Abstract
Sexually antagonistic genetic variation can pose limits to the independent evolution and adaptation of the sexes. The extent of sexually antagonistic variation is reflected in the intersex genetic correlation for fitness (rwFM). Previous estimates of this correlation have been mostly limited to populations in environments to which they are already well adapted, making it difficult to gauge the importance of sexually antagonistic genetic variance during the early stages of adaptation, such as that occurring following abrupt environmental change or upon the colonization of new habitat. Here we assayed male and female lifetime fitness in a population of Drosophila serrata in four novel laboratory environments. We found that rwFM varied significantly across environments, with point estimates ranging from positive to negative values of considerable magnitude. We also found that the variability among estimates was because, at least in part, of significant differences among environments in the genetic variances of both male and female fitness, with no evidence of any significant changes in the intersex covariance itself, although standard errors of these estimates were large. Our results illustrate the unpredictable nature of rwFM in novel environments and suggest that, although sexually antagonistic genetic variance can be pronounced in some novel environments, it may have little effect in constraining the early stages of adaptation in others.
Keywords: intralocus sexual conflict, sexually antagonistic, environmental dependence, genetic covariance
Introduction
Dioecious populations are typically faced with a fundamental evolutionary problem: sexually homologous traits are frequently subjected to sex-specific selection in which different trait values are favored in males and females, yet trait expression in each sex has shared genetic underpinnings. Under such conditions, alleles at a particular locus may confer opposite fitness consequences when expressed in either sex (Rice and Chippindale, 2001; Bedhomme and Chippindale, 2007; Bonduriansky and Chenoweth, 2009), generating intralocus sexual conflict in which the adaptive evolution of one sex is impeded or limited by the other (Lande, 1980; Rice, 1984). Recent reviews suggest that sexually antagonistic selection may be common in natural populations (Cox and Calsbeek, 2009) and that the intersex genetic covariance for phenotypic traits is generally positive (Poissant et al., 2009), indicating considerable potential for ongoing intralocus conflict. Consistent with this, it is becoming increasingly clear that populations often harbor sexually antagonistic genetic variance for fitness (e.g., Chippindale et al., 2001; Fedorka and Mousseau, 2004; Pischedda and Chippindale, 2006; Qvarnström et al., 2006).
The intersex genetic correlation for fitness (rwFM) is calculated as the ratio of the additive genetic covariance for fitness between sexes to the geometric average of male- and female-specific additive genetic variance for fitness, thus providing a standardized measure of the similarity of the additive effects of alleles on the fitness of the sexes. Empirical estimates of the intersex genetic correlation/covariance for total fitness are limited to a few studies, in general providing evidence for a negative genetic covariance (e.g., Chippindale et al., 2001; Qvarnström et al., 2006; Foerster et al., 2007; Delcourt et al., 2009). To date, however, little is known regarding how the genetic architecture for fitness is manifested in novel environments—that is, the role of sexually antagonistic genetic variation during the initial stages of adaptation. Novel environments can result in displacement of phenotypic optima of (i.e., changes in selection on) one or both sexes, as well as affect the genetic basis of trait expression. Consequently, for alleles that are segregating in a population in a novel environment and upon which the initial stages of adaptation depend, additive effects may vary in one or both sexes and this may cause changes in the additive genetic covariance.
It is important to note that the magnitude (but not the sign) of intersex genetic correlation within a particular environment depends also on the sex-specific genetic variances. That is, observed environmental differences in the magnitude of the correlation could be simply attributable to the environmental effects on the respective (sex-specific) genetic variances. Predicting the effects of a novel environment on the expression of genetic variance is difficult because there are numerous theoretical (and biologically plausible) possibilities and empirical data are equivocal, with documented instances of both inflation and contraction of variances attributable to environmental effects (e.g., Sgrò and Hoffmann, 1998, Sgrò and Hoffmann, 2004; Hoffmann and Merilä, 1999; Fowler and Whitlock, 2002; Charmantier and Garant, 2005). However, a simple expectation is that rwFM may vary as a function of absolute fitness, with the extent of sexually concordant genetic variance being positively related to distance from the optimum (e.g., a positive rwFM when a population is far from a fitness peak, and hence at low fitness, characteristic of the early stages of adaptation). Lande (1980) showed how, in dioecious populations where males and females have separate optima, intersex genetic correlations can slow the rate at which mean fitness (i.e., male and female, jointly) increases. Although adaptation is initially rapid due to alleles with sexually concordant effects, as these alleles become fixed the response slows and the remaining genetic variance is characterized by negative intersex correlations (Chapman et al., 2003; Bonduriansky and Chenoweth, 2009). By extension, the rwFM observed in novel environments might also be expected to vary with the absolute fitness of the population in each, whereby relatively harsh environments tend to reveal sexually concordant genetic variation, whereas in relatively benign ones sex-specific (and potentially sexually discordant) genetic variance figures more prominently.
Formal examinations of environmental effects on the rwFM are rare, although a recent study (Delcourt et al., 2009) showed a significant negative correlation in a population of laboratory-adapted fruit flies (Drosophila serrata), irrespective of whether it was assayed in the standard laboratory conditions to which it was adapted or in a novel laboratory environment. Whether this result reflects a fundamental property of sexually antagonistic variation, or merely documents rwFM peculiar to this population and/or these two environments, is not clear. The present study extends this work to provide additional insight into the degree of environmental dependence in the expression of rwFM. This was achieved by employing a set of inbred lines, founded from a wild D. serrata population, to evaluate the genetic correlation between male and female fitness in four different novel laboratory environments.
Methods
Source populations and inbred lines
Inbred lines of D. serrata were derived from a population in St Lucia, Brisbane, Australia. Lines were created by imposing single-pair full-sibling mating for 10–16 generations (inbreeding coefficient, F⩾0.886, Falconer and Mackay, 1996). To corroborate that the inbreeding protocol was successful, these lines were screened and conservatively scored at three microsatellite loci, Dser15, Dser72 and Dser16, using the protocol described in Frentiu and Chenoweth (2008). Lines that exhibited heterozygosity at two loci (none were heterozygous at all three) were excluded from the subsequent fitness assays.
From the remaining lines we created a set of unique and repeatable F1 genotypes to recover the range of the genetic variation present in the sampled wild population (from which the lines originated) while avoiding the extreme levels of homozygosity that result from the inbreeding protocol. This was achieved by crossing males from each of 42 inbred lines with females from three other (arbitrarily chosen) ‘reference' inbred lines. Because the population was maintained as separate isofemale lines upon collection, and inbreeding was initiated soon thereafter, these inbred line crosses largely recover wild genotypes with little opportunity for adaptation to laboratory conditions.
For the purpose of the fitness assays (described below), we also used an outbred laboratory-adapted stock population (described in Rundle et al., 2006 and Chenoweth et al., 2008) into which a visible recessive orange-eyed mutation had been introgressed (see Delcourt et al., 2009). Populations of these mutants were reared in bottles (containing 100 ml of food medium) corresponding to each environment and from these, virgin flies were drawn for use as competitors in the fitness assays described below.
Fitness assays and environments
Fitness was assayed in four environments, three involving different food media consisting primarily of either yeast, corn flour or rice flour (following the recipes in Rundle et al., 2005), and a fourth in which table salt was added (8 g l−1) to the standard yeast medium. Hereafter, the environments are referred to as yeast, corn, rice and salt, respectively. For each of the three reference lines, six females mated to a corresponding inbred line male were allowed to oviposit sequentially for 24 h in each environment. The F1 offspring resulting from these inbred line × reference line crosses were collected as virgins (i.e., within 24 h of eclosion) using light CO2 anesthesia and maintained separately by sex under their respective environmental conditions (i.e., corn, rice, yeast or salt) before assaying male and female fitness. Likewise, orange-eyed male and female mutants were reared in each of the four environments, collected as virgins and maintained in vials of the corresponding medium until use in assays.
Fitness was measured in a competitive assay by aspirating a single male or female F1 individual into a vial containing 10 ml of the appropriate medium. Within 1 min, two orange-eyed mutant competitors of the same sex and two virgin mutants of the opposite sex (all reared in the same environment and of the same age range) were added and flies were allowed to interact (i.e., mate and oviposit). After 48 h, all adults were discarded and a small (approximately 2 × 5 cm) piece of blotting paper was added to facilitate pupation. All resulting offspring that emerged from these vials were removed and counted for 14 days following the first eclosion.
The fitness assays were conducted in two blocks (block 1: n=18 lines; block 2: n=24 lines). Owing to low absolute fitness in the rice environment in the first block, and because our goal was to estimate environment effects on genetic (co)variances (as opposed to absolute fitness), we permitted the interaction to last an extra 24 h in the rice environment in the second block. Five replicate vials were set up for each combination of genotype, environment and sex (see Figure 1). This measure of fitness combines, in a competitive environment, adult reproductive success (e.g., male pre- and post-copulatory reproductive success, female fecundity) and the subsequent survival to emergence of their offspring. In all, 30% of the vials failed to yield any offspring (wild type and mutant) and were thus discarded, resulting in a total of 3542 individual fitness measures included in the analyses (2265 for females and 1277 for males). Competitive fitness was calculated as the number of wild-type offspring produced minus the number of orange-eyed offspring produced. The distributions of sex- and environment-specific competitive fitness were approximately normal (Supplementary Materials S1); these measures were therefore not transformed, preserving their original scale (Houle et al., 2011) and thus facilitating comparisons across environments.
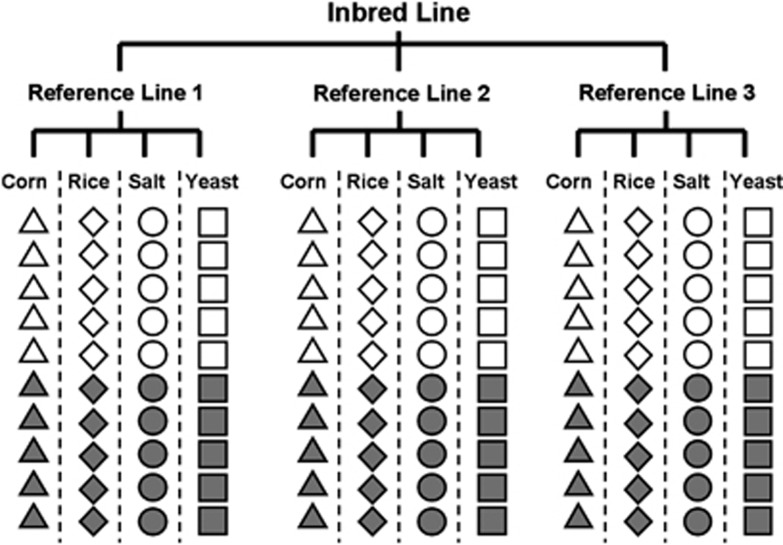
Figure 1.
Schematic representation of the experimental design. Each inbred line was crossed to three reference lines and produced offspring in each of four novel environments (represented by different shapes) for which competitive fitness was measured for five female (open) and male (shaded) offspring. This was performed for a total of 42 inbred lines.
Statistical analyses and estimating genetic covariances
The additive genetic (co)variance matrix for male and female fitness (i.e., G) was estimated at the inbred line level by considering fitness of each sex in each environment as separate traits. Our initial analysis provided an overall test for genetic variance in fitness across sexes and environments, and assessed the effects of the reference lines (i.e., the genetic backgrounds) on these estimates. Accordingly, observed variation was partitioned into components attributable to various effects via the following mixed model, fit using restricted maximum likelihood via the Mixed procedure in SAS v. 9.2 (Cary, NC, USA):
where μ is the population mean, Yijk corresponds to the fitness of the ith offspring, of sex (S) from block (B) and environment (E), and derived from the jth maternal reference line (R) and the kth paternal inbred line (X). ɛ is unexplained error. Inbred line was treated as a random effect and all other terms were fixed. Reference line and vial effects were intentionally confounded as our goal was to quantify variance at the inbred line level.
Following Hine and Blows (2006), we used a factor-analytic (FA) modeling approach to test the dimensionality of G, thereby providing an overall test for genetic variance in fitness. The FA approach finds the major (orthogonal) axes of G and estimates (co)variance parameters along each, allowing reduced rank models to be specified. The utility of the FA approach for the estimation of genetic parameters in multiple traits across multiple environments is reviewed in Meyer (2009) and its advantages include guaranteeing a positive semidefinite G (i.e., avoiding negative variances; Kirkpatrick and Meyer, 2004), as well as providing a straightforward approach to testing dimensionality. Using the FA approach, the dimensionality of G was evaluated via a series of nested likelihood ratio tests that constrained the covariance matrix at the inbred line level to be from zero through eight dimensions, in each case evaluating whether including that particular dimension significantly improved the fit of the model (Hine and Blows, 2006). The possibility of an inbred line × reference line interaction was also evaluated, again using the FA approach, via a likelihood ratio test of the first dimension of this interaction when included in Equation 1. The inbred line × reference line interaction was significant (see Results) and a reduced-rank covariance matrix at this level was therefore included in all models (likelihood convergence problems in some models prevented the fitting of an unconstrained covariance matrix at this level). This reduced-rank matrix was fixed at five dimensions in all cases, corresponding to the number of dimensions with nonzero eigenvalues at this level, to avoid problems that can arise from fitting fewer dimensions than exist in the data (Meyer and Kirkpatrick, 2008).
Genetic variances for male and female fitness in each environment, and the genetic correlations among all of these traits, were estimated at the inbred-line level from a model employing an unstructured (i.e., full rank, non-FA) covariance matrix at this level that was parameterized in terms of correlations (i.e., using the ‘type=unr' statement in the Mixed procedure in SAS). Genetic covariances among all of these traits were similarly estimated using the same model except that the covariance matrix was parameterized in terms of covariances (i.e., using the type=un statement in the Mixed procedure in SAS). Asymptotic standard errors were obtained for all parameters estimates. We should point out that the estimates and errors obtained using Mixed are based on restricted maximum likelihood and subject to the usual assumptions (e.g., large sample sizes and normally distributed errors); thus, bounded parameters (correlations and variances) are likely to be anticonservative compared with alternative approaches that have less restrictive assumptions (e.g., a Bayesian framework, see Hadfield, 2010). For the sake of consistency with the other statistical models used in the present paper, we report the restricted maximum likelihood-based errors but note that the errors should be interpreted with some caution.
Our primary interest is with rwFM within each environment, along with the underlying genetic variances and intersex covariance that determine it. Therefore, to improve parameter stability and decrease computation times when testing hypotheses concerning variation in these parameters, we restricted estimation to these parameters by employing the ‘group' statement at the environment level in the Mixed procedure in SAS. This eliminated the cross-environment (within and between sex) covariances, estimating only the genetic variances and covariance for male and female fitness within each of the four environments within the context of a single model.
To test whether the rwFM varied among environments, we first estimated a single best-fit (i.e., global) rwFM by pooling both male and female fitness estimates from all environments and fitting a single unconstrained 2 × 2 covariance matrix at the inbred line level, employing the ‘type=unr' command to directly estimate the intersex correlation. A likelihood ratio test was then used to compare the fit of an unconstrained model, allowing separate estimates of the sex-specific genetic variances and the intersex genetic correlation for each of the four environments, with one that constrained only the intersex correlation within each environment to the same global estimate. If allowing environment-specific estimates of rwFM significantly improve the fit of the model, this provides direct evidence that rwFM varies among these environments. Finally, to identify potential sources for the among-environment variation in rwFM, we performed three separate analyses, analogous to those above, that in each case compared the fit of an unconstrained model that allowed separate estimates of the sex-specific genetic variances and their covariance within each environment with a model in which either the genetic variance in female fitness, male fitness or their intersex covariance, were respectively fixed to their respective single global estimate. This provides direct tests for among-environment variation in the genetic variance of female fitness, male fitness and their covariance, all of which contribute to rwFM.
Results
The additive genetic covariance (G) matrix of environment-specific male and female fitness (Supplementary Material S2 and S3) was estimated from the competitive fitness of each sex in each environment (Table 1). Significant genetic variance was detected overall, with FA modeling providing statistical support for the first genetic dimension underlying male and female fitness across these environments (Table 2), explaining 45% of the total genetic variance in fitness. The FA approach also found near-significant evidence of a second and third genetic dimension (Table 2), explaining an additional 22% and 21% of the genetic variance, respectively.
Table 1. Phenotypic means and standard deviations (parentheses) of competitive fitness of female and male F1 offspring in four novel environments, calculated as the number of wild-type offspring produced minus the number of competitor (i.e., orange-eye) offspring produced.
| Environment | Female | Male | ||
|---|---|---|---|---|
| |
Mean (s.d.) |
N |
Mean (s.d.) |
N |
| Corn | 10.96 (32.46) | 557 | −1.32 (27.98) | 312 |
| Rice | 16.24 (36.32) | 549 | 1.85 (31.21) | 315 |
| Salt | 21.80 (48.86) | 574 | 6.44 (34.57) | 287 |
| Yeast | 5.21 (50.36) | 585 | 5.97 (41.12) | 363 |
Table 2. Model fit statistics for a nested series of factor-analytic models testing the dimensionality of the inbred line-level covariance matrix (i.e., G).
| Genetic dimensions | −2LL | AIC | Parameters | P-value | % Variance |
|---|---|---|---|---|---|
| 8 | 35 769.7 | 35 855.7 | 44 | 1.000 | 0 |
| 7 | 35 769.7 | 35 855.7 | 43 | 0.819 | 0 |
| 6 | 35 770.1 | 35 848.1 | 41 | 0.978 | 0 |
| 4, 5a | 35 770.3 | 35 846.3 | 38 | 0.544 | 13 |
| 3 | 35 778.2 | 35 834.2 | 29 | 0.082 | 21 |
| 2 | 35 789.4 | 35 835.4 | 23 | 0.088 | 22 |
| 1 | 35 801.8 | 35 833.8 | 16 | 0.001 | 45 |
| 0 | 35 826.9 | 35 842.9 | 8 | — | — |
Percent genetic variance explained was estimated from the eigen values of the full-rank G-matrix. LL and AIC refer to log-likelihood and Akaike's Information Criterion, respectively.
Significance testing of the fourth dimension was not possible because of a non-positive definite Hessian matrix during the likelihood estimation procedure. A combined test of dimensions 4 and 5 was therefore performed.
The genetic basis of male and female fitness, as estimated at the inbred line level, also differed across the genetic backgrounds of the three reference lines, as revealed by a significant inbred line × reference line interaction (likelihood ratio test (LRT): χ2=21.0, df=8, P=0.007). Because our primary aim was to address among-environment variation in the genetic covariance structure of fitness in general (i.e., as opposed to within a specific genetic background), and analyses conducted separately by reference line demonstrated significant or near-significant genetic variance for fitness in each of these genetic backgrounds despite the much smaller data sets (Supplementary Material S4), we proceeded with characterizing and testing for differences in the genetic covariance structure at the inbred line level while retaining the inbred line × reference line interaction in all of the models.
The rwFM differed among environments, as indicated by a significant improvement in the fit of a model that allowed environment-specific estimates of rwFM as compared with one that constrained these correlations to their single best-fit global estimate (χ2=10.37, df=4, P=0.035). Point estimates were highly variable across environments, with both negative (corn) and positive (salt and rice) correlations of fairly large magnitude being observed, although standard errors were also substantial (Table 3). This among-environment variation in rwFM was driven by significant differences in the genetic variances of both male (χ2=13.83, df=4, P=0.008) and female (χ2=24.21, df=4, P<0.001) fitness, whereas there was no evidence of any significant difference in the intersexual covariances (χ2=4.71, df=4, P=0.318).
Table 3. Environment-specific point estimates (±asymptotic s.e.) of genetic (co)variances and corresponding intersex genetic correlations for fitness (r w FM ), as estimated from an unstructured covariance matrix at the inbred-line level.
| Environment | Female variance | Male variance | Intersex covariance | rwFM |
|---|---|---|---|---|
| Corn | 152.76±120.44 | 276.65±194.07 | −128.12±108.33 | −0.60±0.60 |
| Rice | 217.99±183.82 | 81.78±83.53 | 48.92±129.85 | 0.51±1.06 |
| Salt | 396.51±286.34 | 70.17±107.25 | 81.89±184.82 | 0.63±1.27 |
| Yeast | 152.67±179.98 | 442.68±301.73 | 37.80±194.56 | 0.22±0.74 |
Discussion
Sexually antagonistic genetic variance has important implications for adaptation and persistence of a population, yet surprisingly little is known regarding how alleles with potentially sexually discordant fitness effects are expressed in different environments.
Here, we measured male and female competitive fitness to evaluate rwFM arising from alleles naturally segregating in an outbred, wild-collected population in four novel environments. We detected significant genetic variance overall for male and female competitive fitness. We also observed significant environmental differences in rwFM due, at least in part, to environmental differences in the magnitudes of the sex-specific genetic variances. These results illustrate the variable nature of intersex genetic correlations as well as the profound influence of environment on the relative contribution of sexually antagonistic genetic variance to total fitness. We discuss these results and their implications in more detail below.
Genetic variance in male and female environment-specific fitness
We estimated genetic variance for male and female environment-specific fitness by employing controlled crosses among inbred lines. The strengths of this approach include capturing naturally segregating genetic variance from a wild population, and the ability to generate repeatable, but not unusually inbred, offspring (i.e., paternal line × maternal reference line) genotypes from which additive genetic (co)variances can be readily estimated. Among-line variance was assayed across three reference lines to minimize the risk of generating estimates that were peculiar to a single genetic background. We detected significant additive genetic variance for male and female environment-specific fitness, indicated by the variance among inbred lines but also a significant interaction between paternal inbred line and maternal reference line. The latter possibly reflect epistatic effects for fitness, which can have important implications for adaptation (Whitlock et al., 1995). The apparent importance of maternal genetic effects, as well as paternal-by-maternal genetic interactions, suggest that inferences regarding additive effects need to be made across a wide range of genetic backgrounds representative of the population in question.
Environmental differences in the intersex genetic correlation
Estimates of rwFM were highly variable across environment, ranging from moderately positive to negative values of comparable magnitude (Table 3). Differences were attributable, at least in part, to significant variation among environments in the sex-specific genetic variances (i.e., the denominator of the correlation coefficient), with quite low variances in male fitness in rice and salt in particular causing fairly strong positive values of rwFM in these environments. In contrast to the significant differences in genetic variances in both sexes, there was no evidence of significant changes in the intersex covariance (i.e., the numerator). Although some of the point estimates of rwFM differed in sign, thereby suggesting concordant differences in the sign of the intersex covariances, standard errors were large (Table 3), and these differences in sign were not supported in specific tests of the point estimates of the covariances against zero (not shown), likely due to insufficient power. Therefore, although there is some suggestion that the covariances may differ, further exploration of this will require additional studies.
There are a number of possible explanations for the observed variation in rwFM among environments. Theory suggests that the evolutionary history of a population may have strong bearing on the covariance between male and female fitness, and more specifically, that rwFM may relate to the extent of adaptation to a particular environment (Lande, 1980), as previously discussed. Unfortunately our present data do not allow for an explicit test of this, as the extremely low productivity in one environment (i.e., rice) necessitated longer laying periods and prevented quantitative comparisons of absolute fitness among the environments. The positive estimates of rwFM in the two environments (i.e., rice and salt) that, according to a previous study by Rundle et al. (2005) conferred relatively low fitness, are qualitatively consistent with this, as is the negative rwFM in the relatively higher-fitness corn environment. However, yeast, the standard and assumed most productive environment for laboratory-adapted stocks, did not yield a strongly negative estimate. It is possible that the wild population from which inbred lines were derived could have higher absolute fitness in corn. Alternatively, because all lines were created and maintained in the laboratory in a yeast environment, some adaptation to this environment may have occurred before, and during, the inbreeding procedure, despite our attempts to minimize this. Such adaptation may have reduced rwFM in this environment. Additional evidence of sexually antagonistic variance being more apparent in well-adapted populations comes from a recent study by Long et al. (2012), who detected primarily sexually discordant fitness variation in experimental populations allowed to become well adapted and, conversely, sexually concordant variation in populations into which maladaptive alleles were continuously introduced.
It is also likely that the nature of rwFM bears some specificity to the population and/or environments being considered (Poissant et al., 2009). Interestingly, a previous study on D. serrata by Delcourt et al. (2009) used a paternal half-sibling/full-sibling breeding design in a laboratory population long adapted to yeast. Employing two of the same environments as used here (corn and yeast), they detected a negative rwFM in both the adapted yeast environment and the novel corn environment. We also observed a negative rwFM in corn, possibly reflecting sexually antagonistic effects peculiar to this environment. Although it is unclear as to the underlying proximate causes of such effects, they may correspond to the variation in the availability of (or ability to assimilate) limited resources; sex-specific nutritional and energetic requirements can generate sex differences in allocation to components of fitness that is ultimately environment dependent (Hunt et al., 2004; Maklakov et al., 2008; Punzalan et al., 2008; also see Judge et al., 2008). That rwFM was more positive in the yeast environment in our case (as compared to Delcourt et al. (2009), who used a population long adapted to this environment) is also consistent with an increasing proportion of sexually antagonistic genetic variation as adaptation proceeds.
Environmental differences in rwFM could also arise from differences in the relative contributions of various components of fitness to total fitness. Chippindale et al. (2001) showed, using a laboratory population of D. melanogaster, that adult-specific fitness was negatively related between the sexes despite exhibiting a positive intersex correlation during the juvenile stage. Following this example, if the different assayed environments primarily affected the contribution of juvenile fitness to total fitness, this could have strong bearing on the estimate of rwFM for lifetime fitness. Although the relative contributions of fitness components have not been determined for D. serrata, the sexually concordant genetic variance during the D. melanogaster larval stage (Chippindale et al., 2001) suggests that our estimates based on lifetime reproduction may bias our estimates of rwFM toward more positive values.
Conclusions
Collectively, these data suggest that although sexually antagonistic genetic variance might be pervasive, negative genetic correlations are not necessarily characteristic of the genetic architecture underlying male and female fitness. Rather, the intersex genetic correlation can vary considerably depending on environmental conditions, the population in question, and its evolutionary history. Outstanding questions include the degree to which standing intersex genetic correlations might influence the rate of adaptation and how rwFM itself changes during adaptation. With respect to the latter, of much interest will be empirical approaches that estimate changes in intersex genetic (co)variances as populations adapt. One possibility is to estimate rwFM in a population before experimental exposure to a new environment, and then in a series of temporal cross-sections in subsequent generations. This would provide ‘snap-shots' of the genetic architecture of a population at various stages of adaptation. Similarly, one could estimate rwFM in a particular population when exposed to a gradient of environments that reduced absolute fitness (e.g., increasing concentrations of salt). For some study systems, it may be possible to track both the pedigree and individual trait values in a population subject to artificial selection, thereby allowing changes in genetic (co)variances across generations to be estimated via the animal model (Kruuk, 2004) and related to changes in absolute fitness.
Data Archiving
Data deposited in the Dryad repository: doi:10.5061/dryad.5892n.
Acknowledgments
We are grateful to S. Chenoweth for providing the original collection of isofemale lines and for conducting the microsatellite analyses. D. Arbuthnott, M. Charette, P. Harris, G. Moreau, S. Reilly, V. Rotondo and G. Schroeder assisted with various stages of this work. We thank S. Chenoweth and J. Stinchombe for helpful discussions concerning data analyses. We are also grateful to three anonymous reviewers for comments on previous versions of this manuscript. DP and MD were supported by grants to HDR from the Natural Sciences and Engineering Research Council of Canada and the Ontario Ministry of Economic Development and Innovation.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Bedhomme S, Chippindale AK.2007Irreconcilable differences: when sexual dimorphism fails to resolve sexual conflictIn: Fairbairn DJ, Blanckenhorn WU, Székely T (eds)Sex, Size and Gender Roles: Evolutionary Studies of Sexual Size Dimorphism Oxford University Press; 185–194. [Google Scholar]
- Bonduriansky R, Chenoweth SF. Intralocus sexual conflict. Trends Ecol Evol. 2009;24:280–288. doi: 10.1016/j.tree.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Chapman T, Arnqvist G, Bangham J, Rowe L. Sexual conflict. Trends Ecol Evol. 2003;18:41–47. [Google Scholar]
- Charmantier A, Garant D. Environmental quality and evolutionary potential: lessons from wild populations. Proc R Soc Lond B. 2005;272:1415–1425. doi: 10.1098/rspb.2005.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenoweth SF, Rundle HD, Blows MW. Genetic constraints and the evolution of display trait sexual dimorphism by natural and sexual selection. Am Nat. 2008;171:22–34. doi: 10.1086/523946. [DOI] [PubMed] [Google Scholar]
- Chippindale AK, Gibson JR, Rice WR. Negative genetic correlation for adult fitness between sexes reveals ontogenetic conflict in Drosophila. Proc Natl Acad Sci USA. 2001;98:1671–1675. doi: 10.1073/pnas.041378098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RM, Calsbeek R. Sexually antagonistic selection, sexual dimorphism, and the resolution of intralocus sexual conflict. Am Nat. 2009;173:176–187. doi: 10.1086/595841. [DOI] [PubMed] [Google Scholar]
- Delcourt M, Blows MW, Rundle HD. Sexually antagonistic genetic variance for fitness in an ancestral and a novel environment. Proc R Soc Lond B. 2009;276:2009–2014. doi: 10.1098/rspb.2008.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer DS, MacKay TFC. Longman: Essex; 1996. Introduction to quantitative genetics. [Google Scholar]
- Fedorka KM, Mousseau TA. Female mating bias results in conflicting sex-specific offspring fitness. Nature. 2004;429:65–67. doi: 10.1038/nature02492. [DOI] [PubMed] [Google Scholar]
- Foerster K, Coulson T, Sheldon BC, Pemberton JM, Clutton-Brock TH, Kruuk LEB. Sexually antagonistic genetic variation for fitness in red deer. Nature. 2007;447:1107–1111. doi: 10.1038/nature05912. [DOI] [PubMed] [Google Scholar]
- Fowler K, Whitlock MC. Environmental stress, inbreeding and the nature of phenotypic and genetic variance in Drosophila melanogaster. Proc R Soc Lond B. 2002;269:677–683. doi: 10.1098/rspb.2001.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frentiu F, Chenoweth SF. Polyandry and paternity skew in natural and experimental populations of Drosophila serrata. Mol Ecol. 2008;17:1589–1596. doi: 10.1111/j.1365-294X.2008.03693.x. [DOI] [PubMed] [Google Scholar]
- Hadfield J.2010MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package J Stat Softw 331–22.20808728 [Google Scholar]
- Hine E, Blows MW. Determining the effective dimensionality of the genetic variance–covariance matrix. Genetics. 2006;173:1135–1144. doi: 10.1534/genetics.105.054627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Merilä J. Heritable variation and evolution under favourable and unfavourable conditions. Trends Ecol Evol. 1999;14:96–101. doi: 10.1016/s0169-5347(99)01595-5. [DOI] [PubMed] [Google Scholar]
- Houle D, Pelabon C, Wagner GP, Hansen TF. Measurement and meaning in biology. Q Rev Biol. 2011;86:1–32. doi: 10.1086/658408. [DOI] [PubMed] [Google Scholar]
- Hunt J, Brooks R, Jennions MD, Smith MJ, Bentsen CL, Bussiere LF. High-quality male field crickets invest heavily in sexual display but die young. Nature. 2004;432:1024–1027. doi: 10.1038/nature03084. [DOI] [PubMed] [Google Scholar]
- Judge KA, Ting JJ, Gwynne DT. Condition dependence of male life span and calling effort in a field cricket. Evolution. 2008;62:868–878. doi: 10.1111/j.1558-5646.2008.00318.x. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M, Meyer K. Direct estimation of genetic principal components: simplified analysis of complex phenotypes. Genetics. 2004;168:2295–2306. doi: 10.1534/genetics.104.029181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruuk L. Estimating genetic parameters in natural populations using the ‘animal model'. Phil Trans R Soc Lond B. 2004;359:873–890. doi: 10.1098/rstb.2003.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R. Sexual dimorphism, sexual selection, and adaptation in polygenic characters. Evolution. 1980;34:292–305. doi: 10.1111/j.1558-5646.1980.tb04817.x. [DOI] [PubMed] [Google Scholar]
- Long TAF, Agrawal AF, Rowe L. The effect of sexual selection on offspring fitness depends on the nature of genetic variation. Curr Biol. 2012;22:204–208. doi: 10.1016/j.cub.2011.12.020. [DOI] [PubMed] [Google Scholar]
- Maklakov AA, Simpson SJ, Zajitschek F, Hall M, Dessman J, Clissold F, et al. Sex-specific fitness effects of nutrient intake on reproduction and lifespan. Curr Biol. 2008;18:1062–1066. doi: 10.1016/j.cub.2008.06.059. [DOI] [PubMed] [Google Scholar]
- Meyer K, Kirkpatrick M. Perils of parsimony: properties of reduced rank estimates of genetic covariances. Genetics. 2008;180:1153–1166. doi: 10.1534/genetics.108.090159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K. Factor-analytic models for genotype X environment type problems and structured covariance matrices. Genet Sel Evol. 2009;49:21–33. doi: 10.1186/1297-9686-41-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pischedda A, Chippindale AK. Intralocus sexual conflict diminishes the benefits of sexual selection. PLoS Biology. 2006;4:e356. doi: 10.1371/journal.pbio.0040356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poissant J, Wilson AJ, Coltman DW. Sex-specific genetic variance and the evolution of sexual dimorphism: a systematic review of cross-sex genetic correlations. Evolution. 2009;64:97–107. doi: 10.1111/j.1558-5646.2009.00793.x. [DOI] [PubMed] [Google Scholar]
- Punzalan D, Cooray M, Rodd FH, Rowe L. Condition dependence of sexually dimorphic coloration and longevity in the ambush bug Phymata Americana. J Evol Biol. 2008;21:1297–1306. doi: 10.1111/j.1420-9101.2008.01571.x. [DOI] [PubMed] [Google Scholar]
- Qvarnström A, Brommer JE, Gustafsson L. Testing the genetics underlying the co-evolution of mate choice and ornament in the wild. Nature. 2006;441:84–86. doi: 10.1038/nature04564. [DOI] [PubMed] [Google Scholar]
- Rice WR. Sex chromosomes and the evolution of sexual dimorphism. Evolution. 1984;38:735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. [DOI] [PubMed] [Google Scholar]
- Rice WR, Chippindale AK. Intersexual ontogenetic conflict. J Evol Biol. 2001;14:685–693. [Google Scholar]
- Rundle HD, Chenoweth SF, Doughty P, Blows MW. Divergent selection and the evolution of signal traits and mating preferences. PLoS Biology. 2005;3:e368. doi: 10.1371/journal.pbio.0030368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundle HD, Chenoweth SF, Blows MW. The roles of natural and sexual selection during adaptation to a novel environment. Evolution. 2006;60:2218–2225. [PubMed] [Google Scholar]
- Sgrò CM, Hoffmann AA. Effects of stress combinations on the expression of additive genetic variation for fecundity in Drosophila melanogaster. Genet Res Camb. 1998;72:13–18. doi: 10.1017/s0016672398003310. [DOI] [PubMed] [Google Scholar]
- Sgrò CM, Hoffmann AA. Genetic correlations, tradeoffs and environmental variation. Heredity. 2004;93:241–248. doi: 10.1038/sj.hdy.6800532. [DOI] [PubMed] [Google Scholar]
- Whitlock MC, Phillips PC, Moore FBG, Tonsor SJ. Multiple fitness peaks and epistasis. Ann Rev Ecol Syst. 1995;26:601–629. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.