Summary
Detection of a 17p13.1 deletion (loss of TP53) or 11q22.3 deletion (loss of ATM), by fluorescence in situ hybridization (FISH), in chronic lymphocytic leukaemia (CLL) patients is associated with a poorer prognosis. Because TP53 and ATM are integral to the TP53 pathway, we hypothesized that 17p13.1- (17p-) and 11q22.3- (11q-) occurring in the same cell (clonal 17p-/11q-) would confer a worse prognosis than either 17p- or 11q-. We studied 2184 CLL patients with FISH (1995–2012) for the first occurrence of 17p-, 11q-, or clonal 17p-/11q-. Twenty (1%) patients had clonal 17p-/11q-, 158 (7%) had 17p- (including 4 with 17p- and 11q- in separate clones), 247 (11%) had 11q-, and 1759 (81%) had neither 17p- nor 11q-. Eleven of 15 (73%) tested patients with clonal 17p-/11q- had dysfunctional TP53 mutations. Overall survival for clonal 17p-/11q- was significantly shorter (1.9 years) than 17p- (3.1 years, p = 0.04), 11q- (4.8 years, p = <0.0001), or neither 17p- nor 11q- (9.3 years, p = <0.0001). Clonal 17p-/11q- thus conferred significantly worse prognosis, suggesting that loss of at least one copy of both TP53 and ATM causes more aggressive disease. Use of an ATM/TP53 combination FISH probe set could identify these very-high risk patients.
Keywords: Chronic lymphocytic leukaemia, prognostic factors, fluorescencein situ hybridization (FISH), TP53, ATM
Introduction
Chronic lymphocytic leukaemia/small lymphocytic lymphoma (CLL) is a heterogeneous disease with a diverse clinical presentation, response to treatment and survival. In addition to clinical staging, there are now well-validated biological prognostic markers that can be used in the management of CLL (Damle, et al 1999, Dohner, et al 2000, Hamblin, et al 1999). One of the most useful tests in current clinical practice is interphase fluorescence in situ hybridization (FISH) to detect chromosomal defects. The hierarchical risk model of FISH-detected chromosomal defects in CLL considers patients with chromosome 17p13.1 deletions (17p-), which includes loss of the TP53 gene locus, to have the highest risk of disease progression and the worst prognosis followed by those patients with deletion of 11q22.3 (11q-), including loss of the ATM gene locus (Dohner, et al 2000). These lesions are nearly always monoallelic in patients with CLL (Zenz, et al 2008).
The prognostic consequences of either 17p- or 11q- are considerably influenced by the integrity of the remaining allele of TP53 or ATM, respectively. In patients with progressive CLL, there is a dysfunctional mutation in the remaining TP53 allele in the majority of patients with 17p- (80 – 90%) and in the remaining ATM allele in approximately one-third of patients with 11q- (Austen, et al 2007, Dohner, et al 1995, Pospisilova, et al 2012, Zenz, et al 2010). Because TP53 and ATM are integral components of the TP53 DNA damage response pathway, we hypothesized that patients with both 17p- and 11q- in the same CLL cells would have a worse prognosis than patients with either of these deletions alone. To test this hypothesis, we compared the clinical outcome of patients with 17p- and 11q- in the same cells, 17p- alone, 11q- alone and neither 17p- or 11q-.
Materials and methods
Patients
This study was performed with approval of the Mayo Clinic Foundation Institutional Review Board according to the principles of the Declaration of Helsinki. Of the 3711 CLL patients seen in the Division of Hematology between 1 January 1995 and 29 March 2012, 2184 (59%) had at least one FISH assay and were included in this study. We used the Mayo Clinic CLL and Cytogenetics Laboratory databases to collect data on demographics, date and stage at diagnosis, prognostic factors (IGHV somatic hypermutation, CD38 and ZAP70 expression, and FISH), treatment, vital status, and last known alive date. The detection of first occurrence of 17p-, 11q-, or 17p- and 11q- by FISH was considered the sentinel event. Patients were grouped into four cohorts based on the sentinel event and the hierarchical classification of FISH defects (Dohner, et al 2000): 1) 17p- and 11q- in the same clonal CLL cells (“double hit”), 2) 17p-, 3) 11q-, and 4) neither 17p- nor 11q-. Patients with 17p- and 11q- in different clones were considered to have 17p-, and patients with either 17p- or 11q- who were subsequently found to have clonal evolution with both 17p- and 11q- in the same CLL cells were considered to have 17p- and 11q-.
FISH
Interphase FISH studies were performed on blood or bone marrow specimens processed by standard methods for uncultured samples (Dewald, et al 2003). All samples were studied with the standard Mayo Clinic CLL FISH panel that includes probes for D6Z1 (centromere 6) and MYB (6q23.3), D11Z1 (centromere 11) and ATM (11q22.3), D12Z3 (centromere 12) and MDM2 (12q15), D13S319 (13q14.3) and LAMP1 (13q34), D17Z1 (centromere 17) and TP53 (17p13.1) and translocations involving IGH at 14q32. Two technologists each independently scored 100 nuclei, for a total of 200 nuclei per probe set. All patients positive for both 17p and 11q deletions were reflexed to testing with the Abbott Molecular (Des Plaines, IL) ATM/TP53 combination probe set to determine whether the 17p and 11q deletions were present in the same cells or different cells (Figure 1). A validation study was performed using the combination ATM/TP53 probe set to determine normal cutoffs for the various patterns seen, including the 1 red, 1 green (1R1G) pattern seen when a deletion of both probes is detected. Blood or bone marrow fixed cell pellets from 25 cytogenetically normal patients were each scored by two independent technologists. Each scorer analysed 100 consecutive nuclei for a total of 200 nuclei per sample. The maximum number of false positive cells for each signal pattern was put into the beta inverse formula using a 95% confidence interval and rounded to the next highest half number to determine the normal cutoff value. Normal cutoffs values include; 17p- <7.0%, 11q- <6.0% and 17p- and 11q- together <2.5%.
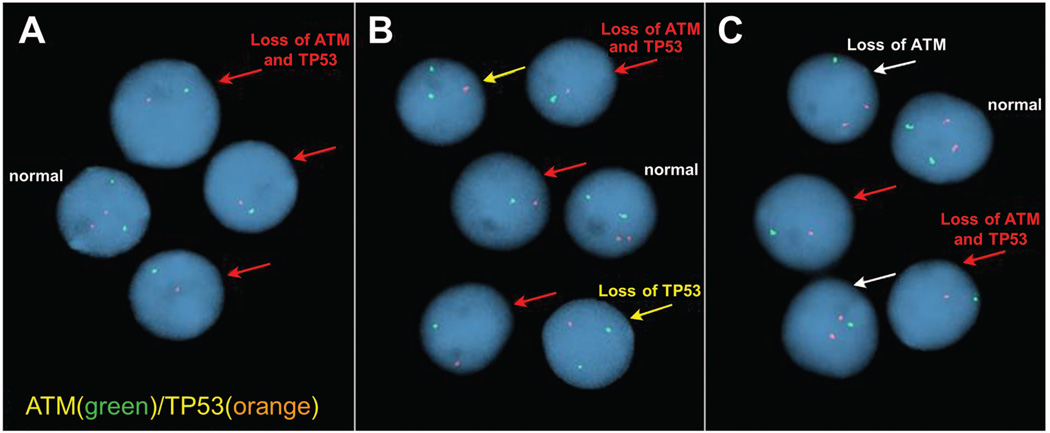
Figure 1. Representative FISH images from three patient samples hybridized with the Abbott Molecular ATM/TP53 combination probe set.
A) Patient with 17p- and 11q- in the same cells (“double hit”). B) Patient with isolated 17p- as well as 17p- and 11q- (“double hit”) in a sub-clone. C) Patient with isolated 11q- as well as 17p- and 11q- (“double hit”) in a sub-clone. Nuclei identified with a red arrow have loss of both TP53 and ATM. The yellow arrows show nuclei with loss of TP53 only. The white arrows indicate nuclei with loss of ATM only. All images were captured at room temperature with a Leica DM5000B microscope at 1000× magnification (100×/1.40-0.70 oil immersion APO objective) and processed using Cytovision software version 4.5.2.Probes were labelled with Spectrum Orange (TP53) and Spectrum Green (ATM) and cells were counterstained with 4',6-diamidino-2-phenylindole (DAPI).
TP53 Sequencing
TP53 was sequenced in 15 of the 20 patients in the cohort with clonal deletions of both 17p and 11q and available DNA. Frozen white blood cell pellets or methanol-acetic acid fixed cell pellets were washed two times with phosphate-buffered saline before extracting DNA with the EZ1 tissue cartridges (Qiagen GmbH, Hilden, Germany). Genomic DNA was amplified by polymerase chain reaction (PCR) with Go Taq Hot Start Polymerase (Promega, Madison, WI) using 4 sets of PCR primers to cover TP53 exons 4 through 9 (Figure S1). Products were analysed on the QIAxcel (Qiagen), treated with Exo SAP-IT (USB, Cleveland, OH) and sequenced with the BigDye v1.1 sequencing kit (Applied Biosystems, Grand Island, NY). The same primers used for amplification were also used for sequencing. Sequencing reactions were run on the ABI 3130×l Genetic Analyzer (Applied Biosystems) and analysed in the Sequencher™ software (Gene Codes, Ann Arbor, MI). All mutations found were confirmed with a second PCR and sequencing reaction. Mutations were also analysed for the predicted effect using the UMD TP53 database: http://p53.fr/Database_download/Database_download.html.
Statistical Considerations
For the four groups, descriptive statistics and comparisons were computed using the appropriate statistical test (Chi-square, Fisher’s exact, or Kruskal-Wallis tests). Overall survival (OS) and time to next treatment (TTT) were calculated from the CLL sentinel FISH event until last known alive date or death date for OS and until next treatment or last known untreated date for TTT. OS and TTT are displayed using Kaplan-Meier curves, and significance between the four groups was determined using log-rank tests. A second measurement of OS, from time of CLL diagnosis, was run on patients who ultimately acquired a “double hit”. Only treatments following the sentinel FISH date were considered for calculation of TTT. Descriptive characteristics of the 20 individuals who had both mutations in the same clone were reviewed and updated as of 29 August 2012. P-values ≤0.05 were considered statistically significant, and all statistical analyses were performed using SAS 9.2 (SAS Institute; Cary, NC).
Results
Of the 2184 CLL patients studied (median follow-up from diagnosis of 5.1 years, range 0– 30.4 years; median follow-up from sentinel event of 3.4 years, range 0 – 17.6 years), 20 (1%) had both 17p- and 11q- in the same cells (“double hit”) at some point during the course of their disease, 158 (7%) had 17p-, 247 (11%) had 11q-, and 1759 (81%) had neither 17p- nor 11q- (Table I). Four patients had both 17p- and 11q- in separate clones and were included in the 17p- group for analysis. The 20 patients with clonal 17p- and 11q- comprised 11% of all patients with 17p- and 7% of all patients with 11q-. Only 2/20 (10%) had never received therapy for CLL before the censor date (Table I). An additional cell population with only 17p- (15–65% of interphase nuclei) was found in 7/20 patients and 3/20 had a cell population with only 11q- (8–20% of interphase nuclei). In 1/20 patients there were additional cell populations with 17p- (10% of interphase nuclei) and 11q- (10% of nuclei) (Table II).
Table I.
Patient Characteristics
| 11q- Only (N=247) |
17p- Only (N=158) |
Both (N=20) |
Neither (N=1759) |
Total (N=2184) |
|
|---|---|---|---|---|---|
| Gender | |||||
| female | 67 (27.1%) | 66 (41.8%) | 5 (25.0%) | 571 (32.5%) | 709 (32.5%) |
| male | 180 (72.9%) | 92 (58.2%) | 15 (75.0%) | 1188 (67.5%) | 1475 (67.5%) |
| Survival Status | |||||
| Alive | 150 (60.7%) | 77 (48.7%) | 6 (30.0%) | 1322 (75.2%) | 1555 (71.2%) |
| Dead | 97 (39.3%) | 81 (51.3%) | 14 (70.0%) | 437 (24.8%) | 629 (28.8%) |
| Clinical Stage (Rai) at Diagnosis | |||||
| Missing | 22 | 14 | 1 | 117 | 154 |
| Rai 0: Low risk | 93 (41.3%) | 73 (50.7%) | 5 (26.3%) | 917 (55.8%) | 1088 (53.6%) |
| Rai 1 or 2: Intermediate risk | 114 (50.7%) | 59 (41.0%) | 9 (47.4%) | 653 (39.8%) | 835 (41.1%) |
| Rai 3 or 4: High risk | 18 (8.0%) | 12 (8.3%) | 5 (26.3%) | 72 (4.4%) | 107 (5.3%) |
| CD38 | |||||
| Missing | 21 | 11 | 1 | 176 | 209 |
| Negative | 102 (45.1%) | 87 (59.2%) | 9 (47.4%) | 1151 (72.7%) | 1349 (68.3%) |
| Positive | 124 (54.9%) | 60 (40.8%) | 10 (52.6%) | 432 (27.3%) | 626 (31.7%) |
| IGHV Mutation Status | |||||
| Missing | 72 | 46 | 4 | 672 | 794 |
| Mutated | 31 (17.7%) | 31 (27.7%) | 4 (25.0%) | 671 (61.7%) | 737 (53.0%) |
| Unmutated | 144 (82.3%) | 81 (72.3%) | 12 (75.0%) | 416 (38.3%) | 653 (47.0%) |
| ZAP70 | |||||
| Missing | 61 | 32 | 4 | 434 | 531 |
| Negative | 84 (45.2%) | 49 (38.9%) | 8 (50.0%) | 859 (64.8%) | 1000 (60.5%) |
| Positive | 102 (54.8%) | 77 (61.1%) | 8 (50.0%) | 466 (35.2%) | 653 (39.5%) |
| Treatment Status* | |||||
| Not treated | 64 (25.9%) | 29 (18.4%) | 2 (10.0%) | 1027 (58.4%) | 1122 (51.4%) |
| Treated | 183 (74.1%) | 129 (81.6%) | 18 (90.0%) | 732 (41.6%) | 1062 (48.6%) |
| Age at Diagnosis | |||||
| N | 247 | 158 | 20 | 1759 | 2184 |
| Median (years) | 59.9 | 61.5 | 63.6 | 62.6 | 62.2 |
| Q1, Q3 (years) | 52.6, 67.7 | 54.0, 70.2 | 54.4, 68.8 | 54.3, 70.3 | 54.1, 70.0 |
| Range (years) | (21.9–89.1) | (33.2–84.7) | (35.3–78.0) | (28.0–92.9) | (21.9–92.9) |
| Years Between Diagnosis & Sentinel FISH | |||||
| N | 247 | 158 | 20 | 1759 | 2184 |
| Median | 1.8 | 3.2 | 3.5 | 0.3 | 0.5 |
| Q1, Q3 | 0.1, 5.8 | 0.2, 7.0 | 1.2, 7.8 | 0.0, 4.0 | 0.1, 4.4 |
| Range | (0.0–36.3) | (0.0–31.0) | (0.0–26.1) | (0.0–43.8) | (0.0–43.8) |
Treatment status was assessed at time of censor date
FISH, fluorescence in situ hybridization
Table II. Characteristics of 20 patients found to have “double hit” chronic lymphocytic leukaemia.
Patients with both 17p- and 11q- (“double hit”) were more likely to be males who presented with more advanced stage disease, unmutated IGHV, and to be previously treated at the time of the sentinel FISH event. Overall, 4/20 patients (20%) had at least one clonal evolution event prior to acquisition of the double hit.
| Patient demographics | FISH Testing (%) | Laboratory Results | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient | Sex | Age at Diagnosis (years) |
Survival Status |
OS | Tx Status |
TTT | Rai stage |
11q- (ATM) |
17p- (TP53) |
17p- & 11q- |
Clonality | CD38 | ZAP70 |
IGHV Mut |
TP53 Mut |
| 1 | F | 76 | Dead | 3.57 | − | 0.04 | na | 10 | 11 | 58 | “Double Hit” | + | na | na | na |
| 2 | M | 42 | Dead | 0.86 | + | 0.05 | IV | − | − | 94 | “Double Hit” | + | + | − | + |
| 3 | M | 65 | Dead | 0.28 | + | 0.02 | I | − | − | 37 | “Double Hit” | na | na | na | na |
| 4 | F | 49 | Dead | 2.98 | + | 1.28 | I | − | 15 | 35 | “Double Hit”* | − | − | + | + |
| 5 | M | 59 | Dead | 2.14 | + | 0.10 | I | − | 55 | 8 | “Double Hit”* | − | − | + | + |
| 6 | M | 75 | Dead | 2.45 | + | 0.05 | 0 | − | 62 | 30 | “Double Hit”* | + | − | − | + |
| 7 | M | 64 | Dead | 1.41 | + | 0.04 | 0 | − | − | 86 | “Double Hit” | − | na | − | + |
| 8 | F | 70 | Dead | 2.55 | + | 0.01 | III | − | − | 88 | “Double Hit” | + | + | − | + |
| 9 | M | 63 | Dead | 0.06 | + | 0.01 | III | − | 65 | 32 | “Double Hit”* | + | − | − | − |
| 10 | F | 63 | Dead | 1.62 | + | 0.05 | 0 | − | − | 92 | “Double Hit” | − | + | − | + |
| 11 | M | 42 | Dead | 1.85 | + | 0.25 | I | 20 | − | 64 | “Double Hit”^ | + | − | − | +# |
| 12 | M | 67 | Dead | 1.45 | + | 0.04 | IV | 8 | − | 59 | “Double Hit ^ | + | na | na | na |
| 13 | M | 62 | Alive | 2.69 | + | 0.07 | I | − | − | 67 | “Double Hit” | + | − | na | na |
| 14 | M | 35 | Alive | 1.60 | + | 1.57 | I | − | 52 | 47 | “Double Hit”* | + | − | + | + |
| 15 | M | 76 | Dead | 1.03 | + | 0.12 | I | 14 | − | 77 | “Double Hit”^ | − | + | − | + |
| 16 | M | 60 | Alive | 1.97 | + | 1.50 | I | − | − | 71 | “Double Hit” | − | + | − | + |
| 17 | M | 56 | Alive | 1.21 | + | 0.07 | IV | − | − | 46 | “Double Hit” | − | + | − | − |
| 18 | M | 52 | Dead | 0.88 | + | 0.06 | I | − | 22 | 17 | “Double Hit”* | − | + | − | − |
| 19 | F | 78 | Alive | 0.43 | − | 0.43 | 0 | − | − | 84 | “Double Hit” | − | − | + | na |
| 20 | M | 64 | Alive | 0.00 | + | 0.00 | 0 | − | 52 | 42 | “Double Hit”* | + | + | − | − |
OS, overall survival; Tx, treatment; TTT, time to treatment from sentinel fluorescence in situ hybridization (FISH); Normal cutoffs for FISH testing: 11q- (6.0%), 17p- (7.0%), 17p- and 11q- (2.5%); IGHV Mut, results for IGHV somatic hypermutation testing; TP53 Mut, results for TP53 mutation testing;
double hit with separate 17p- clone;
double hit with separate 11q- clone;
2 different TP53 mutations seen in this patient;
na, not available.
Patients with both 17p- and 11q- were more likely to be males diagnosed with more advanced stage disease and 60% had received treatment prior to the discovery of their “double hit” FISH event (Table II). A “double hit” was observed in the diagnostic FISH study of 14 (70%) patients and was documented to be a result of clonal evolution in six patients, as detailed in Table III. Four of these six patients (Patients 7, 17, 18, and 20) experienced serial evolution with at least one clonal evolution event prior to acquisition of a “double hit”. Two of the six patients (Patients 11 and 16) had at least one negative FISH study prior to acquiring a “double hit”. Median time from diagnosis to the sentinel FISH event was 3.5 years (range 0 – 26 years) for 17p- and 11q-, 3.2 years for 17p-, 1.8 years for 11q- and 0.3 years for neither 17p- nor 11q-.
Table III. Serial FISH results in six patients with clonal evolution.
Patients 11 and 16 evolved from a negative first fluorescence in situ hybridization (FISH) to acquisition of a “double hit” (DH). Patient 7 had two clonal evolution events, progressing from an isolated 11q- to a “double hit” and then retention of the “double hit” with acquisition of 13q- and loss of chromosome 12 centromere. Patient 17 evolved from isolated +12 to negative FISH and then acquired a “double hit” which was not observed, in the 6th FISH study, after the patient had undergone an allogeneic bone marrow transplant. Patient 18 had a homozygous 13q deletion at first FISH and evolved to a “double hit” with homozygous 13q deletion. Patient 20 had both 13q and 17p deletions, later acquired a 6q deletion and finally acquired an 11q deletion for the “double hit”.
| Patient | 1st FISH Days since Dx |
2nd FISH Days since Dx |
3rd FISH Days since Dx |
4th FISH Days since Dx |
5th FISH Days since Dx |
6th FISH Days since Dx |
Clonal Evolution Summary |
|---|---|---|---|---|---|---|---|
| 7 | 11q- (81%) | 11q- > DH > DH + 13q- + −12cen | |||||
| 13q- (11%) | |||||||
| 11q- (83%) | 11q- (94%) | 17p- (80%) | |||||
| 11q- (45%) | 11q- (57%) | 11q- (11%) | 17p- (74%) | 17p- (97%) | −12cen (10%) | ||
| 975 | 1099 | 1263 | 1781 | 1920 | 2215 | ||
| 11 | 11q- (70%) | 11q- (66%) | neg > DH | ||||
| negative | negative | negative | negative | 17p- (51%) | 17p- (66%) | ||
| 1394 | 2021 | 2703 | 2821 | 3218 | 3490 | ||
| 16 | 11q- (70%) | 11q- (65%) | 11q- (58%) | neg > DH | |||
| negative | 17p- (61%) | 17p- (56%) | 11q- (8%) | 17p- (54%) | |||
| 26 | 1500 | 1647 | 1897 | 1989 | |||
| 17 | 11q- (49%) | +12 > neg > DH | |||||
| +12 (14%) | negative | negative | 17p- (49%) | negative | |||
| 48 | 157 | 214 | 903 | 1126 | |||
| 18 | 11q- (15%) | 13q- > DH + 13q- | |||||
| 13q-x2 (96%) | 13q-x2 (28%) | ||||||
| 13q-x2 (80%) | 13q-x2 (4%) | 13q-x2 (24%) | 17p- (32%) | 17p- (29%) | |||
| 148 | 349 | 645 | 2233 | 2460 | |||
| 20 | 6q- (27%) | 13q- + 17p- > 6q- +13q- + 17p- > DH + 6q- +13q- | |||||
| 6q- (26%) | 11q- (47%) | ||||||
| 13q- (86%) | 13q- (72%) | 13q- (79%) | 13q- (88%) | 13q- (48%) | |||
| 17p- (87%) | 17p- (84%) | 13q- (8%) | 17p- (81%) | 17p- (85%) | 17p- (92%) | ||
| 29 | 103 | 195 | 528 | 720 | 2490 | ||
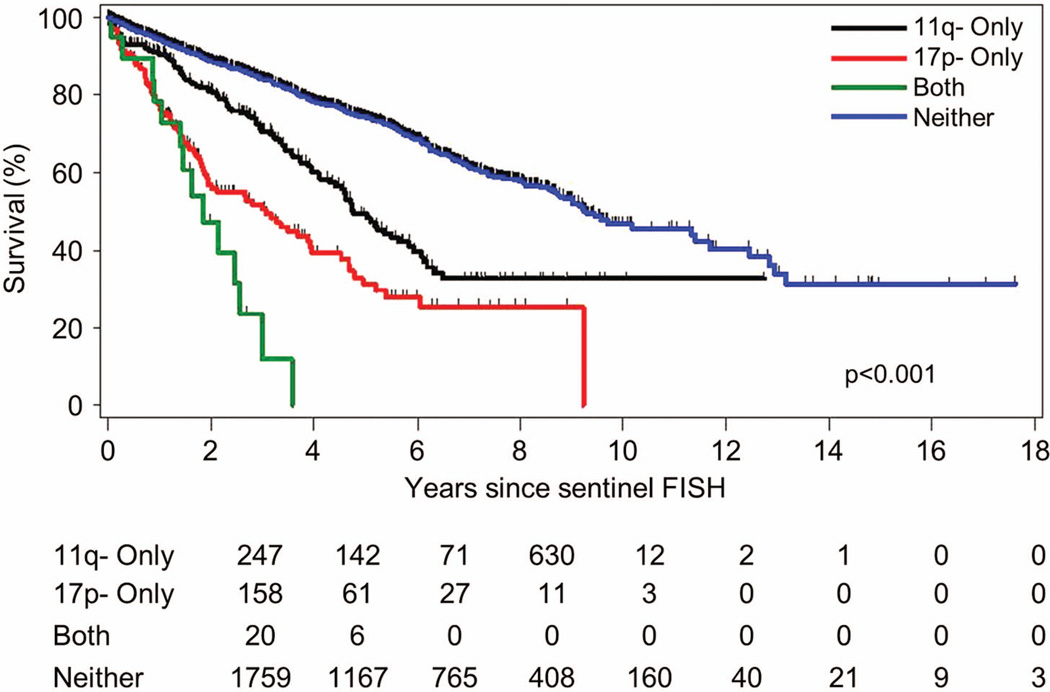
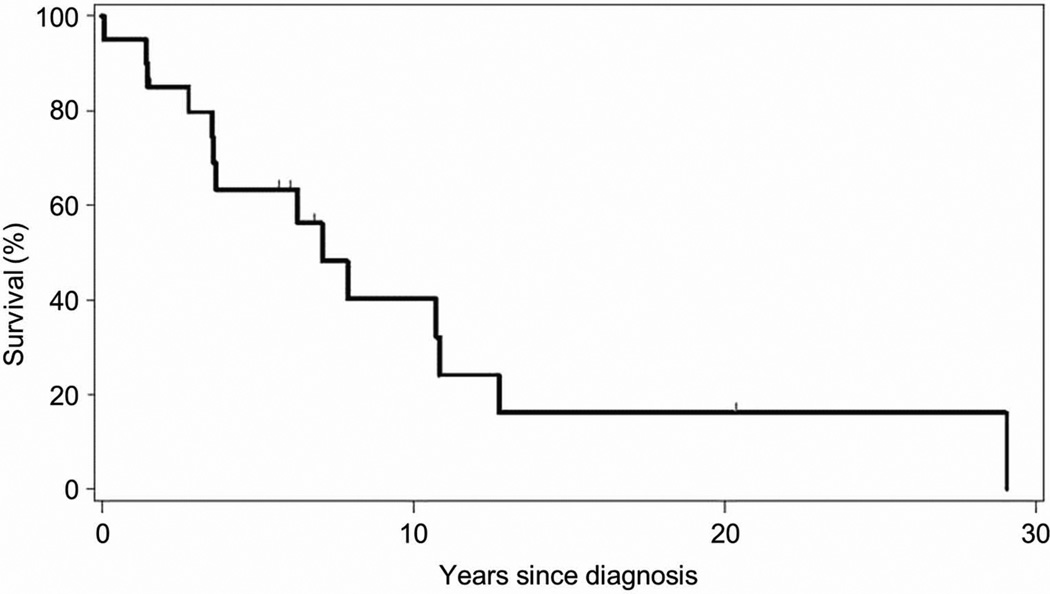
Median OS from the sentinel FISH event was significantly shorter for the patients with 17p- and 11q- (1.9 years) compared to those with 17p- (3.1 years, p = 0.04), 11q- (4.8 years, p = <0.0001), and neither 17p- nor 11q- (9.3 years, p = <0.0001) (Figure 2). The median OS from the time of diagnosis of their CLL for the patients who developed a “double hit” lesion was 7.1 years (Figure 3). Because repeat FISH analysis is usually performed when patients have progressive disease requiring therapy, TTT from the sentinel FISH event was not significantly shorter for patients with 17p- and 11q- (0.07 years) compared to those with 17p- (0.2 years, p = 0.36), or 11q- (0.6 years, p = 0.15). TTT in patients with neither 17p- nor 11q- was 5.2 years (Figure S2).
Figure 2. Overall survival (OS) stratified by the 4 FISH cohorts.
OS was significantly different between the four groups (p <0.0001) with median survival of 1.9 years in the both 17p- and 11q- group (“double hit”), 3.1 years in 17p-, 4.8 years in 11q-, and 9.3 years (p <0.0001) in the neither 17p- nor 11q- group. OS was significantly shorter in the patients with both 17p- and 11q- in the same cells (“double hit”) compared to those with 17p- (p = 0.04).
Figure 3. Median overall survival (OS) from diagnosis of CLL for patients who ultimately acquire “double hit” is poor (7.1 years).
Patients who ultimately acquire both 17p and 11q deletions (“double hit”) have a poor prognosis from time of diagnosis of CLL with a median OS of 7.1 years. When median survival for these patients is considered from the time-point of sentinel FISH event (when 17p- and 11q- are acquired), their prognosis is very poor (median OS 1.9 years). See Figure 2.
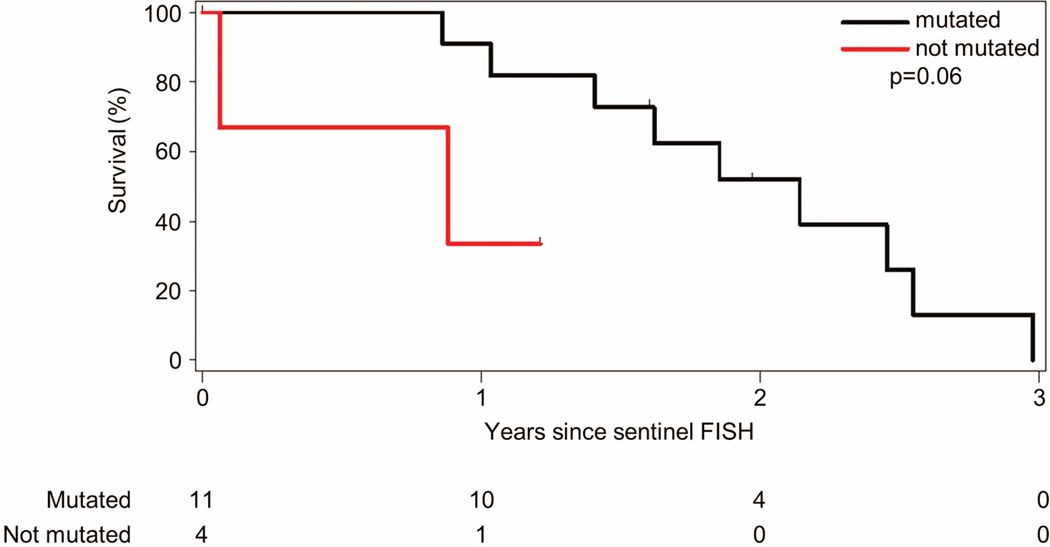
TP53 exons 4–9 were sequenced in 15 of the patients in the 17p- and 11q- cohort. Eleven (73%) patients had DNA changes predicted to be deleterious to normal TP53 protein function (http://p53.free.fr/Database/p53_database.html). These included 9 patients with somatic point mutations. Five of these point mutations involved the TP53 DNA binding motifs (4 within the major groove binding region, 1 in the minor groove region), and 4 were located near these structural motifs. Two patients had frameshift alterations resulting from partial exon deletions, including one patient with a concurrent point mutation. One patient had a point mutation that altered the splice site after exon 9 and is of unknown significance. Median OS (Figure 4) for patients with TP53 mutations were not significantly different to patients without TP53 mutations (2.1 years vs. 0.9 years, p = 0.06) (Table SI).
Figure 4. Overall survival stratified by TP53 mutation analysis.
TP53 analysis performed in samples from 15 patients with clonal 17p- and 11q- (“double hit”) showed that 11 patients had mutations in the remaining TP53 allele predicted to be deleterious to normal TP53 protein function. Median overall survival was 2.1 years for those with TP53 mutations and 0.9 years for those without mutations (p = 0.06).
Discussion
We found that clonal 17p-and 11q-, which occurs in 1% of patients with CLL, predicts a significantly shorter survival than either 17p- or 11q- alone. These data support our hypothesis and show the value of using the dual ATM/TP53 FISH probe set to examine CLL cell populations that have 17p- and 11q-. In CLL cells, 17p- or 11q- only rarely involve both chromosomes, and the biological consequences of these lesions are largely dependent in part on the integrity of the remaining allele. In patients with previously untreated progressive CLL, approximately 80% of those with 17p- have a dysfunctional mutation in their remaining TP53, (Dohner, et al 1995, Pospisilova, et al 2012, Zenz, et al 2010) and 36% of those with 11q- have a dysfunctional mutation in ATM (Austen, et al 2007, Skowronska, et al 2012). These patients have poor responses to purine analogue-containing chemoimmunotherapy and, thus, shorter survival (Austen, et al 2007). In addition, patients without 17p- or 11q- who have TP53 or ATM mutations predicted to cause gene dysfunction also have a poor response to chemoimmunotherapy and shorter survival (Dicker, et al 2009, Rossi, et al 2009, Skowronska, et al 2012, Zenz, et al 2010, Zenz, et al 2008). Our data shows that CLL patients with clonal 17p- and 11q- have a very poor prognosis. In addition, we were able to sequence exons 4–9 of TP53 in 15 of these patients: 11 had mutations predicted to result in a dysfunctional TP53 protein. In this small sample, patients with TP53 mutations, unexpectedly, did not have a worse outcome. This is an intriguing finding and if validated, generates several hypotheses on the relationship between TP53 and ATM abnormalities in the pathobiology of CLL. The poor prognosis of patients with clonal 17p- and 11q- could imply an additive or synergistic effect of haploinsufficiency for both genes. Alternatively, the poor prognosis of these patients could be attributable to loss of other genes included in these deletions. The data on the poorer prognosis of patients with 11q- without mutations in the remaining ATM allele compared to patients without 11q- and a monoallelic ATM mutation suggests a role for loss of additional genes within this deletion (Ouillette, et al 2012). These hypotheses could be tested in future studies using cells from patients with clonal 17p- and 11q-.
The value of the data from this study derives from the size of the population studied, the prospective collection of data from 1995 to 2012, and the ability to include all FISH studies done at a single large centre over this period of time. The weaknesses of the study include the retrospective collection of some data, the non-uniform timing of FISH assays, and the incomplete set of prognostic data. For the population of patients with clonal 17p- and 11q-, we had insufficient high quality DNA to conduct TP53 mutation analysis on 5 patients and were not able to perform ATM mutation analysis on any of the samples. The small size of this population also limited our ability to perform subgroup analysis. One potential limitation of the clinical value of our finding that clonal 17p- and 11q- is associated with very poor prognosis could be that this is a late event in CLL, and it reflects genomic instability rather than the adverse biological consequences of both deletions in the same cells. While the finding of clonal 17p- and 11q- was a late event in several patients in our study, we have recently detected clonal 17p- and 11q- in a patient with newly diagnosed clinical monoclonal B-cell lymphocytosis (MBL) with CLL immunophenotype who has subsequently progressed to CLL. This patient was not included in our study cohort because she was diagnosed after our analysis end date. In this patient, FISH done at diagnosis showed that 50% of circulating blood nuclei had a 17p deletion and 30% had clonal 17p- and 11q- (“double hit”). TP53 exon 4–9 sequencing showed a TP53 mutation (insertion/deletion with frameshift). This case shows that development of clonal 17p- and 11q- can occur in clinically early stage CLL. Finally, our practice of repeating FISH analysis to detect clonal evolution in patients with progression of disease who require treatment could have biased our study population outcome by selection of patients with clinically more aggressive disease. However, 14 patients in our study were found to have the “double hit” at time of first FISH analysis, and this underscores that clonal 17p- and 11q- can occur in clinically early stage CLL.
In conclusion, we report the novel finding that clonal 17p- and 11q- in CLL is associated with a very poor prognosis and suggest that this is important for both understanding the biology of the disease and management of patients with CLL. Although there is currently no specific treatment for this group of patients, further examination of the contributing genetic defects could provide important data for future targeted therapy strategies. In addition, our study suggests that in a clone of CLL cells, loss of both a single TP53 and a single ATM gene could have similar consequences to the previously described complete loss of function of TP53 or ATM (Austen, et al 2007, Rossi, et al 2009, Skowronska, et al 2012, Stilgenbauer and Zenz 2010) and further molecular analysis of these “double hit” cells could provide novel insights into the biology of CLL.
Acknowledgments
The authors thank Neil E. Kay, M.D., for helpful discussions and Cherish Grabau for administrative assistance in the preparation of this manuscript.
P.T.G., S.A.S., K.G.R., and C.S.Z. designed the research; P.T.G., S.A.S., D.S.V., L.S.F., K.G.R., R.G.S., and C.S.Z. performed the research; P.T.G., S.A.S., D.S.V., L.S.F., K.G.R., R.G.S., D.L.V., R.C.T., and T.D.S. contributed vital new reagents or analytical tools; P.T.G., S.A.S., D.S.V., L.S.F., K.G.R., R.G.S., S.L.S., D.L.V., T.D.S., and C.S.Z. analysed data and wrote the paper.
This work was supported by the National Institutes of Health, Mayo Clinic Cancer Center (CA15083) (P.T.G.) and University of Iowa/Mayo Clinic Lymphoma SPORE (CA CA97274) (C.S.Z.).
Footnotes
The authors declare no competing financial interests.
Presented at the 54th Annual Meeting of the American Society of Hematology, Atlanta, GA, December 2012, Abstract 2486.
References
- Austen B, Skowronska A, Baker C, Powell JE, Gardiner A, Oscier D, Majid A, Dyer M, Siebert R, Taylor AM, Moss PA, Stankovic T. Mutation status of the residual ATM allele is an important determinant of the cellular response to chemotherapy and survival in patients with chronic lymphocytic leukemia containing an 11q deletion. Journal of Clinical Oncology. 2007;25:5448–5457. doi: 10.1200/JCO.2007.11.2649. [DOI] [PubMed] [Google Scholar]
- Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, Buchbinder A, Budman D, Dittmar K, Kolitz J, Lichtman SM, Schulman P, Vinciguerra VP, Rai KR, Ferrarini M, Chiorazzi N. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–1847. [PubMed] [Google Scholar]
- Dewald GW, Brockman SR, Paternoster SF, Bone ND, O'Fallon JR, Allmer C, James CD, Jelinek DF, Tschumper RC, Hanson CA, Pruthi RK, Witzig TE, Call TG, Kay NE. Chromosome anomalies detected by interphase fluorescence in situ hybridization: correlation with significant biological features of B-cell chronic lymphocytic leukaemia. British Journal of Haematology. 2003;121:287–295. doi: 10.1046/j.1365-2141.2003.04265.x. [DOI] [PubMed] [Google Scholar]
- Dicker F, Herholz H, Schnittger S, Nakao A, Patten N, Wu L, Kern W, Haferlach T, Haferlach C. The detection of TP53 mutations in chronic lymphocytic leukemia independently predicts rapid disease progression and is highly correlated with a complex aberrant karyotype. Leukemia. 2009;23:117–124. doi: 10.1038/leu.2008.274. [DOI] [PubMed] [Google Scholar]
- Dohner H, Fischer K, Bentz M, Hansen K, Benner A, Cabot G, Diehl D, Schlenk R, Coy J, Stilgenbauer S, Volkmann M, Galle PR, Poustka A, Hunstein W, Lichter P. p53 gene deletion predicts for poor survival and non-response to therapy with purine analogs in chronic B-cell leukemias. Blood. 1995;85:1580–1589. [PubMed] [Google Scholar]
- Dohner H, Stilgenbauer S, Benner A, Leupolt E, Kröber A, Bullinger L, Döhner K, Bentz M, Lichter P. Genomic aberrations and survival in chronic lymphocytic leukemia. New England Journal of Medicine. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- Hamblin T, Davis Z, Gardiner A, Oscier D, Stevenson F. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94:1848–1854. [PubMed] [Google Scholar]
- Ouillette P, Li J, Shaknovich R, Li Y, Melnick A, Shedden K, Malek SN. Incidence and clinical implications of ATM aberrations in chronic lymphocytic leukemia. Genes, Chromosomes & Cancer. 2012;51:1125–1132. doi: 10.1002/gcc.21997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospisilova S, Gonzalez D, Malcikova J, Trbusek M, Rossi D, Kater AP, Cymbalista F, Eichhorst B, Hallek M, Dohner H, Hillmen P, van Oers M, Gribben J, Ghia P, Montserrat E, Stilgenbauer S, Zenz T. ERIC recommendations on TP53 mutation analysis in chronic lymphocytic leukemia. Leukemia. 2012;26:1458–1461. doi: 10.1038/leu.2012.25. [DOI] [PubMed] [Google Scholar]
- Rossi D, Cerri M, Deambrogi C, Sozzi E, Cresta S, Rasi S, De Paoli L, Spina V, Gattei V, Capello D, Forconi F, Lauria F, Gaidano G. The prognostic value of TP53 mutations in chronic lymphocytic leukemia is independent of Del17p13: implications for overall survival and chemorefractoriness. Clinical Cancer Research. 2009;15:995–1004. doi: 10.1158/1078-0432.CCR-08-1630. [DOI] [PubMed] [Google Scholar]
- Skowronska A, Parker A, Ahmed G, Oldreive C, Davis Z, Richards S, Dyer M, Matutes E, Gonzalez D, Taylor AM, Moss P, Thomas P, Oscier D, Stankovic T. Biallelic ATM Inactivation Significantly Reduces Survival in Patients Treated on the United Kingdom Leukemia Research Fund Chronic Lymphocytic Leukemia 4 Trial. Journal of Clinical Oncology. 2012 doi: 10.1200/JCO.2011.41.0852. [DOI] [PubMed] [Google Scholar]
- Stilgenbauer S, Zenz T. Understanding and managing ultra high-risk chronic lymphocytic leukemia. Hematology / the Education Program of the American Society of Hematology. 2010;2010:481–488. doi: 10.1182/asheducation-2010.1.481. [DOI] [PubMed] [Google Scholar]
- Zenz T, Krober A, Scherer K, Habe S, Buhler A, Benner A, Denzel T, Winkler D, Edelmann J, Schwanen C, Dohner H, Stilgenbauer S. Monoallelic TP53 inactivation is associated with poor prognosis in chronic lymphocytic leukemia: results from a detailed genetic characterization with long-term follow-up. Blood. 2008;112:3322–3329. doi: 10.1182/blood-2008-04-154070. [DOI] [PubMed] [Google Scholar]
- Zenz T, Eichhorst B, Busch R, Denzel T, Habe S, Winkler D, Buhler A, Edelmann J, Bergmann M, Hopfinger G, Hensel M, Hallek M, Dohner H, Stilgenbauer S. TP53 mutation and survival in chronic lymphocytic leukemia. Journal of Clinical Oncology. 2010;28:4473–4479. doi: 10.1200/JCO.2009.27.8762. [DOI] [PubMed] [Google Scholar]