Abstract
We evaluated the use of an RNA stabilisation buffer, RNAlater® (Ambion, Austin, Texas), as a preservation medium for parasitic coprology analysis of faecal samples collected from chimpanzees living in the wild (Pan troglodytes troglodytes). Thirty faecal samples collected in the forests of south-east Cameroon (Mambele area) from 2003 to 2011 were preserved in RNAlater® at −80 °C and analysed for their parasite content. We identified and counted parasitic elements and assessed their shape, size and morphology in relation to the storage time of the samples. We found that parasite elements were identifiable in RNAlater® preserved samples after as many as 7 years, showing that RNAlater® could be an effective and reliable preservation medium for coprology. Thus, its use could be an interesting way to optimise sample collection for several types of studies (parasitology and bacteriology/virology) at once, especially considering the logistically challenging and time-consuming field campaigns needed to obtain these faecal samples.
Keywords: Wild-living chimpanzees, Endoparasites, Parasitic coprology, RNAlater®
1. Introduction
In the tropical forest of the Congo basin, intensive logging, mining and bushmeat hunting have increased the contact between humans and non-human primates (NHPs). Such close contact may facilitate the zoonotic transmission of potentially pathogenic micro-organisms from NHPs to humans, with consequences for human health, as well as from humans to NHPs, with consequences for wildlife conservation (Calvignac-Spencer et al., 2012). It is therefore essential to implement sustained surveillance programs designed to detect cross-species transmission in order to prevent the potential spread of emerging infectious diseases in both NHP and human populations.
As a non-invasive method, analysis of faecal samples has proven to be a convenient means of assessing the prevalence of pathogens in NHP populations in the wild. The majority of gastrointestinal parasites that infect apes, as well as viral, bacterial and protozoan agents infecting NHPs, can be monitored through faecal analysis (Kooriyama et al., 2012; Gillespie et al., 2010; Bezjian et al., 2008; Gillespie and Chapman 2008).
Analysing fresh stool samples in the field is challenging in many ways, so collected samples are usually preserved in a 10% formalin fixative solution and analysed several weeks or months later. The use of formalin, a formaldehyde containing solution, has several drawbacks. First, formaldehyde has a low vapour pressure point (35 °C), which may be inappropriate under tropical weather conditions. Secondly, it is a known skin, eye, and respiratory tract irritant and has been shown to be a carcinogen; therefore, its use should be limited to adequate lab facilities. Lastly, formaldehyde induces cross-linking and degradation of DNA, which is unsuitable if the samples are intended for molecular biology-based studies (Coombs et al., 1999; Williams et al., 1999).
To preserve nucleic acids (DNA or RNA) and to allow for molecular analyses on faecal samples collected in the wild from NHPs, other storage media have been used, including 70% or 96% ethanol (Nakamura et al., 2011; Uenishi et al., 2007), RNAlater® (Ambion, Inc.) (Ochman et al., 2010; Szekely et al., 2010) and ethanol followed by desiccation using silica (Nsubuga et al., 2004). Freezing the samples in liquid nitrogen is also an effective means of preservation. However, when freezing was not possible, compared to the other methods of storage, RNAlater® proved the most efficient method for preservation of microbial DNA (Vlčková et al., 2012; Nechvatal et al., 2008). RNAlater® is an aqueous, non-toxic storage reagent that rapidly permeates tissues to stabilise and protect cellular RNA and DNA in situ in unfrozen specimens. RNAlater® has been adopted by an increasing number of researchers focusing on NHPs viral infections (Etienne et al., 2012; Liu et al., 2010; Whittier et al., 2010; Locatelli et al., 2008; van heuverswyn et al., 2007; Keele et al., 2006; Santiago et al., 2003). It is also effective for preservation of Plasmodium DNA in faecal samples (Liu et al., 2010).
Considering the problems associated with storing faecal samples in a formalin solution for parasitic coprology, we decided to assess the feasibility of conducting microscopic examination of faecal samples stored in RNAlater®. We selected and analysed chimpanzee faecal samples stored in RNAlater® collected in the rainforest of southeast Cameroon between 2003 and 2011 in the context of a long-term study assessing the prevalence and genetic diversity of Simian Immunodeficiency Viruses (SIV).
2. Materials and methods
2.1. Sample collection and species identification
We randomly selected 30 faecal samples from the stool bank of the UMI 233 “TransVIHMI” laboratory in Montpellier. These samples were collected from chimpanzees (Pan troglodytes troglodytes) living in the forests of southeast Cameroon (Mambele area) between 2003 and 2011. They were stored in RNAlater® at the time of collection then subsequently frozen at −80 °C one to three weeks later. The host species was confirmed by mtDNA analyses, as described previously (Keele et al., 2006; Van Heuverswyn et al., 2006; van der Kuyl et al., 1995). Briefly, a QIAamp stool DNA miniprep kit (Qiagen, Valencia, CA) was used to extract faecal DNA. Two millilitres of faecal sample were used to obtain a final elution volume of 100 μl of faecal DNA. A ~450- to 500-bp fragment spanning the hypervariable D-loop region was amplified using primers L15997 and H16498 and/or a 386-bp fragment spanning the 12S gene was amplified using primers 12S-L1091 and 12S-H1478. The sequences obtained using a 3130xl Genetic Analyser (Applied Biosystems, France) were aligned using the Seqman DNAStar (Lasergene, Madison, USA) software. The species of animal providing the samples was confirmed by neighbour-joining analysis using the CLUSTAL X 2.0 program (Thompson et al., 1997)
2.2. Coprology
Once the species identification of the host was confirmed, we assessed the parasite content of the samples using the following method: for each sample, one millilitre of 50:50 stool:RNAlater® mix was thawed at room temperature. Each sample was then vortexed briefly. One hundred microliters of this solution was mixed with 100 μl of physiological serum to avoid crystallisation or a multiple layer effect on the slides due to the high salt concentration in the RNAlater® buffer. Then, a 50 μl aliquot of the sample-physiological serum mix was smeared on a slide and observed directly under a microscope (Olympus BX41). A second slide was prepared using Para-selles KOP Color II (Fumouze) dye (10 μl of dye for 50 μl of stool/physiological serum mix). When the results of the slides were both negative or a significant discrepancy was found between colored and not colored slides, either in the type or number of parasitic elements, two additional smears were performed. The species, number and shape of parasitic elements, as well as their ability to be dyed, were recorded. A possible effect of the date of collection on the results of the stool examination was considered. It should be noted that we tried concentration methods on the RNAlater® samples (Bailenger method, Iodesin-Color, Para-selles KOP Color II Kit, Fumouze diagnostics, France) but they proved to be unsuccessful due to the high salt concentration of the RNAlater® buffer.
To compare the efficiency of RNAlater® and 10% formalin in preserving the parasitic elements, ten matched samples that were collected in 2011 and preserved in 10% formalin were analysed in parallel.
2.3. Molecular biology assay
To show the utility of RNAlater® versus formalin samples in combined coprology-molecular biology studies, a PCR detection assay for Blastocystis sp. was performed on DNA extracted from ten RNAlater® and 10 matched formalin samples collected in 2011. Blastocytis was chosen for this test because of the difficulties in detecting it by direct microscopy, especially in stool samples rich in plant debris. Briefly, a QIAamp stool DNA miniprep kit (Qiagen, Valencia, CA) was used to extract faecal DNA from each sample under the conditions specified by the manufacturer for parasites. Two milliliters of faecal sample were used to obtain a final elution volume of 100 μl of faecal DNA. PCR was performed as described by Grabensteiner and Hess, 2006 with the following conditions: forward primer BLF 5′-TAACCGTAGTAATTCTAGGGC-3′, reverse primer BLR 5′-AACGTTAATATACGCTATTGG-3′, denaturation at 94 °C for 5 min, 40 cycles of 94 °C for 1 min, 53 °C for 1 min, and 72 °C for 1 min, followed by a final extension step of 72 °C for 10 min. PCR products were separated on a 1% agarose-ethidium bromide gel and visualised under UV. Positive samples were sent to Beckman Coulter Genomics UK for sequencing.
3. Results
Parasitic coprology performed on our RNAlater® sample set allowed us to detect parasitic elements (helminth eggs and protozoa cysts) in faeces that were several years old.
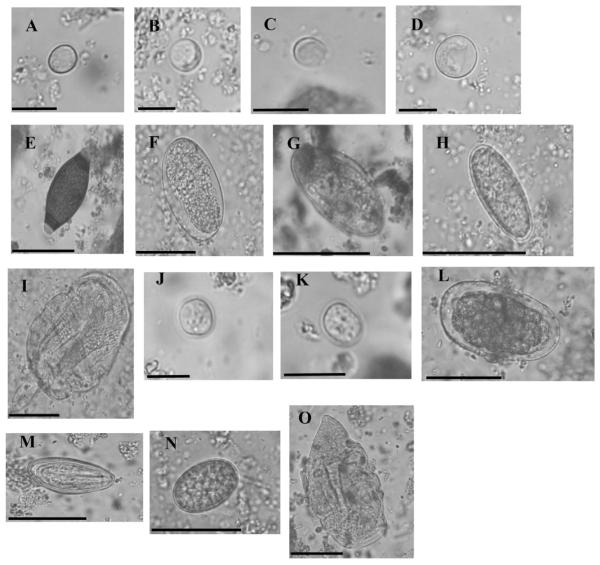
These analyses showed that samples collected in 2003 contained few identifiable parasitic elements, and those that were observed had degraded morphological aspects (Table 1). Samples from 2005 to 2011 were less degraded, and we could identify the parasitological content more easily (Table 1). In this sample set, we were able to observe the presence of several protozoan cysts belonging to the Endolimax and Entamoeba genera (with diameters ranging from 6–8 μm to 16–20 μm, so possibly from different species) as well as Blastocystis cells (Table 1 and Fig. 1A–D).
Table 1.
Type and number of parasite elements found in samples collected from 2003 to 2011 in the Mambele area of Cameroon.
| Sample | Year of collection | Method of preservation | Observed elements, helminths | Observed elements, protozoas |
|---|---|---|---|---|
| Chimp-1 | 2003 | RNAlater® | 4–5 larvae | Small amoeba cysts and Blastocystis |
| Chimp-2 | 2003 | RNAlater® | 1 larvae, 1 Trichuris egg | 0 |
| Chimp-3 | 2003 | RNAlater® | 1 larvae | 1 Troglodytella trophozoite |
| Chimp-4 | 2003 | RNAlater® | 0 | 0 |
| Chimp-5 | 2003 | RNAlater® | 0 | 0 |
| Chimp-6 | 2005 | RNAlater® | 2 strongylid eggs | Endolimax cysts |
| Chimp-7 | 2005 | RNAlater® | 1 strongylid egg, 4–6 unknown eggs | 0 |
| Chimp-8 | 2005 | RNAlater® | 0 | >10 Troglodytella |
| Chimp-9 | 2005 | RNAlater® | 3–4 strongylid eggs | 0 |
| Chimp-10 | 2005 | RNAlater® | 2 strongylid eggs | >30 Troglodytella |
| Chimp-11 | 2007 | RNAlater® | 2–3 strongylid eggs | 0 |
| Chimp-12 | 2007 | RNAlater® | 10–12 strongylid eggs | 0 |
| Chimp-13 | 2007 | RNAlater® | 3–4 strongylid eggs | 0 |
| Chimp-14 | 2007 | RNAlater® | 4–5 strongylid eggs | Blastocystis |
| Chimp-15 | 2007 | RNAlater® | 6–10 strongylid eggs | Blastocystis |
| Chimp-16 | 2010 | RNAlater® | 3–8 strongylid eggs, 1 Enterobius egg | Endolimax, 4–5 Entamoeba cysts, Blastocystis |
| Chimp-17 | 2010 | RNAlater® | 0 | >10 Troglodytella |
| Chimp-18 | 2010 | RNAlater® | 4–5 strongylid eggs | 1–2 Troglodytella |
| Chimp-19 | 2010 | RNAlater® | 1 strongylid egg | 2 Troglodytella |
| Chimp-20 | 2010 | RNAlater® | 2 strongylid eggs | 0 |
| Chimp-21 | 2011 | RNAlater® | 1 larvae | 0 |
| Chimp-22 | 2011 | RNAlater® | 12 larva, 2–3 strongylid eggs | small amoebas cysts |
| Chimp-23 | 2011 | RNAlater® | 1–2 strongylid eggs | 0 |
| Chimp-24 | 2011 | RNAlater® | 2 strongylid eggs | 4–5 Troglodytella |
| Chimp-25 | 2011 | RNAlater® | 2–3 strongylid eggs | >30 Troglodytella |
| Chimp-26 | 2011 | RNAlater® | 1 larvae, 1 strongylid egg | 0 |
| Chimp-27 | 2011 | RNAlater® | 2–3 strongylid eggs | 7–10 Troglodytellas |
| Chimp-28 | 2011 | RNAlater® | 1 larvae, 3 strongylid eggs | 5 Troglodytella |
| Chimp-29 | 2011 | RNAlater® | 3 strongylid eggs, unidentified eggs (+) | 14 Troglodytella |
| Chimp-30 | 2011 | RNAlater® | 2 strongylid eggs, unidentified eggs (+) | 5 Troglodytella |
| Chimp-21* | 2011 | Formaline | 0 | Blastocystis |
| Chimp-22* | 2011 | Formaline | 11 larva, 6–7 strongylid eggs | Endolimax |
| Chimp-23* | 2011 | Formaline | 2 strongylid eggs, 1 Enterobius, egg | lodamoeba cysts |
| Chimp-24* | 2011 | Formaline | 0 | 11 Troglodytella, Endolimax, lodamoeba |
| Chimp-25* | 2011 | Formaline | 1–2 strongylid eggs | >30 Troglodytella, Endolimax, lodamoeba |
| Chimp-26* | 2011 | Formaline | 2–3 strongylid egg | 7–9 Troglodytella |
| Chimp-27* | 2011 | Formaline | 4 strongylid eggs | 5 Troglodytella |
| Chimp-28* | 2011 | Formaline | 1 strongylid eggs | 4 Troglodytella |
| Chimp-29* | 2011 | Formaline | 2 strongylid eggs, unidentified eggs (+) | >20 Troglodytella |
| Chimp-30* | 2011 | Formaline | 1 strongylid eggs, unidentified eggs (+) | 5 Troglodytella |
The samples numbered from Chimp-1 to Chimp-30 are samples preserved in RNAlater at −80 °C.
The samples numbered Chimp-21–30* are the same samples as Chimp-21–30 but preserved in 10% formalin solution and at 4 °C.
The number of parasite elements was counted for each sample and is shown in the table.
The + sign indicates a very high density of unidentified helminth eggs on the slide (2–3 elements on average per microscope field at 40×).
Fig. 1.
Parasite elements observed by microscopy in stools collected from wild chimpanzees. Elements in A to I were found in chimpanzee stools collected in Cameroon from 2003 to 2011 and preserved in RNAlater buffer at −80 °C. Elements in J–O were found in samples collected in the same area in 2011 and preserved in 10% formaldehyde buffer at 4 °C. A: Endolimax cyst. B: small unidentified amoeba cyst (Entamoeba histolytica/E. hartmanni ?) C: Blastocystis cell. D: Entamoeba cyst (Entamoeba histolytica/ E.coli ?) E: Trichuris egg. F: Strongylid egg. G:Enterobius egg. H: unidentified helminth egg. I: Troglodytella trophozoite. J: Iodamoeba cyst. K: Endolimax cyst. L: Strongylid egg. M: Enterobius egg. N: unidentified helminth egg. O: Troglodytella trophozoite. Scale bars indicate 10 μm for elements A–D, J, and K and 50 μm for elements E–I, L–O.
Moreover, we detected Trichuris eggs, Enterobius eggs and strongylid eggs of different sizes, which likely represent different genera (Table 1 and Fig. 1E–G). We also observed eggs of an undetermined helminth species (Fig. 1 H) and were able to detect Troglodytella trophozoites (Table 1 and Fig. 1I).
The types of parasitic elements observed in the matched samples (RNAlater® vs. 10% formalin) were similar in both sets of samples (Table 1: Chimp-21–30 vs. Chimp-21*–30*, respectively). However, parasites preserved in RNAlater® displayed more degraded cellular content (retracted cytoplasm for the protozoa cysts and lysed blastomere cells for the helminth eggs; Fig. 1A–H) than those from the formalin preserved samples, with the cytoplasmic content being better preserved in formalin (Fig. 1J–O). The increased degradation of the parasitic content in the RNAlater® buffer may be due to the high salt hyper-osmotic content of this medium combined with storage at −80 °C. Thus, we were able to determine the genera of well-described parasites (helminth eggs, amoeba cysts) stored in RNAlater® but this induced degradation was a limiting factor for species identification relying on fine internal structures (number of nuclei, position of the chromatin for the Entamoeba species, number of blastomeres for Ancylostoma duodenale etc.). The detection and identification of smaller elements (Endolimax and Iodamoeba cysts) was also more difficult in the samples preserved in RNAlater® than in 10% formalin. Finally, as the high salt concentration of RNAlater® buffer modifies the density of the sample, faecal floatation and sedimentation concentration methods were inappropriate. As such, the samples were observable only by direct smear as described above.
With respect to molecular detection assays for parasite elements (Blastocystis sp.), as expected, none of the formalin-preserved samples (Chimp21*–30* gave a positive result, whereas the same matched samples preserved in RNAlater® were all positive by PCR (Fig. 2). Sequencing of products and sequence analyses by BLAST confirmed that 8 of these PCR fragments (Chimp-23–30) indeed matched Blastocystis sp (Blastocystis subtype 1 Chimp) whereas the sequences obtained from the Chimp-21 and Chimp-22 individuals matched undetermined uncultured alveolate eukaryotes (data not shown).
Fig. 2.
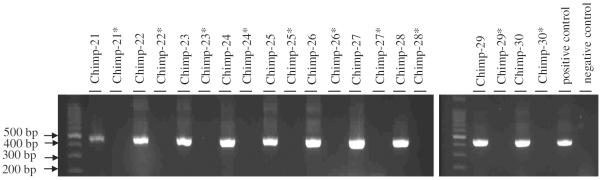
Blastocystis sp. PCR detection assay. Detection of Blastocystis sp was performed by PCR on total DNA extracted from chimpanzee stools with specific Blastocystis primers. Chimp-21–30 correspond to PCR profiles obtained with DNA extracted from stool samples preserved in RNAlater® Chimp-21*–30* correspond to PCR profiles obtained with DNA extracted from samples obtained from the same animals as Chimp-21–30 but preserved in formalin. The positive control PCR profile was obtained using DNA extracted from a stool sample tested positive for Blastocystis sp. by direct microscopy that had been discarded from the study because the shedding species was not chimpanzee but Gorilla gorilla. The negative control PCR profile was obtained using water instead of DNA.
4. Discussion
In this study, we observed that RNAlater® has some minor limitations when used as a preservation method for parasite coprology. In fact, we could not obtain any data using concentration methods because of the high salinity of the buffer. As a consequence, we were only able to rely on direct smears for observation. Moreover, RNAlater® is less efficient than formalin for the preservation of fine internal structures of some fragile parasite eggs or cysts.
Nevertheless, we were able to identify Endolimax cysts, Entamoeba cysts, Troglodytella trophozoites, Blastocystis cells, Trichuris eggs, Enterobius eggs, strongylid eggs of different sizes (possibly Trichostrongylus, Oesophagostomum, Strongyloides and Ancylostoma), and several nematode larvae (not shown), as well as eggs from an unknown helminth in RNAlater®-preserved samples. The fact that we were able to observe nematode eggs, ranging from resistant (Trichuris eggs, Sanguinetti et al., 2005) to more fragile (strongylid eggs, Waruiru et al., 1998), supports the efficacy of RNAlater® as a preservation method. The observation of Troglodytella was also particularly interesting, demonstrating that it is possible to identify a ciliate trophozoite (which is a more fragile cellular element compared to parasite cysts or thick-walled helminth eggs) in samples stored in RNAlater®. Moreover, we were able to identify these parasites in samples aged up to 7 years old that had been kept at 80 °C and which, in some cases, were already thawed and frozen several times during previous studies. Given our results, RNAlater® stool samples stored for up to seven years in laboratory banks worldwide could therefore represent an interesting source for retrospective studies on gastrointestinal parasites of endangered animals. Finally, a probe molecular assay showed that combined molecular biology studies (detection of Blastocystis sp. by PCR in this article) and parasitic coprology studies can be performed on RNAlater® samples in parallel, something that is impossible with formalin-preserved samples.
Until now, NHP samples stored in RNAlater® were only used for molecular biology analyses such as species identification, genotyping, and viral/bacterial/fungal molecular diagnoses. We show here for the first time that they can also be used to measure in parallel the extent and diversity of gastrointestinal endoparasites, thereby providing a broader spectrum of information regarding the pathogens infecting rare or endangered NHP species.
Finally, these results showed that, in the future, faecal sample collection in the field can be simplified by working with a single preservation buffer, thus avoiding issues related to the use of formaldehyde.
HIGHLIGHTS
We evaluated if RNAlater® could preserve parasites in wild-living chimpanzees stools.
RNAlater® is suited for samples collections in logistically challenging areas.
Parasites were identifiable in samples preserved at −80 °C for as many as 7 years.
RNAlater® may facilitate combined studies (molecular analyses and coprology together).
Acknowledgments
We thank the staff and the SIV team of Innocent Ndong Bass, Aimé Mebenga and Joseph Moudindo from PRESICA for logistical support in Cameroon and the Cameroonian Ministries of Health, Environment and Forestry, and Research for permission to collect samples in Cameroon. We also thank Dr L. Mondolot, Pr. Y. Pelissier and Dr. S. Bertout for their useful help with the identification of false positives.
Footnotes
Conflicts of interest
The authors declare no conflict of interest.
References
- Bezjian M, Gillespie TR, Chapman CA, Greiner EC. Coprologic evidence of gastrointestinal helminths of forest baboons, Papio anubis, in Kibale National Park, Uganda. J. Wildl. Dis. 2008;44:878–887. doi: 10.7589/0090-3558-44.4.878. [DOI] [PubMed] [Google Scholar]
- Calvignac-Spencer S, Leendertz SA, Gillespie TR, Leendertz FH. Wild great apes as sentinels and sources of infectious disease. Clin. Microbiol. Infect. 2012;18:521–527. doi: 10.1111/j.1469-0691.2012.03816.x. [DOI] [PubMed] [Google Scholar]
- Coombs NJ, Gough AC, Primrose JN. Optimisation of DNA and RNA extraction from archival formalin-fixed tissue. Nucleic Acids Res. 1999;27 doi: 10.1093/nar/27.16.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne L, Locatelli S, Ayouba A, Esteban A, Butel C, Liegeois F, Aghokeng A, Delaporte E, Mpoudi Ngole E, Peeters M. Noninvasive follow-up of simian immunodeficiency virus infection in wild-living nonhabituated western lowland gorillas in Cameroon. J. Virol. 2012;86:9670–9672. doi: 10.1128/JVI.01186-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie TR, Chapman CA. Forest fragmentation, the decline of an endangered primate, and changes in host-parasite interactions relative to an unfragmented forest. Am. J. Primatol. 2008;70:222–230. doi: 10.1002/ajp.20475. [DOI] [PubMed] [Google Scholar]
- Gillespie TR, Lonsdorf EV, Canfield EP, Meyer DJ, Nadler Y, Raphael J, Pusey AE, Pond J, Pauley J, Mlengeya T, Travis DA. Demographic and ecological effects on patterns of parasitism in eastern chimpanzees (Pan troglodytes schweinfurthii) in Gombe National Park, Tanzania. Am. J. Phys. Anthropol. 2010;143:534–544. doi: 10.1002/ajpa.21348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabensteiner E, Hess M. PCR for the identification and differentiation of Histomonas meleagridis, Tetratrichomonas gallinarum and Blastocystis spp. Vet. Parasitol. 2006;142:223–230. doi: 10.1016/j.vetpar.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, Bibollet-Ruche F, Chen Y, Wain LV, Liegeois F, Loul S, Ngole EM, Bienvenue Y, Delaporte E, Brookfield JF, Sharp PM, Shaw GM, Peeters M, Hahn BH. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooriyama T, Hasegawa H, Shimozuru M, Tsubota T, Nishida T, Iwaki T. Parasitology of five primates in Mahale Mountains National Park, Tanzania. Primates. 2012;53:365–375. doi: 10.1007/s10329-012-0311-9. [DOI] [PubMed] [Google Scholar]
- Liu W, Li Y, Learn GH, Rudicell RS, Robertson JD, Keele BF, Ndjango JB, Sanz CM, Morgan DB, Locatelli S, Gonder MK, Kranzusch PJ, Walsh PD, Delaporte E, Mpoudi-Ngole E, Georgiev AV, Muller MN, Shaw GM, Peeters M, Sharp PM, Rayner JC, Hahn BH. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature. 2010;467:420–425. doi: 10.1038/nature09442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locatelli S, Liegeois F, Lafay B, Roeder AD, Bruford MW, Formenty P, Noë R, Delaporte E, Peeters M. Prevalence and genetic diversity of simian immunodeficiency virus infection in wild-living red colobus monkeys (Piliocolobus badius badius) from the Taï forest, Côte d'Ivoire SIVwrc in wild-living western red colobus monkeys. Infect. Genet. Evol. 2008;8:1–14. doi: 10.1016/j.meegid.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Amato KR, Garber P, Estrada A, Mackie RI, Gaskins HR. Analysis of the hydrogenotrophic microbiota of wild and captive black howler monkeys (Alouatta pigra) in Palenque National Park, Mexico. Am. J. Primatol. 2011;73:909–919. doi: 10.1002/ajp.20961. [DOI] [PubMed] [Google Scholar]
- Nechvatal JM, Ram JL, Basson MD, Namprachan P, Niec SR, Badsha KZ, Matherly LH, Majumdar AP, Kato I. Fecal collection, ambient preservation, and DNA extraction for PCR amplification of bacterial and human markers from human feces. J. Microbiol. Methods. 2008;72:124–132. doi: 10.1016/j.mimet.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Nsubuga AM, Robbins MM, Roeder AD, Morin PA, Boesch C, Vigilant L. Factors affecting the amount of genomic DNA extracted from ape faeces and the identification of an improved sample storage method. Mol. Ecol. 2004;13:2089–2094. doi: 10.1111/j.1365-294X.2004.02207.x. [DOI] [PubMed] [Google Scholar]
- Ochman H, Worobey M, Kuo CH, Ndjango JB, Peeters M, Hahn BH, Hugenholtz P. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol. 2010;8:e1000546. doi: 10.1371/journal.pbio.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanguinetti GS, Tortul C, García MC, Ferrer V, Montangero A, Strauss M. Investigating helminth eggs and Salmonella sp. in stabilisation ponds treating septage. Water Sci. Technol. 2005;51:239–247. [PubMed] [Google Scholar]
- Santiago ML, Lukasik M, Kamenya S, Li Y, Bibollet-Ruche F, Bailes E, Muller MN, Emery M, Goldenberg DA, Lwanga JS, Ayouba A, Nerrienet E, McClure HM, Heeney JL, Watts DP, Pusey AE, Collins DA, Wrangham RW, Goodall J, Brookfield JF, Sharp PM, Shaw GM, Hahn BH. Foci of endemic simian immunodeficiency virus infection in wild-living eastern chimpanzees (Pan troglodytes schweinfurthii) J. Virol. 2003;77:7545–7562. doi: 10.1128/JVI.77.13.7545-7562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekely BA, Singh J, Marsh TL, Hagedorn C, Werre SR, Kaur T. Fecal bacterial diversity of human-habituated wild chimpanzees (Pan troglodytes schweinfurthii) at Mahale Mountains National Park, Western Tanzania. Am. J. Primatol. 2010;72:566–574. doi: 10.1002/ajp.20809. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uenishi G, Fujita S, Ohashi G, Kato A, Yamauchi S, Matsuzawa T, Ushida K. Molecular analyses of the intestinal microbiota of chimpanzees in the wild and in captivity. Am. J. Primatol. 2007;69:367–376. doi: 10.1002/ajp.20351. [DOI] [PubMed] [Google Scholar]
- van der Kuyl AC, Kuiken CL, Dekker JT, Goudsmit J. Phylogeny of African monkeys based upon mitochondrial 12S rRNA sequences. J. Mol. Evol. 1995;40:173–180. doi: 10.1007/BF00167111. [DOI] [PubMed] [Google Scholar]
- van Heuverswyn F, Li Y, Bailes E, Neel C, Lafay B, Keele BF, Shaw KS, Takehisa J, Kraus MH, Loul S, Butel C, Liegeois F, Yangda B, Sharp PM, Mpoudi-Ngole E, Delaporte E, Hahn BH, Peeters M. Genetic diversity and phylogeographic clustering of SIVcpzPtt in wild chimpanzees in Cameroon. Virology. 2007;368:155–171. doi: 10.1016/j.virol.2007.06.018. [DOI] [PubMed] [Google Scholar]
- van Heuverswyn F, Li Y, Neel C, Bailes E, Keele BF, Liu W, Loul S, Butel C, Liegeois F, Bienvenue Y, Ngolle EM, Sharp PM, Shaw GM, Delaporte E, Hahn BH, Peeters M. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature. 2006;444:164. doi: 10.1038/444164a. [DOI] [PubMed] [Google Scholar]
- Vlčková K, Mrázek J, Kopečný J, Petrželková KJ. Evaluation of different storage methods to characterise the fecal bacterial communities of captive western lowland gorillas (Gorilla gorilla gorilla) J Microbiol. Methods. 2012;91:41–45. doi: 10.1016/j.mimet.2012.07.015. [DOI] [PubMed] [Google Scholar]
- Waruiru RM, Munyua WK, Thamsborg SM, Nansen P, Bøgh HO, Gathuma JM. Development and survival of infective larvae of gastrointestinal nematodes of cattle on pasture in central Kenya. Vet. Res. Commun. 1998;22:315–323. doi: 10.1023/a:1006112802459. [DOI] [PubMed] [Google Scholar]
- Whittier CA, Cranfield MR, Stoskopf MK. Real-time PCR detection of Campylobacter spp. In free-ranging mountain gorillas (Gorilla beringei beringei) J. Wildl. Dis. 2010;46:791–802. doi: 10.7589/0090-3558-46.3.791. [DOI] [PubMed] [Google Scholar]
- Williams C, Pontén F, Moberg C, Söderkvist P, Uhlén M, Pontén J, Sitbon G, Lundeberg J. A high frequency of sequence alterations is due to formalin fixation of archival specimens. Am. J. Pathol. 1999;155:1467–1471. doi: 10.1016/S0002-9440(10)65461-2. [DOI] [PMC free article] [PubMed] [Google Scholar]