Abstract
Induced pluripotent stem cells (iPSCs) are capable of unlimited self-renewal and can give rise to all three germ layers, thereby providing a new platform with which to study mammalian development and epigenetic reprogramming. However, iPSC generation may result in subtle epigenetic variations, such as the aberrant methylation of the Dlk1-Dio3 locus, among the clones, and this heterogeneity constitutes a major drawback to harnessing the full potential of iPSCs. Vitamin C has recently emerged as a safeguard to ensure the normal imprinting of the Dlk1-Dio3 locus during reprogramming. Here, we show that vitamin C exerts its effect in a manner that is independent of the reprogramming kinetics. Moreover, we demonstrate that reprogramming cells under 2i conditions leads to the early upregulation of Prdm14, which in turn results in a highly homogeneous population of authentic pluripotent colonies and prevents the abnormal silencing of the Dlk1-Dio3 locus.
Keywords: 2i conditions, Dlk1-Dio3, induced pluripotent stem cells, Prdm14
INTRODUCTION
The unique properties of embryonic stem cells (ESCs) can be reestablished in differentiated cell types through nuclear reprogramming (Yamanaka and Blau, 2010). The ectopic expression of four transcription factors - Oct4 (O), Sox2 (S), Klf4 (K) and c-Myc (M) (Yamanaka factors) - can induce somatic cells to become ESC-like cells, which are referred to as induced pluripotent stem cells (iPSCs) (Yamanaka and Blau, 2010). The ability to reprogram somatic cells into iPSCs using defined factors holds tremendous potential for regenerative medicine due to its relative simplicity; additionally, iPSCs are not subject to the same ethical concerns associated with using human embryos for medical and scientific research (Saha and Jaenisch, 2009). However, iPSC generation is a slow, inefficient process, and not all of the resulting iPSC clones are functionally equivalent. For example, the aberrant methylation of the Dlk1-Dio3 locus, an imprinted region on mouse chromosome 12qF, has been reported to be a common error in mouse iPSC generation, and the activation of this region is correlated with the degree of iPSC pluripotency (Stadtfeld et al., 2010).
It has been suggested that the extracellular environment regulates key rate-limiting events during reprogramming (Chen et al., 2013). In the present report, we focus on the relationship between the culture conditions and the quality of the resulting iPSCs. Here, we report that different culture environments can result in different reactivation and/or upregulation patterns for endogenous factors that are the primary determinants of obtaining authentic iPSCs.
MATERIALS AND METHODS
Cell culture
Mouse embryonic fibroblast cells (MEFs) carrying an Oct4-GFP reporter were derived from day 13.5 embryos of the OG2 transgenic mouse strain and were cultured in Fibro Gro Low Serum (F.Gro) medium (Millipore). iPSCs and mESCs were maintained in 2i medium (Leitch et al., 2013) on gelatin-coated plates or on a mitomycin C-inactivated MEF feeder (Millipore) layer in mESC medium, which consisted of 85% KnockOut Dulbecco’s Modified Eagle’s Medium (KO DMEM; Gibco), 15% FBS (Gibco), 1 mM L-glutamine, 0.1 mM non-essential amino acids, 0.1 mM 2-mercaptoethanol and 1,000 U/ml ESGRO.
iPSC generation
The pMXs Oct4, pMXs Sox2, pMXs Klf4 and pMXs Nanog plasmids were obtained from Addgene. Dppa2, Sall4 and lin28 were amplified from a mESC cDNA library via PCR and were cloned into the pMXs vector, which resulted in the addition of an HA tag at the C terminus of the protein. Plat E cells were transfected with the pMXs vectors. The cells were then incubated overnight, and the medium was replaced with fresh medium. The virus-containing supernatants were collected 48 h after transfection and were concentrated using Retro-Concentin (SBI). Low-passage MEFs (p 1–3) were seeded 12 h prior to infection. The infections were performed in F.Gro medium, without vitamin C, that was supplemented with 4 mg/ml polybrene (Millipore) and equal amounts of each viral concentrate. After overnight incubation, the cells were washed with PBS. According to the mES protocol, 3 ml of mESC medium was added. According to the LS/2i protocol, the infected cells were maintained in F.Gro medium without vitamin C, and the medium was replaced with 2i medium 3 days after treatment. The iPSC colonies were isolated based on the expression of Oct4-GFP and ESC morphology.
Immunofluorescence
miPSCs were fixed and permeabilized. The fixed samples were incubated for 24 h at 4°C with the anti-Nanog (Abcam) or anti-SSEA-1 (Millipore) primary antibodies. The samples were then washed and incubated in TRITC-conjugated secondary antibodies (Molecular Probes) for 2 h, and the nuclei were counterstained with DAPI (Vector Laboratories). Finally, the slides were photographed using an LSM 510 META confocal microscope (Carl Zeiss).
Alkaline phosphatase staining
Alkaline phosphatase staining was performed using the Alkaline Phosphatase Staining Kit II (Stemgent) according to the manufacturer’s instructions. The cells were photographed using a Nikon Eclipse Ti camera (Nikon).
Real-time PCR
Total RNA was extracted from the cells using the RNeasy Mini Kit (QIAGEN) according to the manufacturer’s instructions. cDNA synthesis was performed using the SuperScript® Vilo cDNA Synthesis Kit (Invitrogen). The qRT-PCR assays were performed in triplicate using SYBR Green I Master Mix (Roche). The primer sequences used in these assays are listed in Supplementary Table S1.
Whole-genome expression analysis
RNA samples for the microarray analysis were prepared using QIAGEN RNeasy columns and were analyzed by Macrogen Inc. using the MouseWG-6 v2 expression Bead-Chips (Illumina).
Teratoma formation and immunohistochemical analyses
iPSCs were injected into the testicular region of NOD/SCID mice (The Jackson Laboratory, USA), and the resulting teratomas were explanted 8 weeks later. The teratoma samples were histologically examined using hematoxylin and eosin (H&E) staining of the gut epithelium and using the following special stains: PAS for the secretory epithelium, Alcian blue for cartilage and Masson’s trichrome for muscle fibers. Images were obtained and analyzed using an inverted microscope (Nikon) (Moon et al., 2011).
RESULTS AND DISCUSSION
Adding vitamin C to the reprogramming medium helps maintain the normal imprinting of the Dlk1-Dio3 locus (Stadtfeld et al., 2012). Furthermore, vitamin C substantially reduces the reprogramming time during OSKM (4F)-mediated iPSC generation (Esteban et al., 2010). Reprogramming under chemically defined conditions revealed that vitamin C promotes iPSC growth and survival (Chen et al., 2011). Considering all of these observations, we generated iPSCs using a small number of reprogramming factors to test whether the presence of vitamin C can help maintain the normal gene expression of the Dlk1-Dio3 locus by accelerating iPSC formation. We introduced cDNAs encoding the transcription factors Oct4 and Klf4 (2F) into MEFs derived from day 13.5 OG2 transgenic strain embryos, which carry the GFP gene under the control of the Oct4 promoter (Oct4-GFP), using a retrovirus-mediated delivery system. After infection, the MEFs were cultured for 3 weeks in the presence of vitamin C, and the iPSCs were subsequently isolated and individually propagated based on Oct4-GFP expression and ESC morphology. We successfully established 6 2F iPSC lines. The 2F iPSCs were stably maintained over 20 passages and were indistinguishable from mESCs (Figs. 1A and 1B). To determine whether the 2F-iPSCs maintained normal Dlk1-Dio3 imprinting, qRT-PCR analysis was performed to detect the expression levels of the Gtl2 and Rian genes, which are located within this region. The expression levels of these genes in all of the tested clones were similar to the expression levels in ESCs, which suggests that the normal imprinting of the Dlk1-Dio3 locus was maintained (Fig. 1C). The reprogramming rate of the 2F-mediated iPSCs was much slower than the 4F system with vitamin C, in which the GFP-positive colonies were first detected approximately 6 to 8 days post-infection (data not shown). Many groups have reported that other factors can also reduce the reprogramming time under mESC conditions. Therefore, we tested whether including these factors in the reprogramming cocktail would yield a homogeneous population of iPSCs by accelerating the establishment of pluripotency. In these tests, OG2 MEFs were infected with OSK coupled with Nanog, Esrrb, Sall4 and lin28, which are factors that can reprogram somatic cells in the absence of the Yamanaka factors (Buganim et al., 2012) (Fig. 2A). The infected cells were cultured under standard mESC culture conditions, and the first GFP-positive colonies (designated 7F iPSCs) appeared within a time interval comparable to that of 4F iPSC generation in the presence of vitamin C. The 7F iPSC clones displayed ESC characteristics (Fig. 2B); however, these clones also exhibited variable Gtl2 and Rian expression patterns (Fig. 2C). These results suggest that vitamin C ensures the maintenance of normal imprinting through a mechanism that is independent of the reprogramming kinetics.
Fig. 1.
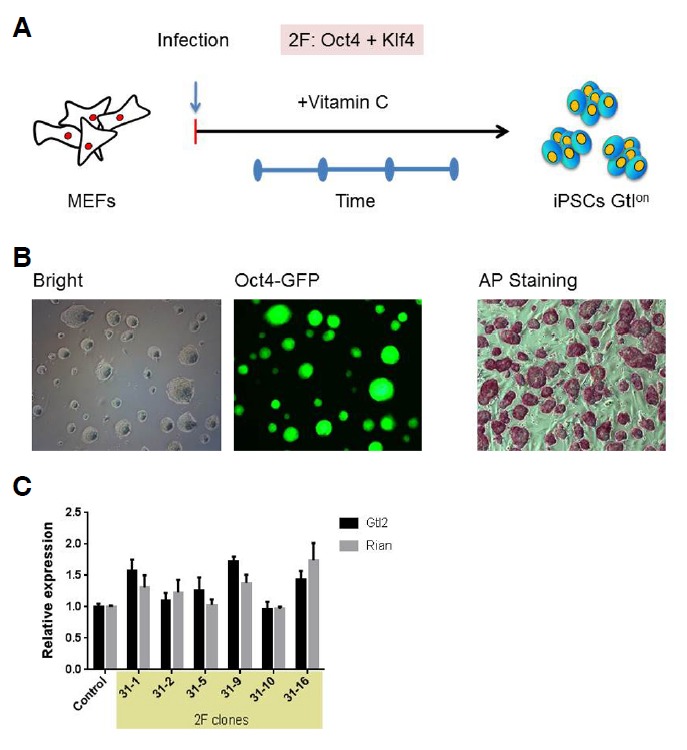
Generation and characterization of 2F-iPSCs. (A) A schematic diagram of the process of iPSC generation from OG2 MEFs using 2 factors. Infected MEFs were cultured in the presence of vitamin C, and individual colonies were isolated 3 weeks post-infection. iPSCs with an active Dlk1-Dio3 locus were designated Gtlon. (B) The morphology, Oct4-GFP expression pattern and AP staining of the 2F-OG2 iPSCs grown in 2i medium are shown. (C) The results of the qRT-PCR analysis of Gtl2 and Rian expression in the 2F-OG2 iPSCs clones are shown. The gene expression levels are shown as relative expression levels compared to the OG2 ESC expression levels, which were set to 1.
Fig. 2.
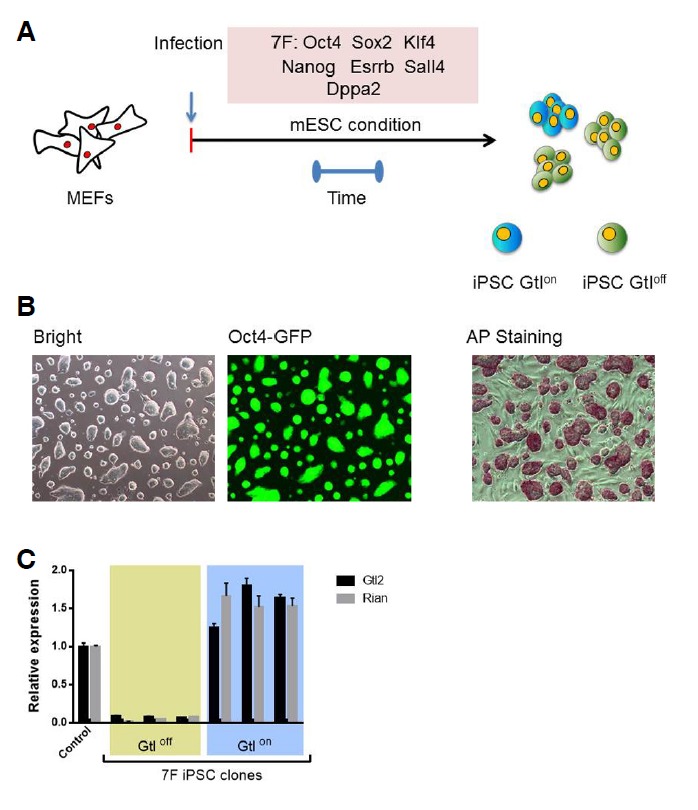
Generation and characterization of 7F-iPSCs. (A) A schematic diagram of iPSC generation from OG2 MEFs using seven factors. Following infection, the MEFs were cultured under standard mESC conditions in the absence of vitamin C, and individual colonies were isolated 8 days post-infection. iPSCs with silenced Dlk1-Dio3 loci were designated as Gtloff. (B) The morphology, Oct4-GFP expression pattern and AP staining of the 7F-OG2 iPSCs grown in 2i medium are shown. (C) The results of the qRT-PCR analysis of Gtl2 and Rian expression in the 7F-OG2 iPSC clones are presented. The gene expression levels are shown as relative expression levels compared to the OG2 ESC expression levels, which were set to 1.
Recent studies have revealed that the type of culture medium used during reprogramming is associated with key rate-limiting events. Pei and colleagues demonstrated that FBS has an inhibitory effect and prevents the activation of pluripotency markers during iPSC generation (Chen et al., 2013). Furthermore, it has been reported that chemically defined conditions supplemented with a mixture of two selective small-molecule inhibitors against GSK3β and MEK1/2 (the 2i or ground-state condition) promote global DNA hypomethylation, which is associated with the naïve state of pluripotency in mESCs. Whereas Prdm14 is upregulated under 2i conditions, Dnmt3a and Dnmt3b are downregulated, leading to DNA hypomethylation (Leitch et al., 2013). Dnmt3a and Dnmt3b are known to be responsible for the aberrant silencing of the Dlk1-Dio3 locus (Stadtfeld et al., 2012). Therefore, we investigated whether reprogramming under 2i conditions results in fully pluripotent iPSCs.
We infected OG2 MEFs with OSK and Nanog. Following infection, the MEFs were maintained under low-serum conditions, and the medium was replaced with 2i medium 3 days after infection (Fig. 3A, LS/2i method). ES-like colonies were picked approximately 8 days post-infection and were individually maintained (Fig. 3B). Consistent with the inhibitory effect of FBS (Chen et al., 2013), GFP-positive cells appeared as early as 3 days post-treatment under the low-serum conditions. However, infection with these factors did not result in GFP-positive cells 3 days post-infection under standard mESC conditions, including 15% FBS (data not shown). The resulting iPSC clones (named OSKN) exhibited stable growth and remained pluripotent in 2i medium (Fig. 3C). Immunocytochemical analysis showed that the OSKN iPSCs expressed the pluripotency markers Nanog and SSEA-1 (Supplementary Fig. S1A). To obtain a more global view of the molecular properties of the OSKN iPSCs, gene expression profiles were compared using pairwise scatter plots of the DNA microarray data. These plots revealed that the OSKN iPSCs were highly similar to mESCs (Fig. 3D). The sub-cutaneous injection of the OSKN iPSCs into immunodeficient mice resulted in the formation of teratomas consisting of tissues derived from each germ layer (Supplementary Fig. S1B). These results show that the OSKN iPSCs are pluripotent and are capable of differentiating into derivatives from all three germ layers. Most importantly, the results from the qRT-PCR experiments demonstrated that all of the tested clones contained an active Dlk1-Dio3 locus (Fig. 3E).
Fig. 3.
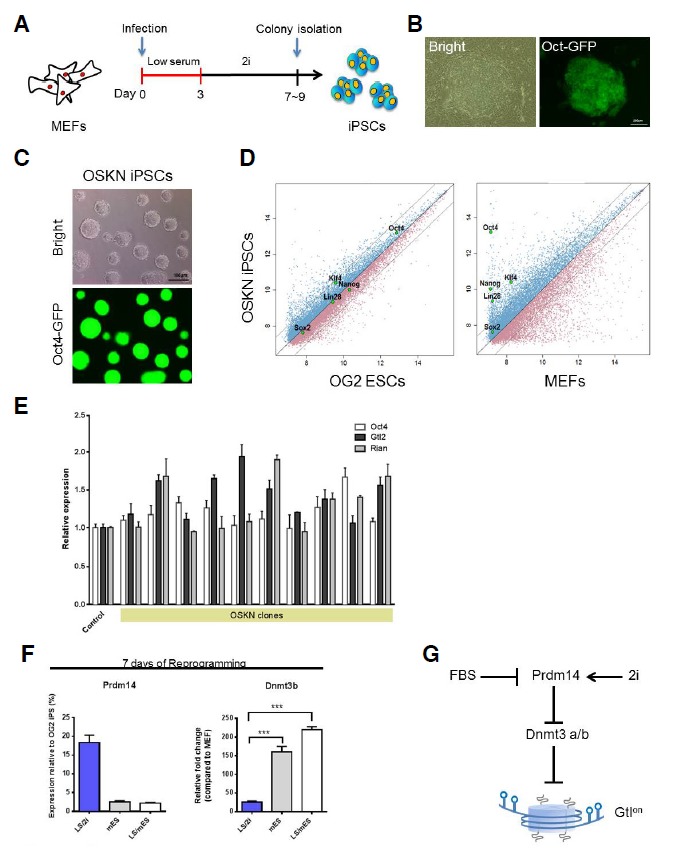
Generation and characterization of iPSCs. (A) The timeline for OSKN-mediated reprogramming of MEFs is shown. After infection, the cells were maintained in low-serum medium for the indicated number of days, and the medium was then replaced with 2i medium. (B) Representative images of a primary OSKN-iPSC colony are shown. (C) The morphology of the OSKN-iPSCs grown in 2i medium on gelatin-coated plates without feeder cells and the Oct4-GFP expression in these cells are shown. (D) Pairwise scatter plots comparing the global gene expression patterns observed in the OSKN-iPSCs based on cDNA microarray analysis with the expression patterns of the OG2 ESCs and MEFs are shown. (E) The expression levels of Gtl2 and Rian, which are expressed from the Dlk1-Dio3 locus, and Oct4 were analyzed using qRT-PCR analysis. The gene expression levels are presented as relative expression levels compared to the expression levels in the OG2 ESCs, which were set to 1. (F) The results of the qRT-PCR analysis of Prdm14 and Dnmt3b expression in the OSKN-infected cells cultured according to the indicated methods are shown. (G) A proposed model displaying the effects of the different culture environments on reprogramming. Prdm14 is differentially reactivated under the different culture conditions. The 2i medium promotes the robust reactivation of endogenous Prdm14, which represses Dnmt3a/b expression and yields Gtlon iPSCs.
Next, we compared the expression profiles of the genes associated with DNA hypomethylation and pluripotency from iPSCs generated using the different reprogramming culture conditions (Supplementary Fig. S2A). We observed that Oct4 and Prdm14 were expressed at higher levels when the LS/2i method was used compared to the results obtained following the mES method. Furthermore, the initial maintenance of the cells in low-serum medium had no significant effect on Prdm14 expression when the medium was replaced with mES medium on the third day of reprogramming (the LS/mES method). The expression levels of Dnmt3a and Dnmt3b were inversely correlated with the Prdm14 expression level, which is consistent with previous reports (Leitch et al., 2013; Yamaji et al., 2013) (Fig. 3F and Supplementary Fig. S2B). These results suggest that reprogramming under 2i conditions may support the robust reactivation of Prdm14, which can result in fully pluripotent iPSCs (Fig. 3G).
iPSCs can be potentially used in disease modeling, drug screening and autologous cell therapy. However, the subtle epigenetic variations that have been observed among the iPSC clones generated in the same dish raise safety concerns and call into question their suitability for clinical applications. The activation of the Dlk1-Dio3 region was recently recognized as a marker of authentic pluripotency in miPSCs (Liu et al., 2010). In the present study, we successfully generated homogeneous populations of miPSCs with an active Dlk1-Dio3 locus using the chemically defined 2i medium. Consistent with a recent report (Leitch et al., 2013), we found that reprogramming in 2i medium resulted in the early upregulation of Prdm14, which represses Dnmt3a and Dnmt3b expression. Our results suggest that the endogenous factors regulating and/or modifying the epigenome are differentially reactivated under different culture conditions, and these variations may affect the pluripotency of the resulting iPSCs. Because the heterogeneity among cells undergoing reprogramming may prevent the accurate recapitulation of the process, it is enticing to think that defined conditions may provide the most ideal model for investigating nuclear reprogramming.
Supplementary information
Acknowledgments
This research was supported by a grant (2012-0006107) from the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean government [The Ministry of Education, Science and Technology (MEST)].
Note:
Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Buganim Y., Faddah D.A., Cheng A.W., Itskovich E., Markoulaki S., Ganz K., Klemm S.L., van Oudenaarden A., Jaenisch R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Gulbranson D.R., Hou Z., Bolin J.M., Ruotti V., Probasco M.D., Smuga-Otto K., Howden S.E., Diol N.R., Propson N.E., et al. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Liu H., Liu J., Qi J., Wei B., Yang J., Liang H., Chen Y., Wu Y., Guo L., et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat. Genet. 2013;45:34–42. doi: 10.1038/ng.2491. [DOI] [PubMed] [Google Scholar]
- Esteban M.A., Wang T., Qin B., Yang J., Qin D., Cai J., Li W., Weng Z., Chen J., Ni S., et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6:71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Leitch H.G., McEwen K.R., Turp A., Encheva V., Carroll T., Grabole N., Mansfield W., Nashun B., Knezovich J.G., Smith A., et al. Naive pluripotency is associated with global DNA hypomethylation. Nat. Struct. Mol. Biol. 2013;20:311–316. doi: 10.1038/nsmb.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Luo G.Z., Yang W., Zhao X., Zheng Q., Lv Z., Li W., Wu H.J., Wang L., Wang X.J., et al. Activation of the imprinted Dlk1-Dio3 region correlates with pluripotency levels of mouse stem cells. J. Biol. Chem. 2010;285:19483–19490. doi: 10.1074/jbc.M110.131995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S.H., Kim S.W., Kim J.S., Park S.J., Do J.T., Lee D.R., Chung H.M. Gene expression profiles in CHA3 and CHA4 human embryonic stem cells and embryoid bodies. Mol. Cells. 2011;31:315–326. doi: 10.1007/s10059-011-0039-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha K., Jaenisch R. Technical challenges in using human induced pluripotent stem cells to model disease. Cell Stem Cell. 2009;5:584–595. doi: 10.1016/j.stem.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M., Apostolou E., Akutsu H., Fukuda A., Follett P., Natesan S., Kono T., Shioda T., Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M., Apostolou E., Ferrari F., Choi J., Walsh R.M., Chen T., Ooi S.S., Kim S.Y., Bestor T.H., Shioda T., et al. Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat. Genet. 2012;44:398–405. S391–392. doi: 10.1038/ng.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji M., Ueda J., Hayashi K., Ohta H., Yabuta Y., Kurimoto K., Nakato R., Yamada Y., Shirahige K., Saitou M. PRDM14 ensures naive pluripotency through dual regulation of signaling and epigenetic pathways in mouse embryonic stem cells. Cell Stem Cell. 2013;12:368–382. doi: 10.1016/j.stem.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Yamanaka S., Blau H.M. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465:704–712. doi: 10.1038/nature09229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


