Abstract
Jumonji domain-containing proteins (JMJD) catalyze the oxidative demethylation of a methylated lysine residue of histones by using O2, α-ketoglutarate, vitamin C, and Fe(II). Several JMJDs are induced by hypoxic stress to compensate their presumed reduction in catalytic activity under hypoxia. In this study, we showed that an H3K27me3 specific histone demethylase, JMJD3 was induced by hypoxia-inducible factor (HIF)-1α/β under hypoxia and that treatment with Clioquinol, a HIF-1α activator, increased JMJD3 expression even under normoxia. Chromatin immunoprecipitation (ChIP) analyses showed that both HIF-1α and its dimerization partner HIF-1β/Arnt occupied the first intron region of the mouse JMJD3 gene, whereas the HIF-1α/β heterodimer bound to the upstream region of the human JMJD3, indicating that human and mouse JMJD3 have hypoxia-responsive regulatory regions in different locations. This study shows that both mouse and human JMJD3 are induced by HIF-1.
Keywords: Arnt, JMJD3, HIF-1α, histone demethylase, hypoxia
INTRODUCTION
In response to hypoxia, cells express several genes to maintain homeostasis. More than 500 genes, including vascular endothelial growth factor (VEGF), erythropoietin (EPO), phosphoglycerate kinase-1 (PGK-1), glucose transporter-1 (glut-1), and BCL-2/adenovirus E1B 19 kDa interacting protein 3 (Bnip3) are induced under hypoxia in MCF7 cells (Elvidge et al., 2006). These diverse target genes are transcribed by a common transactivator, the hypoxia-inducible factor 1-α/β (HIF-1α/β) heterodimer (Semenza, 2012). Under normoxic conditions, HIF-1α is ubiquitinated and rapidly degraded whereas HIF-1β is constitutively expressed. HIF-1β is identical to the Aryl hydrocarbon receptor nuclear translocator (Arnt) that is a dimerization partner protein of the dioxin receptor, Aryl hydrocarbon receptor. The 403rd and 564th proline residues of human HIF-1α are hydroxylated by prolyl hydroxylase domain protein (PHD) (Bruick and McKnight, 2001). The hydroxylated prolines are recognized by the E3 ubiquitin ligase von Hippel-Lindau protein (pVHL), which triggers the ubiquitination-dependent degradation of HIF-1α (Maxwell et al., 1999). The 803rd asparagine residue of HIF-1α is also hydroxylated by an oxygen-dependent asparagine hydroxylase, referred to as Factor-Inhibiting HIF-1 (FIH-1). The hydroxylated asparagine residue hinders the recruitment of the CREB-binding protein (CBP)/p300 coactivator. PHD and FIH-1 require molecular oxygen, α-ketoglutarate, vitamin C, and Fe(II) for their catalytic activities. The trans-active form of HIF-1α is stabilized under hypoxia because the activities of these two oxygen-dependent hydroxylases decrease (Hewitson et al., 2002).
FIH-1 has unique structural and catalytic properties compared to PHD (Hewitson et al., 2007). The catalytic region of FIH-1 (145–340 amino acids in human FIH-1) shows structural similarity to the Jumonji (Jmj) domain, which is often found in histone demethylases (Chen et al., 2006; Lee et al., 2003). JmjC domain containing proteins (JMJDs) catalyze the oxidative demethylation of a methylated lysine residue of histones. Although JMJDs and FIH-1 hydroxylate different target proteins, both utilize O2, α-ketoglutarate, vitamin C, and Fe(II) and produce succinate and CO2.
More than 100 JMJDs have been identified, and most of them have demethylase activity with different substrate specificities (Shi and Whetstine, 2007). JMJD1A/JHDM2A/KDM3A demethylate H3K9me2 and me1, JMJD2 isozymes demethylate H3K9me3, Jarid1 isozymes demethylate H3K4me3, and JMJD3/KDM6B demethylate H3K27me3. Histone methylation affects gene expression in distinct localized patterns. The tri-methylation of H3K4 and acetylation of H3 in the promoter region are associated with actively transcribed genes, whereas the methylation of H3K9 and H3K27 in the promoter region are associated with inactive genes (Kooistra and Helin, 2012). The fact that JmjC proteins need molecular oxygen for their catalytic activities suggests that their activities can be inhibited under hypoxic conditions. Recent findings that hypoxia induced several JmjC proteins imply that their induction compensates their reduced catalytic activity under hypoxia (Beyer et al., 2008; Pollard et al., 2008; Wellmann et al., 2008; Xia et al., 2009). Thus, in response to hypoxic stress, histone methylation of individual genes can be dynamically regulated by the net change in the activity and expression of the JmjC proteins.
JMJD2s, JMJD1A, and Jarid1B have been identified as HIF-1α target genes (Beyer et al., 2008; Pollard et al., 2008; Xia et al., 2009). This study first demonstrated that hypoxia also induces an H3K27me3 demethylase, JMJD3, by a HIF-1α dependent mechanism. Together with the recent findings that JMJD3 is involved in inflammation, cardiac development, neural development, and senescence, this study suggests that hypoxia can influence these diverse biological processes by changing the expression level of JMJD3.
MATERIALS AND METHODS
Cell culture, reagents, and antibodies
Human adipose-derived stem cells (hADSC, R7788-115), which are isolated from human adipose tissue collected during liposuction procedures, were purchased from Life technology (USA). Cells were plated in a 100-mm plate at a density of 5,000 cells/cm2 in DMEM supplemented with 10% mesenchymal stem cell (MSC)-qualified fetal bovine serum (FBS). Wild-type mouse embryonic fibroblasts (MEFs) were prepared from 13.5 post-coitum (p.c.) embryos of C57BL/6 background and maintained in DMEM supplemented with 10% FBS by using the 3T3 protocol until reaching passage 7. The other mouse cells were maintained as described previously (Choi et al., 2008; Park et al., 2013). For hypoxic treatments, cells were cultured in an anaerobic incubator (< 0.5% O2, Model 1029, Forma Scientific, Inc.) or in InVivo2 200 hypoxia workstation (3% O2, Ruskin). Deferoxamine (DFN), cobalt chloride (CoCl2), and Clioquinol (CQ) were purchased from Sigma-Aldrich (USA). Antibodies against mouse and human JMJD3 were purchased from Abgent (USA) and Abcam (UK), respectively. Anti-mouse and human HIF-1α antibodies were purchased from Novus Biologicals (Littleton, CO) and BD Biosciences (USA), respectively. Anti-HSP70 and anti-β-Actin antibodies were obtained from Enzo Life Sciences (USA) and Sigma-Aldrich, respectively. Anti-Arnt and anti-H3K4 me3 antibodies were obtained from Cell Signaling Technology (USA). Antibodies against H3K9me3, H3K27me3, H3K9/K14Ac, and 14-3-3γ were purchased from Millipore (USA).
RNA isolation and quantitative real-time PCR
Total RNA was isolated using an RNeasy spin column according to the manufacturer’s instructions (Qiagen Inc., USA). cDNA was reverse transcribed from total RNA (2 μg) using M-MLV reverse transcriptase (Promega, USA) with dNTPs and random primers. The qRT-PCR analyses were performed by using the Power SYBR Green PCR Master Mix and ABI 7000 Real-Time PCR System (Applied Biosystems) for MEFs experiments and all hADSC experiments. The iQ™ SYBR Green Supermix and MyiQ single color real-time PCR detection system (Bio-Rad, USA) were used for the analyses of the RNA from the other mouse cells including 3T3-L1 cells, NIH-3T3 cells, Hepa1c1c7 cells, and BpRC1 cells. The primer sets for JMJD3 were following: mJMJD3, (F) 5′-CCC CCA TTT CAG CTG ACT AA-3′, (R) 5′-CTG GAC CAA GGG GTG TGT T-3′, for Figs. 1A, 1B, 1D and 2B (De Santa et al., 2007); mJMJD3, (F) 5′-CCT GCA GTC AAT GAA GCA CTG-3′, (R) 5′-CTC CAC GTC GCA TTC GTT G-3′, for Figs. 1C and 1G; hJMJD3, (F) 5′-GGA GGC CAC ACG CTG CTA C-3′, (R) 5′-GCC AGT ATG AAA GTT CCA GAG CTG-3′. The primer sets for other genes were previously described (Choi et al., 2008; Farrall and Whitelaw, 2009).
Fig. 1.

HIF-1α-dependent hypoxic induction of the mouse JMJD3 gene. (A–C) NIH-3T3 cells, and MEF were incubated under hypoxia (< 0.5% O2) for 48 h. 3T3-L1 cells were incubated under hypoxia (< 0.5% O2) for 36 h. The relative mRNA levels of BNIP3 and JMJD3 were analyzed by qRTPCR and normalized by 18S ribosomal RNA. The mean value of qRT-PCR from normoxic cells was set to 1. The data represent the average and standard deviations of triplicate measurements from qPCR. (D, E) HIF-1α knockdown 3T3-L1 cells and control 3T3-L1 cells were treated with hypoxia for 16 h; (D) qRT-PCR was performed using the primer sets for the indicated mouse genes; (E) Western blot analyses were performed by using the indicated antibodies and lysates from hypoxia treated mouse 3T3-L1 cells. HSP70 and β-Actin protein were used as the loading control. The results of two independent experiments were shown. The numbers indicated the relative intensity of each band which was measured using LAS-3000 luminescent image analyzer and Multi Gauge ver.3.0 imaging software. (F, G) 3T3-L1 cells were treated with DFN (100 μM), CoCl2 (200 μM), or hypoxia for 48 h; (F) Western blot analyses were performed using the indicated antibodies; (G) The RT-PCR analyses of JMJD3 and BNIP3 are shown. The 18S ribosomal RNA was used as the loading control. N, normoxia; H, hypoxia (< 0.5% O2). The p-values were obtained using the Student’s t-test and the significance between the groups is indicated (**p < 0.05, *p < 0.1).
Fig. 2.

Arnt-dependent hypoxic induction of the mouse JMJD3 gene. (A, B) Hepa1c1c7 cells and BpRc1 cells were incubated under hypoxia (< 0.5% O2) for 16 h; (A) Western blot analyses were performed by using the indicated antibodies. The 14-3-3γ protein was used as the loading control; (B) The relative mRNA levels of mouse JMJD3 were analyzed by qRT-PCR. (C) The putative HREs in the mouse JMJD3 gene are identified by using the MatInspector program (www.genomatix.de) (Cartharius, Frech et al., 2005). The bars indicate the positions of the primer sets used for the ChIP analyses. The nucleotide sequences of putative HRE sites (−0.2 Kb, +3.9 Kb, +4.2 Kb) and flanking regions were shown (lower panel). (D) 3T3-L1 cells were incubated under hypoxia (< 0.5% O2) for 16 h. ChIP analyses were performed with the indicated antibodies and primer sets of mouse JMJD3 and mouse Stra13 genes. Input values were obtained from samples treated in the same way as the experimental method, except that no immunoprecipitation steps were performed. The values on the y-axis are presented as the percentage input of the average and standard deviation of triplicate determinations of qPCR. (E) MEF were incubated under < 0.5% O2, 3% O2, and 21% O2 (normoxia) for 48 h. ChIP analyses (upper panel) and Western blot analyses (lower panel) were performed using the indicated antibodies. N, normoxia; H, hypoxia (< 0.5% O2). The p-values were obtained using the Student’s t-test and the significance between the groups is indicated (**p < 0.05, *p < 0.1).
Chromatin Immunoprecipitation
ChIP analyses were performed as described previously with the following modifications (Gao et al., 2011; Park and Park, 2010). Briefly, hADSC, 3T3-L1, and MEF cells were cross-linked in 1% formaldehyde at 37°C for 10 min, and then the nuclei were isolated (Kouskouti et al., 2004). The isolated nuclei of hADSC and 3T3-L1 cells were treated with MNase (300 gel units) for 15 min, and sonicated to shear the chromatin (Woon Kim et al., 2011). The insoluble materials were removed by centrifugation at 10,000 × g for 10 min. We measured the DNA concentration of the supernatant containing soluble chromatin to ensure that the amount of chromatin used in each sample was similar. For each immunoprecipitation, 30–40 μg of soluble chromatin was 10-fold diluted with ChIP dilution buffer [20 mM Tris-HCl (pH 8.0), 0.01% SDS, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl]. The diluted lysates of soluble chromatin (100 μl) were analyzed using qPCR and the resulting quantitative values were used as input. The diluted lysates (1 ml) were immunoprecipitated with 2–4 μg of the indicated antibodies at 4°C overnight. The antibody-chromatin complexes were recovered by incubation with 20 μl of protein G-agarose (50% slurry) and sequentially washed as described previously (Park and Park, 2010). The immuno-complexes were eluted with 150 μl of elution buffer (1% SDS, 0.1 M NaHCO3) and reversed. Target DNA from 3 μl sample were analyzed by PCR. The ChIP primer sets for putative HREs located at upstream and first intron of the JMJD3 gene are following ; mJMJD3 −0.2 kb, (F) 5′-TGG GCC TGA GCC AGA ATA-3′, (R) 5′-AGC AGC CTG GTT CAG GTA GTA A-3′; mJMJD3 +4.2 kb, (F) 5′-TGC GCA AGT AGC CAC GTC A-3′, (R) 5′-TCC TGT ATC CTC TGT GCC CTT T-3′; hJMJD3 −4.1 kb, (F) 5′-ACG TGC ACT CCC ACA CGC T-3′, (R) 5′-CAC AGA CTC CAC GAG GGG AG-3′; hJMJD3 −3.3 kb, (F) 5′-GGG GAT CCT GCA GAT GGA AG-3′, (R) 5′-ACA TTT CTG ACG GGA GTC GG-3′; hJMJD3 +1.9 kb, (F) 5′-AAG AAG CGA GCG TGT AAC CC-3′, (R) 5′-GAG CCC GCA GAT AGG GTT TT-3′; hJMJD3 +3.3 kb, (F) 5′-CTG ATG GAA AGG ATG CTG CC-3′, (R) 5′-ATG GAA GCG CTT GGA CCC TA-3′; hJMJD3 +4.1 kb, (F) 5′-CAT TTA CAC ACA CCG CTT CCG-3′, (R) 5′-TGG CTC GCG TAT CCT CTC T-3′. The ChIP primer sets for the other genes were described previously (Park and Park, 2012).
shRNA mediated knock-down
HIF-1α was knocked down using a retrovirus infection system, pSIREN-RetroQ (BD Biosciences), as described previously (Park and Park, 2010). The short hairpin RNA (shRNA) sequence against mouse HIF-1α was 5′-TGT GAG CTC ACA TCT TGA TT-3′.
RESULTS
Hypoxia induced JMJD3 in HIF-1α-dependent manner
We found that hypoxia increased the expression of JMJD3 in several types of cells including mouse fibroblast NIH3T3 cells, mouse preadipocytes 3T3-L1 cells, and MEFs (Figs. 1A–1C). In order to test whether hypoxia increased the expression of JMJD3 in HIF-1α-dependent manner, we used mouse 3T3-L1 cells infected with a retrovirus encoding shRNA against mouse HIF-1α. The qRT-PCR analyses confirmed that the mRNA levels of either HIF-1α or its target Bnip3 were reduced in the HIF-1α knockdown 3T3-L1 (shHIF-1α 3T3-L1) cells (Fig. 1D). Hypoxia failed to induce JMJD3 mRNA in shHIF-1α 3T3-L1 cells. Western analyses also confirmed that hypoxia stabilized HIF-1α protein in shControl 3T3-L1 cells but not in shHIF-1α 3T3-L1 cells, and that hypoxia failed to increase the levels of JMJD3 protein in shHIF-1α 3T3-L1 cells (Fig. 1E). HIF-1α protein was stabilized by nonhypoxic mimickers such as deferoxamine (DFN) and CoCl2 that inhibit the prolyl hydroxylation-dependent ubiquitination of HIF-1α (Fig. 1F). We also showed that not only hypoxia but also nonhypoxic HIF-1α activators induce JMJD3 (Fig. 1G).
Hypoxia induced JMJD3 in Arnt-dependent manner
The heterodimerization with Arnt is a prerequisite for HIF-1α to interact with the hypoxia-responsive elements (HRE). In order to test whether Arnt is also required for the hypoxic induction of JMJD3, we used BpRC1 cells which are Arnt-defective variants of mouse hepa1c1c7 cells (Ko et al., 1996). Western analyses revealed that Arnt protein is constitutively expressed in wild type hepa1c1c7 cells but not in BpRC1 cells (Fig. 2A). We found that hypoxia failed to induce the mRNA of JMJD3 in Arnt-defective hepa1c1c7 variant cells (Fig. 2B). These findings indicated that hypoxia induced JMJD3 in an HIF-1α/Arnt-dependent mechanism. In macrophages, transcription of mouse JMJD3 was driven by the promoter located in the first intron (De Santa et al., 2007). By using the MatInspector program, we found HRE consensus sequences in the upstream and the first intron of the mouse JMJD3 gene (Fig. 2C) (Cartharius et al., 2005). In order to test whether HIF-1 occupies HREs in the JMJD3 gene, we performed ChIP analyses using the HIF-1α antibody. We confirmed that HIF-1α binds to its target gene Stra13. According to the ChIP analyses, HIF-1α was detected in the first intron rather than in the upstream region of mouse JMJD3 gene in hypoxic 3T3-L1 cells (Fig. 2D). ChIP analyses using an antibody against the 9th and 14th acetylated residues of histone 3 (H3K9/K14Ac) revealed that hypoxia increases the transactive histone maker, H3K9/K14Ac, in both JMJD3 and Stra13 genes. We also found the HIF-1α occupancy on the first intron of JMJD3 in hypoxic MEFs. Additionally, we showed that 3% oxygen slightly increases HIF-1α occupancy on both JMJD3 and Stra13 (Fig. 2E).
Hypoxic induction of human JMJD3 in hADSC
We tested whether hypoxia induces JMJD3 expression in human cells. qRT-PCR analyses revealed that hypoxia increased the mRNA levels of JMJD3 in hADSC. Clioquinol, an HIF-1α activator, also induced JMJD3 in these cells (Fig. 3A). Previous findings showed that Clioquinol inhibits the asparaginyl hydroxylation activity of FIH-1 and also the prolyl hydroxylation-dependent ubiquitination of HIF-1α, thus Clioquinol stabilized the transactive form of HIF-1α protein even under normoxia (Choi et al., 2006). We confirmed that Clioquinol also increased both the mRNA and protein levels of human JMJD3 in hADSC (Fig. 3B). We also found several HREs located in both upstream and the first intron of the human JMJD3 gene (Fig. 3C) (Cartharius et al., 2005). Differently from mouse JMJD3, in hADSC, ChIP analyses revealed that both HIF-1α and Arnt bind HREs in the upstream region rather than on the first intron of human JMJD3 (Figs. 4A–4D). In order to detect any change of histone mark in the upstream region of human JMJD3, we performed ChIP analyses using antibodies detecting the methylation and acetylation of histone 3. We found that repressive histone marks such as H3K9me3 and H3K27me3 were maintained at undetectable low level in the upstream of the human JMJD3 gene, while hypoxia and Clioquinol increased the active histone marks H3K4me3 and H3K9/K14Ac (Figs. 4E–4F).
Fig. 3.
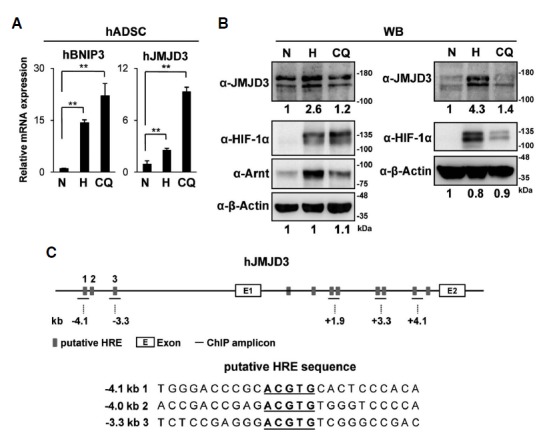
Hypoxic induction of human JMJD3 in hADSC. (A, B) hADSC were treated with hypoxia (<0.5% O2, 48 h) or Clioquinol (50 μM, 16 h); (A) The relative mRNA levels of BNIP3 and JMJD3 were analyzed by qRT-PCR. The data represent the average and standard deviations of two independent experiments; (B) Western blot analyses were performed using the indicated antibodies. The results of two independent experiments were shown. The relative intensity of each band was measured as described in Fig. 1E. (C) The putative HREs in the human JMJD3 gene are identified as described in Fig. 2C. The bars indicate the positions of the primer sets used for the ChIP analyses. The nucleotide sequences of putative HRE sites (−4.1 kb, −4.0 kb, −3.3 kb) and flanking regions were shown (lower panel). The p-values were obtained using the Student’s t-test and the significance between the groups is indicated (**p < 0.05).
Fig. 4.
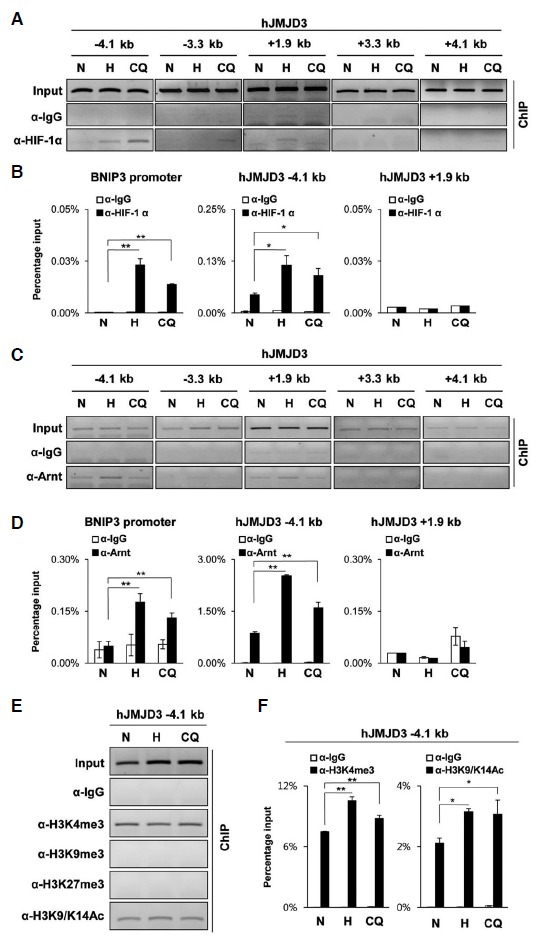
ChIP analyses of the human JMJD3 gene. hADSC were treated with hypoxia (< 0.5% O2, 48 h) or Clioquinol (50 μM, 16 h). (A–F) ChIP analyses were performed using the indicated antibodies and primer sets for the human JMJD3 gene; (A, C, and E) ChIP-eluted DNA was analyzed by PCR and visualized on 2% agarose gel electrophoresis. (B, D, and F) ChIP-eluted DNA was analyzed by qPCR. The values on the y-axis are presented as the percentage input of the average and standard deviation of triplicate determinations of qPCR. The primer set for the human BNIP3. promoter was used as the positive control. The p-values were obtained using the Student’s t-test and the significance between the groups is indicated (**p < 0.05, *p < 0.1).
DISCUSSION
In this study, we showed that hypoxia induced the expression of JMJD3 in an HIF-1α-dependent manner, and that the presence of Clioquinol, an HIF-1α activator, increased JMJD3 expression even under normoxia. ChIP analyses showed that both HIF-1α and HIF-1β/Arnt bound to the first intron region of the mouse JMJD3 gene, but to the upstream region of the human JMJD3, indicating that human and mouse JMJD3 have hypoxia regulatory regions in different locations. HIF-1α binding sites are often found far from the transcription start sites. In ARRDC3 and ERRFI1 genes, HREs are located around −100 kb from the transcription start sites and activate the transcription (Schodel et al., 2011). HIF-1α binds HREs in open chromatin region of which H3K4 residues are tri-methylated (Xia and Kung, 2009). Consistently, our results indicated that HIF-1α bound to chromatin region of human JMJD3 gene which has a high level of H3K4me3.
The fact that JMJD3 uses molecular oxygen as a substrate suggests that the catalytic activity of JMJD3 can be inhibited under hypoxia. Interestingly, many hydroxylases, including collagen-prolyl hydroxylase and the H3K9me2 demethylase JMJD1A, maintain their catalytic activities even under severe hypoxia, because their Km values for O2 are very low. The Km value for O2 associated with JMJD3 remains to be estimated. The facts that HIF-1 can be activated by nonhypoxic signals such as mTOR pathways, oxidative stresses, and inflammatory signals, and that HIF-1 increases JMJD3 expression suggest that some of the nonhypoxic HIF-1-activating stimuli can increase the amounts of JMJD3 without reducing its catalytic activity. The biological importance of JMJD3 primarily relies on the function of its target genes. Since histone modifiers such as histone methyltransferase and demethylases do not have the ability to recognize specific DNA sequences, the targets for histone modifiers depend on their interacting transcription factors.
The tri-methylation of H3K27 (H3K27me3) of a certain gene is determined by the balance between the Polycomb Repressive Complex 2 (PRC2) and histone demethylases JMJD3 and UTX (Agger et al., 2007). H3K27me3 and H3K9me3 on the promoter and enhancer region are generally associated with the transcriptional inactivation, whereas H3K4me3 and the acetylation of H3K27 are associated with the transcriptional activation. A subset of promoters marked with both H3K4me3 and H3K27me3 is also known as bivalent promoters (Bernstein et al., 2006). The bivalent promoters are inactive but able to drive the gene expression with the loss of H3K27me3 and gain of H3K27Ac.
De Santa et al. (2007) showed that in macrophages JMJD3 can be induced by inflammatory stimuli such as lipopolysaccharide (LPS) and interferon (IFN)-γ. JMJD3 is highly expressed in differentiating bone marrow cells, while it is repressed in terminally differentiated macrophages. Upon treat-ment with LPS or IFN-γ, activated nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) occupies the promoter region of JMJD3 and increases its transcription. LPS-induced JMJD3 activates a subset of genes by reducing the levels of the repressive histone mark H3K27me3. In macrophages, Bone morphogenetic protein 2 (Bmp-2) is one of the targets of JMJD3. The promoter of the Bmp-2 gene contains a bivalent chromatin domain with high levels of both H3K4me3 and H3K27me3. LPS-induced JMJD3 demethylates H3K27me3 of the bivalent Bmp-2 promoter to increase the transcription of Bmp-2.
The locus of inhibitor of cyclin-dependent Kinase 4a/Alternative Reading Frame (INK4a/ARF) has been identified as a target of JMJD3. Mouse Ink4a/Arf locus encodes p16INK4a, p15 INK4b, and p19ARF. Both p16Ink4a and p15Ink4b inhibit CDK4/6 kinases leading to the activation of retinoblastoma (Rb) protein, whereas p19Arf (p14Arf in human) inhibits mouse double minute 2 homolog (MDM2) leading to the activation of p53. Therefore, the activation of INK4a/ARF locus triggers cellular senescence and apoptosis. In young proliferating cells, the promoter regions of INK4a and ARF genes are repressed with H3K27me3 marks, whereas in senescent cells this locus is activated by the loss of H3K27me3 marks. The expression of the oncogene H-RASV12G or Δ B-RAF contributes to the rapid induction of JMJD3 through the signal cascade of RAS-RAF-MEK-ERK-AP-1, leading to the demethylation of H3K27me3 on the INK4a/ARF locus (Agger et al., 2009; Barradas et al., 2009).
Together with these findings, our study suggests that the expression of JMJD3 is regulated by many different transcription activators, including AP-1, NF-κB, and HIF-1α that are activated by senescence, inflammatory signals, hypoxia, and nonhypoxic HIF-1α activators. Therefore, hypoxia-induced JMJD3 can influence the demethylation of JMJD3 target genes such as Bmp-2, INK4A, ARF, and late HoxA genes such as HoxA7 and HoxA11.
Acknowledgments
This work was supported by the 2011 Research Fund of the University of Seoul for H. Park.
REFERENCES
- Agger K., Cloos P.A., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A.E., Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- Agger K., Cloos P.A., Rudkjaer L., Williams K., Andersen G., Christensen J., Helin K. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23:1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barradas M., Anderton E., Acosta J.C., Li S., Banito A., Rodriguez-Niedenfuhr M., Maertens G., Banck M., Zhou M.M., Walsh M.J., et al. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev. 2009;23:1177–1182. doi: 10.1101/gad.511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Beyer S., Kristensen M.M., Jensen K.S., Johansen J.V., Staller P. The histone demethylases JMJD1A and JMJD2B are transcriptional targets of hypoxia-inducible factor HIF. J. Biol. Chem. 2008;283:36542–36552. doi: 10.1074/jbc.M804578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruick R.K., McKnight S.L. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- Chen Z., Zang J., Whetstine J., Hong X., Davrazou F., Kutateladze T.G., Simpson M., Mao Q., Pan C.H., Dai S., et al. Structural insights into histone demethylation by JMJD2 family members. Cell. 2006;125:691–702. doi: 10.1016/j.cell.2006.04.024. [DOI] [PubMed] [Google Scholar]
- Choi S.M., Choi K.O., Park Y.K., Cho H., Yang E.G., Park H. Clioquinol, a Cu(II)/Zn(II) chelator, inhibits both ubiquitination and asparagine hydroxylation of hypoxia-inducible factor-1alpha, leading to expression of vascular endothelial growth factor and erythropoietin in normoxic cells. J. Biol. Chem. 2006;281:34056–34063. doi: 10.1074/jbc.M603913200. [DOI] [PubMed] [Google Scholar]
- Choi S.M., Oh H., Park H. Microarray analyses of hypoxia-regulated genes in an aryl hydrocarbon receptor nuclear translocator (Arnt)-dependent manner. FEBS J. 2008;275:5618–5634. doi: 10.1111/j.1742-4658.2008.06686.x. [DOI] [PubMed] [Google Scholar]
- De Santa F., Totaro M.G., Prosperini E., Notarbartolo S., Testa G., Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Elvidge G.P., Glenny L., Appelhoff R.J., Ratcliffe P.J., Ragoussis J., Gleadle J.M. Concordant regulation of gene expression by hypoxia and 2-oxoglutarate-dependent dioxygenase inhibition: the role of HIF-1alpha, HIF-2alpha, and other pathways. J. Biol. Chem. 2006;281:15215–15226. doi: 10.1074/jbc.M511408200. [DOI] [PubMed] [Google Scholar]
- Farrall A.L., Whitelaw M.L. The HIF1alpha-inducible procell death gene BNIP3 is a novel target of SIM2s repression through cross-talk on the hypoxia response element. Oncogene. 2009;28:3671–3680. doi: 10.1038/onc.2009.228. [DOI] [PubMed] [Google Scholar]
- Gao X., Wang Q., Li W., Yang B., Song H., Ju W., Liu S., Cheng J. Identification of nucleolar and coiled-body phosphoprotein 1 (NOLC1) minimal promoter regulated by NF-kappaB and CREB. BMB Rep. 2011;44:70–75. doi: 10.5483/BMBRep.2011.44.1.70. [DOI] [PubMed] [Google Scholar]
- Hewitson K.S., McNeill L.A., Riordan M.V., Tian Y.M., Bullock A.N., Welford R.W., Elkins J.M., Oldham N.J., Bhattacharya S., Gleadle J.M., et al. Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J. Biol. Chem. 2002;277:26351–26355. doi: 10.1074/jbc.C200273200. [DOI] [PubMed] [Google Scholar]
- Hewitson K.S., Lienard B.M., McDonough M.A., Clifton I.J., Butler D., Soares A.S., Oldham N.J., McNeill L.A., Schofield C.J. Structural and mechanistic studies on the inhibition of the hypoxia-inducible transcription factor hydroxylases by tricarboxylic acid cycle intermediates. J. Biol. Chem. 2007;282:3293–3301. doi: 10.1074/jbc.M608337200. [DOI] [PubMed] [Google Scholar]
- Ko H.P., Okino S.T., Ma Q., Whitlock J.P., Jr. Dioxin-induced CYP1A1 transcription in vivo: the aromatic hydrocarbon receptor mediates transactivation, enhancer-promoter communication, and changes in chromatin structure. Mol. Cell. Biol. 1996;16:430–436. doi: 10.1128/mcb.16.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra S.M., Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat. Rev. Mol. Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- Kouskouti A., Scheer E., Staub A., Tora L., Talianidis I. Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol. Cell. 2004;14:175–182. doi: 10.1016/s1097-2765(04)00182-0. [DOI] [PubMed] [Google Scholar]
- Lee C., Kim S.J., Jeong D.G., Lee S.M., Ryu S.E. Structure of human FIH-1 reveals a unique active site pocket and interaction sites for HIF-1 and von Hippel-Lindau. J. Biol. Chem. 2003;278:7558–7563. doi: 10.1074/jbc.M210385200. [DOI] [PubMed] [Google Scholar]
- Maxwell P.H., Wiesener M.S., Chang G.W., Clifford S.C., Vaux E.C., Cockman M.E., Wykoff C.C., Pugh C.W., Maher E.R., Ratcliffe P.J. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- Park Y.K., Park H. Prevention of CCAAT/enhancer-binding protein beta DNA binding by hypoxia during adipogenesis. J. Biol. Chem. 2010;285:3289–3299. doi: 10.1074/jbc.M109.059212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.K., Park H. Differentiated embryo chondrocyte 1 (DEC1) represses PPARgamma2 gene through interacting with CCAAT/enhancer binding protein beta (C/EBPbeta) Mol Cells. 2012;33:575–581. doi: 10.1007/s10059-012-0002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.K., Park B., Lee S., Choi K., Moon Y., Park H. Hypoxia-inducible factor-2alpha-dependent hypoxic induction of Wnt10b expression in adipogenic cells. J. Biol. Chem. 2013;288:26311–26322. doi: 10.1074/jbc.M113.500835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard P.J., Loenarz C., Mole D.R., McDonough M.A., Gleadle J.M., Schofield C.J., Ratcliffe P.J. Regulation of Jumonji-domain-containing histone demethylases by hypoxiainducible factor (HIF)-1alpha. Biochem. J. 2008;416:387–394. doi: 10.1042/BJ20081238. [DOI] [PubMed] [Google Scholar]
- Schodel J., Oikonomopoulos S., Ragoussis J., Pugh C.W., Ratcliffe P.J., Mole D.R. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood. 2011;117:e207–217. doi: 10.1182/blood-2010-10-314427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza G.L. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Whetstine J.R. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Wellmann S., Bettkober M., Zelmer A., Seeger K., Faigle M., Eltzschig H.K., Buhrer C. Hypoxia upregulates the histone demethylase JMJD1A via HIF-1. Biochem. Biophys. Res. Commun. 2008;372:892–897. doi: 10.1016/j.bbrc.2008.05.150. [DOI] [PubMed] [Google Scholar]
- Woon Kim Y., Kim S., Geun Kim C., Kim A. The distinctive roles of erythroid specific activator GATA-1 and NFE2 in transcription of the human fetal gamma-globin genes. Nucleic Acids Res. 2011;39:6944–6955. doi: 10.1093/nar/gkr253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X., Kung A.L. Preferential binding of HIF-1 to transcriptionally active loci determines cell-type specific response to hypoxia. Genome Biol. 2009;10:R113. doi: 10.1186/gb-2009-10-10-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X., Lemieux M.E., Li W., Carroll J.S., Brown M., Liu X.S., Kung A.L. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc. Natl. Acad. Sci USA. 2009;106:4260–4265. doi: 10.1073/pnas.0810067106. [DOI] [PMC free article] [PubMed] [Google Scholar]


