2-CYB5A+ subclones restored proliferation (data not shown) and wound healing (Supplementary Figure 4D, available online). Conversely, no differences were detected for double-strand breaks (Supplementary Figure 5, available online).
Effects of CYB5A on Autophagy
Annexin-V staining showed a statistically significant increase in apoptosis in both SU86.86-CYB5A+ (P = .02, two-sided t test) and PDAC-2-CYB5A+ (P = .02, two-sided t test), as shown in Figure 2D. In SU.86.86-CYB5A+, early apoptosis was increased from 2.3% to 9.1%, whereas late-apoptosis was increased from 4.5% to 9.5%. A similar increase was also observed for mitochondrial damage (Supplementary Figure 6A, available online), with a clear decrease of DiOC6 binding in CYB5A+ cells (P = .02, two-sided t test).
However, the most striking effect of CYB5A transduction was the different morphological appearance of CYB5A+ cells, which showed a round shape and a dramatic increase in cytoplasmic vacuolization. Cytoplasm and nucleoplasm were slightly darker, and ultrastructural analysis revealed double-membrane vesicles containing engulfed cytoplasmic contents and autophagolysosomes (Figure 2E). Quantitative analysis confirmed increased autophagic vacuoles formation and accumulation of residual bodies and autophagolysosomes (+25% and +33% vs. CTR, respectively); this was further induced by chloroquine, (+35% and +41% vs. CTR, respectively), which was also able to inhibit cell proliferation (Supplementary Figure 6B, available online). The ability of CYB5A to induce autophagy was supported by the increase of acridine-orange–stained cells, double-labeling of CYB5A and LC3-II, and immunoblotting (Supplementary Figure 6, C–E, available online).
Effects of CYB5A on Proautophagic Pathways and Oncogenic Kinases
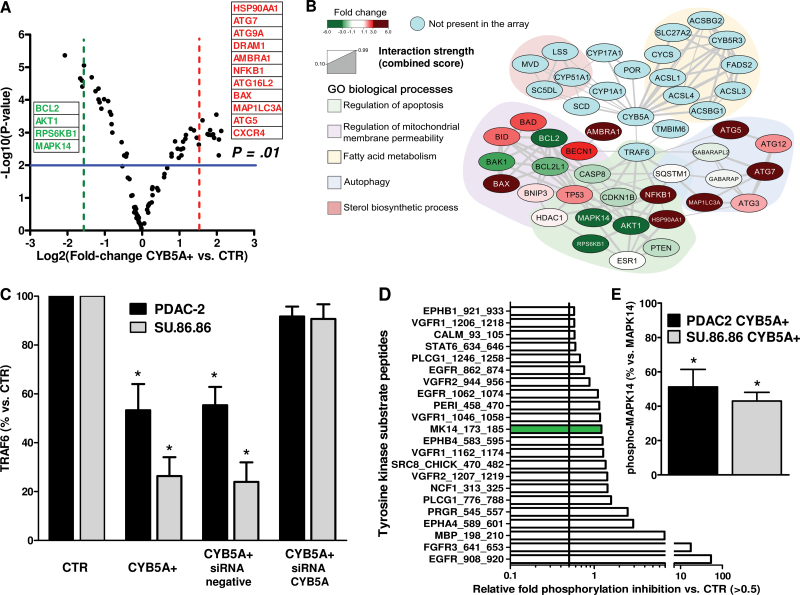
To shed light on the molecular events driving autophagy, we evaluated 84 genes encoding key components of the autophagic machinery. The volcano plot in Figure 3A arranged the genes along differential regulation (up- or downregulation, x-axis) and statistical significance (y-axis) of genes in CYB5A+ compared with CTR cells. Sorting for threefold change cutoff, this analysis identified 11 relevantly upregulated genes, including the ATG genes ATG5, ATG7, ATG9A, and ATG16L2, the ATG8 homolog MAPLC3A, the proautophagic gene AMBRA1, and the coregulators of autophagy and apoptosis BAX and DRAM1 (18). However, we also observed a marked increase of HSP90AA1; NFKB1, which is frequently overexpressed in PDAC (19); and CXCR4, which has been linked to PDAC metastasis (20).
Figure 3.
Key pathways involved in the activity of CYB5A. A) Volcano plot showing the results of the polymerase chain reaction autophagy array. P values were calculated with two-sided Student t test. B) Network analysis of CYB5A interactions retrieved from STRING database using low stringency settings and a number of maximum interactors for each node of 100. Nodes are colored according to the expression fold change. Genes were clustered using the ClusterOne Cytoscape plug-in, and each subnetwork was searched for statistically significant association with gene ontologies biological process categories using a two-sided hypergeometric test with Benjamini−Hochberg correction for multiple comparison. C) Densitometric analysis of immunoblots for TRAF6 protein expression in the PDAC-2 and SU.86.86 cells (CTR and CYB5A+) after transfection with a specific small interfering RNA (siRNA) for CYB5A (representative immunoblots are shown in Supplementary Figure 4A, available online). P values were calculated with two-sided Student t test. D) Relevantly inhibited kinases in the kinase array, with green bar for the MK14_173-185 peptide. E) Densitometric analysis of immunoblots for phospho-MAPK14 vs MAPK14 and in the PDAC-2 and SU.86.86 cells (CYB5A+ vs CTR). P values were calculated with two-sided Student t test. Columns indicate mean values obtained from three independent experiments. Bars indicate standard deviation. *P < .05.
The downregulated genes included the major antiapoptotic players BCL-2 and AKT1, as well as RPS6KB1, which repress BAD proapoptotic function (21). Another striking finding was the downregulation of MAPK14, which is one of the four p38-MAPKs involved in cascades of cellular responses evoked by proinflammatory cytokines or physical stress.
Based on these results, we initially built two separate networks; the first was composed of all possible interactions for CYB5A, whereas the second included interactions retrieved for the genes that had differential expression on the polymerase chain reaction array (Figure 3B). These analyses revealed that the physical interaction of CYB5A with TRAF6 (22) constitutes a molecular bridge for many signals, both upstream and downstream, including Akt and MAP-kinases, as well as for genes involved in cell death.
CYB5A+ cells had a statistically significant reduction of TRAF6 levels (−54% and −69% vs CTR, two-sided t-test P = .02, and P = .005 in PDAC-2 and SU.86.86 cells, respectively), which was restored in rescue experiments with CYB5A small interfering RNA (Figure 3C).
Moreover, we observed a reduction of the phosphorylation of MAPK14 peptide substrate, consistent with statistically significant reduction of phospho-MAPK14 (P < .01, two-sided t test) (Figure 3D–E), as well as a marked reduction of Src and EGFR substrates phosphorylation (in Y908/Y1062/Y962).
Effects of CYB5A In Vivo
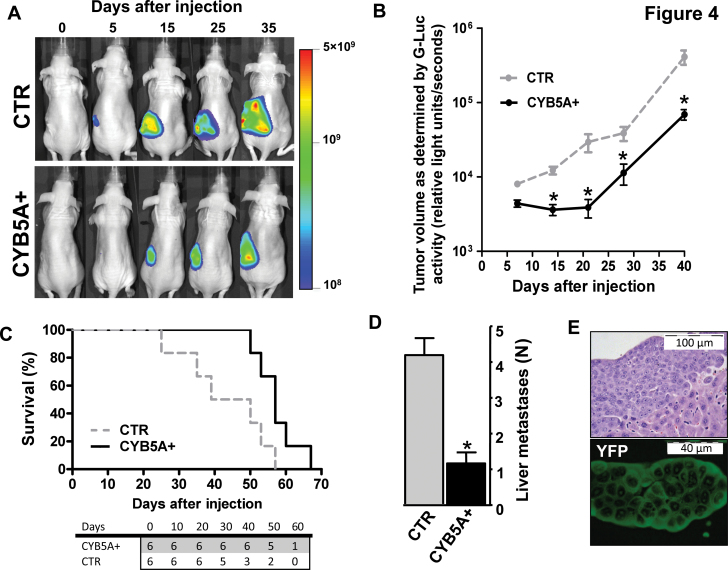
Figure 4A shows Fluc signal in mice injected with PDAC-2-Fluc/mCherry cells, indicating tumor engraftment and growth. This signal was statistically significantly lower in CYB5A+ mice (1.7×1010 vs 4.6×1010 photons/sec/cm2 at day 35; P = .006, two-sided t test). Magnetic resonance imaging confirmed the localization of tumor cells in the mouse pancreas, as well as retroperitoneal invasion, whereas high-frequency ultrasound enabled the assessment of vasculature networks (Supplementary Figure 7, A and B, and video, available online). Gluc signal further supported the reduced tumor burden due to CYB5A transduction (3.8×104 vs 6.9×104 relative light units/seconds at day-35; P = .01, two-sided t test) (Figure 4B).
Figure 4.
Effects of CYB5A in vivo. A) Five days after injection, primary tumors were detectable in all mice (ie, 100% take rate, without surgery-related mortality), and Fluc activity (proportional to the number of cancer cells, carrying Fluc/mCherry) increased over time. B) BLI signal of the pancreatic ductal adenocarcinoma patients (PDAC) tumors as detected in the blood samples with the Gluc (proportional to the number of cancer cells, carrying Gluc/CFP). P values were calculated with two-sided Student t test. C) Survival curves in the two groups of mice. Statistically significant differences were determined by two-sided log-rank test. The number of mice at risk in each group is given below the graph. D) Number of liver metastases in the two groups of mice. All mice were also evaluated for the presence of metastatic localization in other organs, and lung metastases were identified in two of six animals of the control (CTR) group. P values were calculated with two-sided Student t test. E) Immunofluorescence image of a representative section showing a liver metastasis in a mouse from the CYB5A+ group (YFP-positive cells). Points or columns indicate mean values obtained from the analysis of the six mice in each group. Bars indicate standard deviation. *P < .05.
Fluorescence microscopy identified YFP-marked CYB5A+ cells in pancreatic cross-sections (data not shown). The differential expression of CYB5A was confirmed by immunohistochemistry, which also detected its variability in three other models of orthotopic PDACs (Supplementary Figure 7C, available online).
Survival analysis demonstrated that mice of the CYB5A+ group lived longer (median OS = 57 days, 95% CI = 52 to 61; vs. median OS = 44 days, 95% CI = 21 to 57; P = .03, two-sided log-rank test; HR = 0.4, 95% CI = 0.1 to 0.8) (Figure 4C). Numerous macroscopic metastases were observed in all of the livers of CTR mice (Figure 4, D and E), whereas no liver metastases were detected in 33% of CYB5A+ mice.
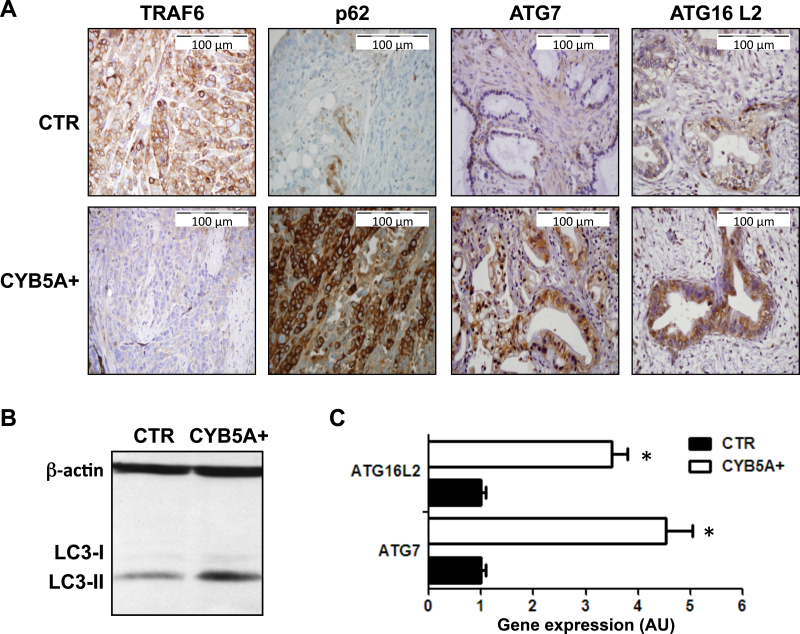
Remarkably, PDACs in the CYB5A+ group demonstrated reduced expression of TRAF6 associated with increased expression of its turnover regulator p62 (23) and of the autophagic markers LC3-II, ATG7, and ATG16L2 (Figure 5, A–C).
Figure 5.
Effects of CYB5A overexpression on autophagy induction in vivo. A) Representative IHC images showing a weak staining for TRAF6 and strong staining for ATG7, ATG16L2, and p62 in PDAC-2-CYB5A+ mice, compared with strong staining for TRAF6 and weak/intermediate staining for ATG7, ATG16L2, and p62 in PDAC-2 control (CTR) mice. B) Immunoblot showing the overexpression of LC3-II in lysates from frozen tissues from the PDAC-2-CYB5A+ mice compared with the PDAC-2 CTR mice. C) Polymerase chain reaction results (arbitrary units [AUs] calculated with the 2-ΔΔCt method) showing the increased expression of the autophagy genes ATG7 and ATG16L2 in lysates from frozen tissues from the PDAC-2-CYB5A+ mice compared with the PDAC-2 CTR mice. P values were calculated with two-sided Student t test. Columns indicate mean values obtained from the analysis of the six mice in each group. Bars indicate standard deviation. *P < .05.
Discussion
Our results support a role for CYB5A as a novel prognostic factor in PDAC and unravel its inhibitory activity of oncogenic phenotypes through modulation of autophagic, apoptotic, and invasive processes.
Because the high rates of genetic alterations and chromosomal instability in core signaling pathways are a major cause for the intrinsic aggressiveness of PDAC (6,24), we recently performed a genome-wide screening supporting the prognostic role of the cytoband 18q22.3 (7). In this study we further explored the clinical relevance of this discovery and reported the key role of CYB5A in both radically resected and metastatic PDAC patients.
In agreement with previous data (25), our multivariable analyses showed the association of grading to patient outcome. However, in a validated PDAC nomogram, pathologic features contribute less than 10% to survival predictions (26), and identification of new prognostic factors appears to be critical to improve the clinical management of PDAC (27). Moreover, prognostic biomarkers provide mechanistic insights into cancer progression and might identify novel molecular targets (28).
Bench-to-bedside research on hundreds of samples improved prognostic capabilities in several tumor types, such as breast cancer (29). Similar studies are difficult in PDACs, which are characterized by high nuclease activity and small amount of tissue. Most candidate biomarkers are based solely on mRNA evidence from tissues that were not microdissected to separate cancer stroma (30). Molecules with both mRNA and protein evidence in independent cohorts of patients are high-priority candidates, and our findings on CYB5A prompt prospective studies for further validation.
Microarray analyses revealed CYB5A downregulation compared with paired and unpaired normal pancreatic tissues (31–34), which is in agreement with the lower expression levels of CYB5A in all of our PDAC cells compared with normal cells. Moreover, a recent study identified CYB5A among genes that had lower copy number in lung squamous cell carcinomas of patients who had recurrence (35), but it is unclear which exact mechanism is responsible for the difference in clinical outcome.
CYB5A is a membrane-bound cytochrome that reduces methemoglobin to ferrous haemoglobin, and defects in this gene cause type IV methemoglobinemia (36). Additionally, it functions as an electron carrier for several oxygenases and plays a key role in the reductive detoxification of arylhydroxylamines (37).
Here for the first time we compared the behaviour of gain- and loss-of-function models of PDAC. In these models, CYB5A upregulation suppressed proliferation and migration/invasion, which were restored by silencing. Moreover, mice with CYB5A+ retrovirus-transduced PDACs had statistically significant increased survival and decreased tumor dimension and metastatic spread.
Studies on molecular basis of these reduced proliferation and invasive/metastatic abilities showed a statistically significant increase in both early and late apoptosis, accompanied by marked accumulation of autophagic vacuoles. The role of autophagy in PDAC remains to be elucidated (38). Several studies suggest that autophagy promotes tumorigenesis and protects cancers (39,40). Other lines of investigation showed that autophagy is detrimental to tumor cells, demonstrating the induction of VMP1-mediated autophagy in response to gemcitabine in PANC-1 and MIA PaCa-2 cells (41). Similarly, sulforaphane caused both autophagy and apoptosis in several PDAC-derived cells (42).
In our in vitro and in vivo models, activation of proautophagic/apoptotic pathways was coupled with a statistically significant upregulation of several ATG genes. Interestingly, we also observed a downregulation in expression and phosphorylation of MAPK14. Networks analysis and modulation of TRAF6 in cells and tumors with CYB5A transduction (or silencing) suggested its pivotal role in CYB5A multipathway regulation. TRAF6 is a member of the TRAF family with unique binding activities that result in signaling responses from interleukin 1 receptor (IL-1R), toll-like receptor (TLR), CD40, and RANK (43). TRAF6 associates directly with CD40 and RANK and indirectly with IL-1R/TLR through IRAK1 and IRAK2. This leads to activation of MAP-kinase signaling through downstream association with TGF-beta activating kinase 1 (TAK1)–binding protein complex (44). Additionally, TRAF6 modulates Src kinases leading to Akt activation (45). This cross-talk might explain our data on apoptosis induction and inhibition of tumor invasion/metastasis, in agreement with previous results in PDAC cells and orthotopic models (46). Of note, Src family members are commonly activated by growth factors such as EGF, with the formation of a complex where Src phosphorylates the associated receptor enhancing its activity (47). This could determine the downregulation of Src and EGFR, which are associated with reduced proliferation in our CYB5A+ models.
Limitations of this study include the fact that CYB5A was tested retrospectively in PDAC tissues and on a small number of primary cells. Further studies in prospective series and additional models are needed. However, our findings raise also the possibility that strategies aimed at restoring CYB5A activity, through gene therapy that replaces the function of this gene or targeted therapy inhibiting its deregulated downstream TRAF6, may constitute a novel strategy that favors cancer cell death while preventing potentially deleterious cross-talk between EGFR, Akt, and Src pathways in specific subgroups of PDAC patients. On the front of cancer therapeutics, TRAF6 has been implicated as an oncogene in lung cancer and as a target in multiple myeloma (48,49). TRAF2, which shares many functional similarities with TRAF6, is overexpressed in pancreatic tumors and cells, promotes invasiveness, and protects from CD95-mediated apoptosis (50). Furthermore, silencing or inhibition of TAK1 emerged as a potential approach to counteract antiapoptotic signaling pathways and PDAC chemoresistance (19). Future studies on inhibitors of TRAF6, such as cell-permeable TRAF6 decoy peptides that can target the TRAF6/binding peptide interaction (43), are warranted.
In conclusion, our clinical data, together with our in vitro and in vivo findings, strongly suggest that PDACs are more aggressive if they have low expression of CYB5A and deregulation of its downstream pathways, which therefore represent promising new tools for prognostic and therapeutic purposes.
Funding
This work was partially supported by grants from Netherlands Organization for Scientific Research ; Veni grant (to EG); Marie Curie International Fellowship (to EG); CCA Foundation 2012 (to EG, AA, GJP); Italian Minister of Research; PRIN-2009 (to UB, NF, EG); Istituto Toscano Tumori (to UB, NF, EV, EG), and National Institutes of Health/National Cancer Institute intramural research funds. Funding for continued development and maintenance of Cytoscape is provided by the U.S. National Institute of General Medical Sciences (GM070743). Cytoscape user support, education and new initiatives are supported by the National Resource for Network Biology (P41 RR031228 and GM103504). Further information is available at http://www.cytoscape.org/.
E. Giovannetti and Q. Wang contributed equally to this study.
The authors have no conflict of interest to disclose. The study sponsors had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
This work was presented previously in abstract form: Giovannetti E, Wang Q, Avan A, et al. Unraveling the role of CYB5A in pancreatic ductal adenocarcinoma (PDAC): correlation with clinical outcome and functional characterization in the modulation of autophagy and oncogenic phenotypes. 104th Annual Meeting American Association Cancer Research (AACR), Washington, DC, 2013, abstract 1143.
We would like to thank Dr Franco Fedeli (Menarini, Florence, Italy), Ing Henk Dekker (VUmc, Amsterdam, The Netherlands), Dr Olaf Van Tellingen (Netherlands Cancer Institute, Amsterdam, The Netherlands), Ing Pien Delis-van Diemen (VUmc, Amsterdam, The Netherlands), Dr Dieter Fuchs (VisualSonics, Amsterdam, The Netherlands), and Dr Elena Galvani and Dr Davide Chiasserini (VUmc, Amsterdam, The Netherlands) for the valuable technical support with the D-Sight acquisition system, PamChip arrays, MR scanning, 3DHistech TMA grand master, Vevo-2100 high-frequency ultrasound, and networks analysis, respectively.
References
- 1. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29 [DOI] [PubMed] [Google Scholar]
- 2. Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet. 2011;378(9791):607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hidalgo M, Von Hoff DD. Translational therapeutic opportunities in ductal adenocarcinoma of the pancreas. Clin Cancer Res. 2012;18(16):4249–4256 [DOI] [PubMed] [Google Scholar]
- 4. Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Olive KP, Jacobetz MA, Davidson CJ, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324(5933):1457–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Biankin AV, Waddell N, Kassahn KS, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491(7424):399–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee JH, Giovannetti E, Hwang JH, et al. Loss of 18q22.3 involving the carboxypeptidase of glutamate-like gene is associated with poor prognosis in resected pancreatic cancer. Clin Cancer Res. 2012;18(2):524–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang P, Chan DW, Zhu Y, et al. Identification of carboxypeptidase of glutamate like-B as a candidate suppressor in cell growth and metastasis in human hepatocellular carcinoma. Clin Cancer Res. 2006;12(22):6617–6625 [DOI] [PubMed] [Google Scholar]
- 9. Giovannetti E, Funel N, Peters GJ, et al. MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 2010;70(11):4528–4538 [DOI] [PubMed] [Google Scholar]
- 10. Funel N, Vasile E, Del Chiaro M, et al. Correlation of basal EGFR expression with pancreatic cancer grading but not with clinical outcome after gemcitabine-based treatment. Ann Oncol. 2011;22(2):482–484 [DOI] [PubMed] [Google Scholar]
- 11. Martino L, Vasile E, Del Chiaro M, et al. Palmitate activates autophagy in INS-1E β-cells and in isolated rat and human pancreatic islets. PLoS One. 2012;7(5):e36188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Giovannetti E, Labots M, Dekker H, et al. Molecular mechanisms and modulation of key pathways underlying the synergistic interaction of sorafenib with erlotinib in non-small-cell-lung cancer (NSCLC) cells. Curr Pharm Des. 2013;19(5):927–939 [PubMed] [Google Scholar]
- 13. von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31(1):258–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nepusz T, Yu H, Paccanaro A. Detecting overlapping protein complexes in protein-protein interaction networks. Nat Methods. 2012;9(5):471–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21(16):3448–3449 [DOI] [PubMed] [Google Scholar]
- 16. Wurdinger T, Badr C, Pike L, et al. A secreted luciferase for ex vivo monitoring of in vivo processes. Nat Methods. 2008;5(2):171–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Avan A, Caretti V, Funel N, et al. Crizotinib inhibits metabolic inactivation of gemcitabine in c-Met-driven pancreatic carcinoma [published online ahead of print October 1, 2013]. Cancer Res. 10.1158/0008-5472.CAN-13–0837 [DOI] [PubMed] [Google Scholar]
- 18. Xie X, Le L, Fan Y, Lv L, Zhang J. Autophagy is induced through the ROS-TP53-DRAM1 pathway in response to mitochondrial protein synthesis inhibition. Autophagy. 2012;8(7):1071–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Melisi D, Xia Q, Paradiso G, et al. Modulation of pancreatic cancer chemoresistance by inhibition of TAK1. J Natl Cancer Inst. 2011;103(15):1190–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marchesi F, Monti P, Leone BE, et al. Increased survival, proliferation, and migration in metastatic human pancreatic tumor cells expressing functional CXCR4. Cancer Res. 2004;64(22):8420–8427 [DOI] [PubMed] [Google Scholar]
- 21. Scholl C, Fröhling S, Dunn IF, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137(5):821–834 [DOI] [PubMed] [Google Scholar]
- 22. Ewing RM, Chu P, Elisma F, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ling J, Kang Y, Zhao R, et al. KrasG12D-induced IKK2/β/NF-κB activation by IL-1α and p62 feedforward loops is required for development of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21(1):105–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yachida S, Xia Q, Paradiso G, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467(7319):1114–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yeo CJ, Cameron JL, Sohn TA, et al. Six hundred fifty consecutive pancreaticoduodenectomies in the 1990s: pathology, complications, and outcomes. Ann Surg. 1997;226(3):248–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brennan MF, Kattan MW, Klimstra D, Conlon K. Prognostic nomogram for patients undergoing resection for adenocarcinoma of the pancreas. Ann Surg. 2004;240(2):293–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Frampton AE, Krell J, Giovannetti E, et al. Defining a prognostic molecular profile for ductal adenocarcinoma of the pancreas highlights known key signaling pathways. Expert Rev Anticancer Ther. 2012;12(10):1275–1278 [DOI] [PubMed] [Google Scholar]
- 28. Kelloff GJ, Sigman CC. Cancer biomarkers: selecting the right drug for the right patient. Nat Rev Drug Discov. 2012;11(3):201–214 [DOI] [PubMed] [Google Scholar]
- 29. van de Vijver MJ, He YD, van’t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009 [DOI] [PubMed] [Google Scholar]
- 30. Winter JM, Tang LH, Klimstra DS, et al. A novel survival-based tissue microarray of pancreatic cancer validates MUC1 and mesothelin as biomarkers. PLoS One. 2012;7(7):e40157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pei H, Li L, Fridley BL, et al. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell. 2009;16(3):259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Badea L, Herlea V, Dima SO, Dumitrascu T, Popescu I. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology. 2008;55(88):2016–2027 [PubMed] [Google Scholar]
- 33. Balagurunathan Y, Morse DL, Hostetter G, et al. Gene expression profiling-based identification of cell-surface targets for developing multimeric ligands in pancreatic cancer. Mol Cancer Ther. 2008;7(9):3071–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Iacobuzio-Donahue CA, Maitra A, Olsen M, et al. Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am J Pathol. 2003;162(4):1151–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sriram KB, Larsen JE, Savarimuthu Francis SM, et al. Array-comparative genomic hybridization reveals loss of SOCS6 is associated with poor prognosis in primary lung squamous cell carcinoma. PLoS One. 2012;7(2):e30398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hegesh E, Hegesh J, Kaftory A. Congenital methemoglobinemia with a deficiency of cytochrome b5. New Engl J Med. 1986; 314(12):757–761 [DOI] [PubMed] [Google Scholar]
- 37. Kurian JR, Chin NA, Longlais BJ, Hayes KL, Trepanier LA. Reductive detoxification of arylhydroxylamine carcinogens by human NADH cytochrome b5 reductase and cytochrome b5. Chem Res Toxicol. 2006; 19(10):1366–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ropolo A, Bagnes CI, Molejon MI, et al. Chemotherapy and autophagy-mediated cell death in pancreatic cancer cells. Pancreatology. 2012;12(1):1–7 [DOI] [PubMed] [Google Scholar]
- 39. Kang R, Tang D, Lotze MT, Zeh HJ., 3rd AGER/RAGE-mediated autophagy promotes pancreatic tumorigenesis and bioenergetics through the IL6-pSTAT3 pathway. Autophagy. 2012;8(6):989–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Monma H, Harashima N, Inao T, Okano S, Tajima Y, Harada M. The HSP70 and autophagy inhibitor pifithrin-μ enhances the antitumor effects of TRAIL on human pancreatic cancer. Mol Cancer Ther. 2013;12(4):341–351 [DOI] [PubMed] [Google Scholar]
- 41. Pardo R, Lo Ré A, Archange C, et al. Gemcitabine induces the VMP1-mediated autophagy pathway to promote apoptotic death in human pancreatic cancer cells. Pancreatology. 2010;10(1):19–26 [DOI] [PubMed] [Google Scholar]
- 42. Naumann P, Fortunato F, Zentgraf H, Büchler MW, Herr I, Werner J. Autophagy and cell death signaling following dietary sulforaphane act independently of each other and require oxidative stress in pancreatic cancer. Int J Oncol. 2011;39(1):101–109 [DOI] [PubMed] [Google Scholar]
- 43. Ye H, Arron JR, Lamothe B, et al. Distinct molecular mechanism for initiating TRAF6 signalling. Nature. 2002;418(6896):443–447 [DOI] [PubMed] [Google Scholar]
- 44. Sorrentino A, Thakur N, Grimsby S, et al. The type I TGF-beta receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat Cell Biol. 2008;10(10):1199–1207 [DOI] [PubMed] [Google Scholar]
- 45. Wong BR, Besser D, Kim N, et al. TRANCE, a TNF family member, activates Akt/PKB through a signaling complex involving TRAF6 and c-Src. Mol Cell. 1999;4(6):1041–1049 [DOI] [PubMed] [Google Scholar]
- 46. Kelber JA, Reno T, Kaushal S, et al. KRas induces a Src/PEAK1/ErbB2 kinase amplification loop that drives metastatic growth and therapy resistance in pancreatic cancer. Cancer Res. 2012;72(10):2554–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bao J, Gur G, Yarden Y. Src promotes destruction of c-Cbl: implications for oncogenic synergy between Src and growth factor receptors. Proc Natl Acad Sci U S A. 2003;100(5):2438–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Starczynowski DT, Lockwood WW, Deléhouzée S, et al. TRAF6 is an amplified oncogene bridging the RAS and NF-κB pathways in human lung cancer. J Clin Invest. 2011;121(10):4095–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu H, Tamashiro S, Baritaki S, et al. TRAF6 Activation in multiple myeloma: a potential therapeutic target. Clin Lymphoma Myeloma Leuk. 2012;12(3):155–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Trauzold A, Röder C, Sipos B, et al. CD95 and TRAF2 promote invasiveness of pancreatic cancer cells. FASEB J. 2005;19(6):620–622 [DOI] [PubMed] [Google Scholar]