Abstract
Combination of divergent genomes in allopolyploids creates genome-wide gene expression changes including many miRNA targets in Arabidopsis allotetraploids relative to the parents Arabidopsis thaliana and Arabidopsis arenosa. Here we report expression and epigenetic changes in a chromosomal region containing the MIR172b locus in the allotetraploids. Although mature miRNA sequences are conserved, A. thaliana and A. arenosa miRNA loci diverge rapidly in sequence and expression. Among four MIR172 loci in Arabidopsis, the level of nucleotide sequence divergence between A. thaliana and A. arenosa MIR172 loci is 15–25%, which is higher than that of protein-coding genes (∼5%). MIR172b locus and its flanking genes in A. arenosa were expressed at low levels relative to that in A. thaliana, which is associated with hypermethylation of this region in the allotetraploids. Consistently with this notion, pri-miR172 transcripts in the allotetraploids were primarily derived from the A. thaliana MIR172b locus. Expression of homoeologous alleles in miR172 target loci is associated with allelic loss, allelic changes in outcrossing A. arenosa or repression of A. thaliana alleles. These data suggest that gene expression changes in this homoeologous region are associated with genetic diversity and epigenetic variation of miRNA genes and their targets in allopolyploids.
Keywords: polyploidy, microRNA, gene expression epigenetics, duplication, Arabidopsis
Introduction
Polyploidy is a predominant feature for the evolution of many plants and some animals (Masterson, 1994; Otto, 2007; Leitch and Leitch, 2008). Autopolyploidy (duplication of a single genome) and allopolyploidy (combination of two or more sets of divergent genomes) often induce genomic instabilities and gene expression changes (Levy and Feldman, 2002; Comai, 2005; Hegarty et al., 2006; Wang et al., 2006b; Chen, 2007; Flagel et al., 2008; Jackson and Chen, 2010; Buggs et al., 2011). In Arabidopsis allotetraploids formed by interspecific hybridization between A. thaliana and A. arenosa autotetraploids, many genes that are highly expressed in A. thaliana or A. arenosa are not expressed additively (as the sum of two parents) in the allotetraploids, coinciding with morphological changes including phenotypic dominance of A. arenosa over A. thaliana (Wang et al., 2006b).
Gene expression changes in resynthesized allotetraploids are partly associated with changes in small RNAs, including microRNAs (miRNAs) and small interfering RNAs (siRNAs) (Baulcombe, 2004; Chapman and Carrington, 2007; Chen, 2009). In a previous study, both siRNAs and miRNAs are nonaddtitively expressed in Arabidopsis allotetraploids (Ha et al., 2009). A subset of siRNAs is absent in F1 allotetraploids but present in late (F8) generations of allotetraploids (Ha et al., 2009). In wheat, some siRNAs are slightly reduced in the allohexaploid (AABBDD) but not in the hybrid (Kenan-Eichler et al., 2011). These siRNAs silence genes and transposable elements, and affect RNA-directed DNA methylation (RdDM) (Law and Jacobsen, 2010) and genomic stability in hybrids and allopolyploids (Ng et al., 2012). Divergent siRNAs between parental alleles could alter allelic patterns of expression and RNA-directed DNA methylation and overall genomic stability in hybrids (Ng et al., 2012). In addition to siRNAs, ∼50% of miRNAs including miR163 and miR172 are expressed differently between A. thaliana and A. arenosa (Ha et al., 2009), and ∼6% of genes are nonadditively expressed in two independently derived resynthesized A. suecica allotetraploids (Wang et al., 2006b). These genes include many miRNA targets that are negatively correlated with miRNA accumulation levels (Ha et al., 2009). Moreover, nonadditive accumulation of these miRNAs explains ∼58% of expression changes in their target mRNAs in the allopolyploids. The miRNA accumulation difference may result from cis- and trans-regulation, as well as nonadditive expression of miRNA biogenesis genes (Ha et al., 2009; Ng et al., 2011). Moreover, there is a preference for degrading homoeologous gene transcripts in Arabidopsis allotetraploids (Ha et al., 2009; Ng et al., 2011). These data suggest that miRNA abundance in the allotetraploids results from transcriptional regulation of miRNA precursor transcripts as well as post-transcriptional regulation through miRNA processing (Ng et al., 2011).
It is unclear how sequence and expression of miRNA loci diverge between species. Here we investigate sequence divergence in the miR172 family and expression changes of MIR172b and its neighboring genes in the allotetraploids and between the parents. We found that allelic polymorphisms and epigenetic changes in a chromosomal region containing MIR172b between the progenitors are associated with expression differences of homoeologous alleles in the resynthesized allotetraploids.
Materials and methods
Plant materials
Arabidopsis allotetraploid lines were resynthesized by pollinating an autotetraploid A. thaliana (Landsberg erecta or Ler, accession #CS3900 in the Arabidopsis Biological Resource Center) (2n=4x=20) with tetraploid A. arenosa (accession #3901) (2n=4x=32), as previously described (Wang et al., 2004a). Individual F1 plants were self-pollinated to produce allotetraploid family lines and maintained by single-seed descent until sixth generations. To minimize developmental variations among progenitors and different lines, we adopted a published protocol (Wang et al., 2006b) for plant care and tissue harvest. The same A. suecica (9502) strain used in the previous study (Wang et al., 2004a) was used in this study.
DNA and RNA isolation
Genomic DNA and total RNA were isolated using Trizol reagents according to the manufacturer's recommendations (Invitrogen, Carlsbad, CA, USA). Messenger RNA was isolated from total RNA using a Micro-FastTrack 2.0 mRNA isolation kit (Invitrogen).
Small RNA blot analysis
Total RNA was isolated from leaves using TRIZOL (Invitrogen) according to the manufacturer's instructions. Twenty micrograms of total RNA was separated on a 15% polyacrylamide-urea gel and blotted onto Hybond-N+ membranes (Amersham, Piscataway, NJ, USA). Probes were made by end-labeling 21- to 24-mer DNA oligonucleotides that corresponded to the antisense strand of microRNAs with T4 polynucelotide kinase. RNA blot analysis was performed using a previously published protocol (Tian and Chen, 2001).
Cloning and sequence analyses of miRNA precursors and upstream regions
Full-length AaMIR172 sequences were obtained by cloning PCR products amplified using the primers listed in Supplementary Table S1.
Genome walking was performed using the BD GenomeWalker Universal Kit following the manufacturer's instructions (Clontech, Mountain View, CA, USA). Total genomic DNA (2.5 μg) was digested with DraI, EcoRV, PvuII and StuI, respectively, and ligated using GenomeWalker Adaptors. Resulting ligation products were used to construct four individual GenomeWalker libraries. Gene or genome-specific primers (GSP) were designed based on the sequence for the genes of interest. Two nested GSP primers (adjacent but non-overlapping) were made for each gene, giving rise to outside (producing larger-size fragments) and inside (producing smaller-size fragments) primers. Primary PCR amplification was performed using a GSP1 primer (outside) and AP1 (adaptor primer 1). After amplification, the PCR products were subjected to electrophoresis in a 1.2% agarose gel, and the band was sliced out and purified with the Qiagen Gel extraction kit, and cloned into pGEM T-easy vector for sequencing. The sequence information of A. arenosa miR172 loci has been deposited in GenBank under accession numbers KF543109-KF543120.
Full-length A. arenosa MIR172 sequences were aligned with the sequences encoding AtMIR172 precursors to predict stem-loop structures in AaMIR172. The stem-loop structures of five AtMIR172 loci were analyzed using the Vienna RNA Package (http://www.tbi.univie.ac.at/~ivo/RNA/).
Phylogenetic analysis
Sequences of MIR172 loci from A. thaliana, A. arenosa and rice (Oryza sativa) were aligned using the CLUSTAL W (v. 1.83) (University College Dublin, Dublin, Ireland). Phylogenetic trees were generated, and branch lengths were calculated using the DNAML in PHYLIP package (University of Washington, Seattle, WA, USA) with 1000 bootstraps. The branch lengths represent the expected numbers of substitutions, including both transitions and transversions and excluding the replacement of a base by itself, per 100 nucleotides.
RNA ligase-mediated-5′ RACE analysis of miRNA target genes
To determine miRNA origins in the synthetic allopolyploids, we performed 5′ and 3′ rapid amplification of cDNA ends (RACE) using the GeneRacer Kit according to the manufacturer's instructions (Invitrogen). The 5′ and 3′ ends of pri-miRNA cDNA were amplified with the GeneRacer primers (Supplementary Table S2), respectively. PCR products were gel-purified and cloned into pGEM T-easy vector, and 50 individual inserts were sequenced to estimate the transcript frequency.
SSCP analysis of homoeologous gene expression
Single-strand conformation polymorphism (SSCP) was performed as described using the 0.5 × Mutation Detection Enhancement (MDE, Cambrex Bio Science, East Rutherford Plaza, NJ, USA) gel containing 7.5% (w/v) urea (Hayashi, 1991; Adams et al., 2003). Primer pairs used to analyze allelic expression of 13 genes in a 50-kb region containing MIR172 were shown in Supplementary Table S3. For some genes when primers had biased amplification for A. thaliana alleles, the primers designed from A. arenosa (Aa) alleles that also matched A. thaliana alleles were used.
Genomic bisulfite sequencing
Two micrograms of genomic DNA prepared from A. thaliana, A. arenosa, synthetic allopolyploids (F1) and A. suecica were digested with BamHI, followed by the bisulfite treatment as previously described (Jacobsen et al., 2000). For each gene, two pairs of nested primers were designed according to the sequences of the plus (Watson) DNA strands (Supplementary Table S4), and the results were quantified as described in the text (Supplementary Table S5).
Results
Sequence divergence between miRNA loci in Arabidopsis and its related species
Many mature miRNAs are conserved among related species in the animal or plant kingdom (Bartel, 2004), and a few are even conserved between animals and plants (Arteaga-Vazquez et al., 2006). However, little is known about sequence conservation and divergence between miRNA loci in closely related species such as A. thaliana and A. arenosa that diverged ∼6 Mya (Koch et al., 2000) (Figure 1). We cloned and analyzed four miRNA loci of the miR172 family in A. arenosa (Figure 2a and Supplementary Figures S1–S5). MIR172 loci are conserved between A. thaliana and A. arenosa with the exception of MIR172c. AtMIR172c diverged from other AtMIR172 loci, and we failed to clone MIR172c locus in A. arenosa.
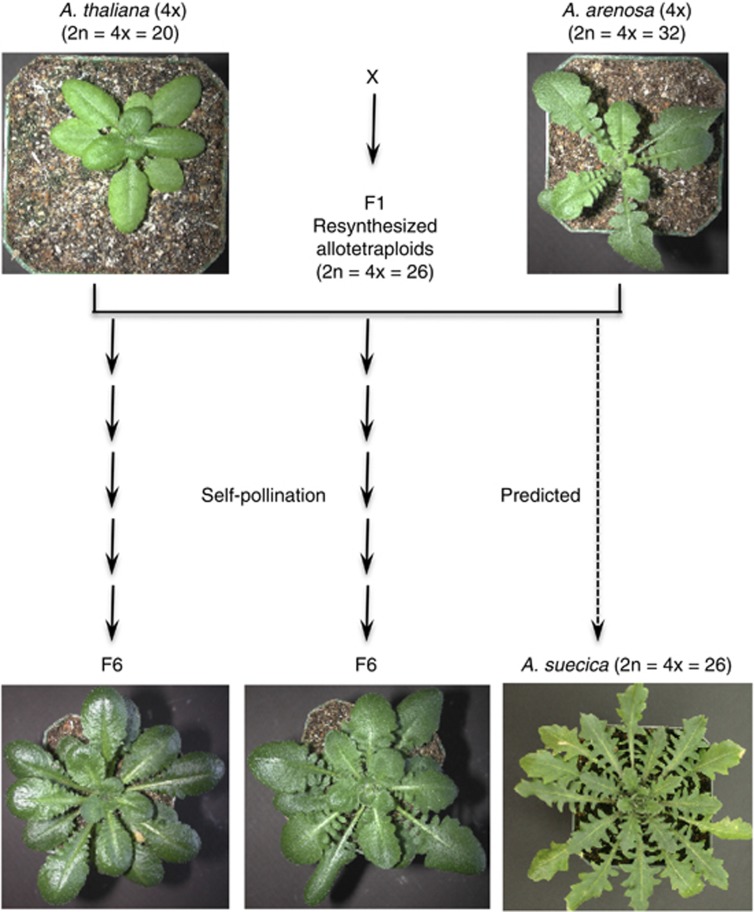
Figure 1.
Arabidopsis allotetraploids were resynthesized by interspecific hybridization between autotetraploid A. thaliana and tetraploid A. arenosa. Individual F1 plants were self-pollinated to produce stable allotetraploids (F6). Two independent allotetraploid lines (Allo733 and Allo738) and A. suecica (#9502) are shown. A. suecica is a natural allotetraploid that is derived from A. thaliana- and A. arenosa-like progenitors (Chen, 2007).
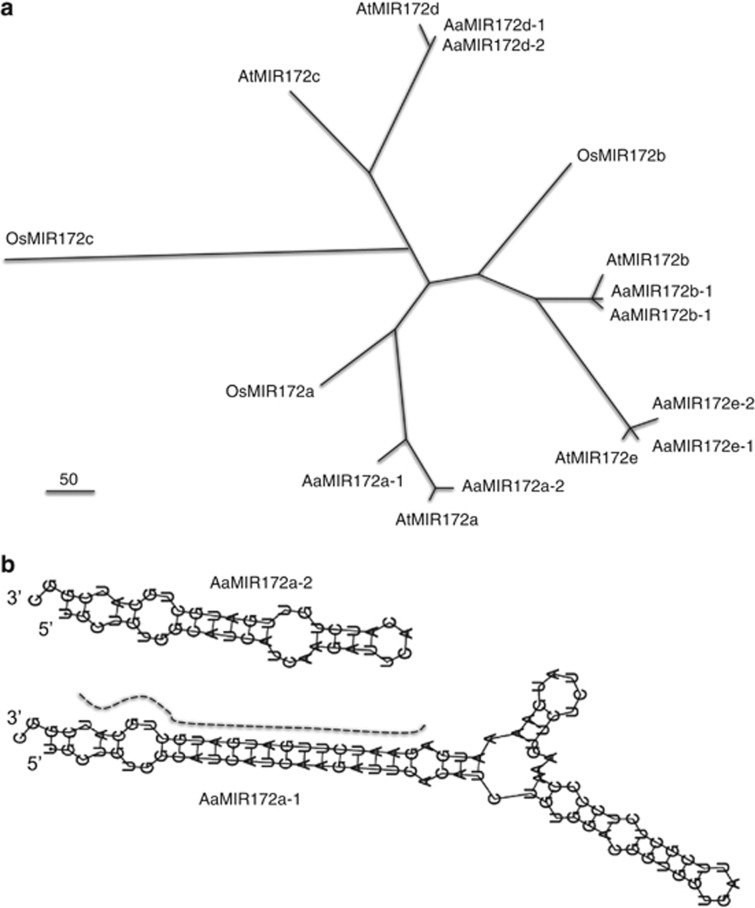
Figure 2.
Sequence divergence of miRNA loci in A. thaliana and A. arenosa. (a) Phylogenetic analysis of five MIR172 loci using miR172 stem-loop sequences originating in A. thaliana (At), A. arenosa (Aa) and three MIR172 loci in rice (Oryza sativa) (Os). Each A. arenosa locus has two corresponding A. thaliana loci, except for MIR172c that was absent in A. arenosa. Two rice MIR172 homologs are related to A. thaliana MIR172b and MIR172c, respectively. Brach lengths were calculated using bootstrap values with 1000 permutations. The unit of branch is indicated in the figure as the number of substitutions per 100 nucleotides. The nucleotide divergence rate between AaMIR172d-1 and AaMIR172d-2 was so low (0.3/100) that their branches were inseparable in the figure. (b) Stem-loop pairing of pre-mature miR172 sequences is mostly conserved between A. thaliana and A. arenosa, except that AaMIR172a-2 contains a deletion in the core miR172a so it is dwarfed. Dashed line indicates mature miR172 in AaMIR172a-1.
There were two MIR172 alleles corresponding to each A. thaliana locus (Figure 2b, Supplementary Figures S1–S5 and Supplementary Table S1), which is consistent with allelic polymorphisms in the outcrossing tetraploid A. arenosa (Comai et al., 2000; Koch et al., 2000). Two A. arenosa alleles of each locus, except for AaMIR172e, were conserved and shared >90% sequence identity. AaMIR172e-1 was more closely related to AtMIR172e than to AaMIR172e-2. The sequence identities were 78 and 83% between AtMIR172a and AaMIR172a-1 and a-2, respectively (Supplementary Figure S2). A. thaliana MIR172b, d and e had ∼84, ∼88 and ∼90% sequence identities with corresponding A. arenosa MIR172b, d and e alleles, respectively (Supplementary Figures S3–S5). Four A. thaliana and A. arenosa MIR172 alleles displayed sequence changes including small insertions and deletions located upstream and downstream of the hairpin molecules but had similar predicted secondary structures (Supplementary Figure S1). AaMIR172a-2 contained a 57-nucleotide deletion including 10-nucleotide from the 5′-end of miR172, generating a dwarf hairpin missing a large portion of the stem and loop (Figure 2b), suggesting that AamiR172a-2 is defective. Like rapid evolution of miRNA genes in A. thaliana and A. lyrata (Fahlgren et al., 2010), A. thaliana and A. arenosa MIR172 loci evolved rapidly over ∼6 million years (Koch et al., 2000), compared with protein-coding genes that share >95% DNA sequence identity (Hanfstingl et al., 1994; Henikoff and Comai, 1998; Lee and Chen, 2001; Wang et al., 2006a). Multiple members of the MIR172 family may also lead to rapid evolution.
Expression changes in miR172 and target genes in allotetraploids and their progenitors
We examined the abundance of miR172 in leaves and flower buds of allotetraploids and their progenitors, including a natural allotetraploid A. suecica (As9502) (Figures 3a and b). miR172 accumulated at higher levels in A. thaliana than in A. arenosa or A. suecica and higher levels in the resynthesized allotetraploids (F1 and F2) than in A. arenosa and in A. suecica in leaves but very similar levels in flowers. Within selfing generations, miRNA accumulation levels in leaves decreased from F1 and F2 to F6, suggesting a trend toward a relatively low level of miR172 accumulation in a natural allotetraploid A. suecica. The data are reminiscent of stochastic changes in some other protein-coding genes during selfing in resynthesized allotetraploids (Wang et al., 2004a). Expression levels of miR172 in flower buds were slightly higher in F6 than in F1 and F2 but low in A. suecica. Expression variation of miR172 among leaves and flower buds may suggest developmental regulation of miR172 in allopolyploids as in diploids (Aukerman and Sakai, 2003; Chen, 2004).
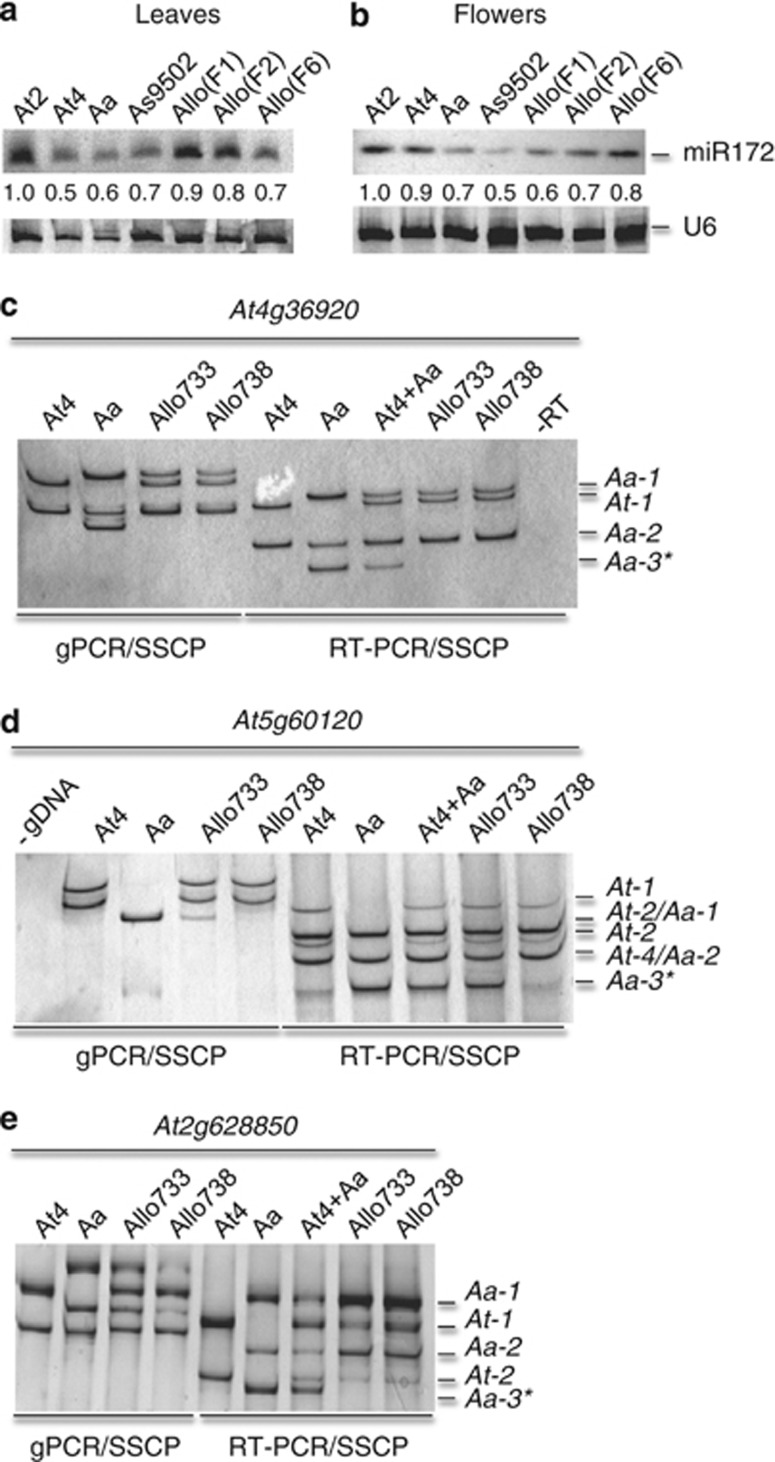
Figure 3.
Differential accumulation of miR172 and their targets in allotetraploids. (a,b) Small RNA blot analyses of miR172 in leaves (a) and flowers (b) of A. thaliana diploid (At2) and tetraploid (A4), A. arenosa (Aa), A. suecica (As9502) and resynthesized allotetraploids (F1, F2, and F6). Hybridization with U6 was used as a loading control. One of three replicates is shown. The relative band intensities were calculated based on U6 and normalized to that in At2 as 1. (c–e) PCR and SSCP analyses of four miR172 targets. Both genomic PCR and RT-PCR were performed, and their PCR products were subjected to SSCP analysis in At4g36920 (c), At5g60120 (d) and At2g28850 (e). PCR using genomic DNA (gPCR) and RNA/cDNA (RT-PCR) plus SSCP analysis were indicated below the gels. No genomic DNA (-gDNA) and no RT (-RT) controls are shown in (c) and (d), respectively. At4 +Aa: an equal amount of RNA mixtures from At4 and Aa. Separated DNA fragments that represent At and Aa alleles of each locus are given in numerical orders from large (1) to small (2 and 3) bands. Asterisks indicate alleles that are not detected, silenced or repressed.
If miRNAs mediate target gene expression, the expression levels of target genes should be inversely correlated with that of miRNA accumulation in the allotetraploids. To test this relationship, we compared miRNA targets identified in A. thaliana (Llave et al., 2002; Reinhart et al., 2002; Wang et al., 2004b; Lu et al., 2005; Xie et al., 2005) with the genes that are nonadditively expressed in resynthesized Arabidopsis allotetraploids (Wang et al., 2006b). A gene is nonadditively expressed if its expression level in an allotetraploid is not equal to the sum of two parents, suggesting activation or silencing (Chen, 2007). We found that miR172 targets are among those that were nonadditively expressed in the allotetraploids (Ha et al., 2009).
miR172 regulates the expression of APETALA2 (AP2)-related genes that mediate flowering time and flower organ identities (Aukerman and Sakai, 2003; Chen, 2004). Four AP2-related targets (At4g36920, At5g60120, At2g28850 and At5g67180) were nonadditively expressed in the allotetraploids (Wang et al., 2006b). We employed reverse-transcriptase PCR (RT-PCR) followed by the SSCP analysis (Hayashi, 1991) to discriminate allelic variation in three of four loci. Qualitative expression changes in Aa-3 allele of At4g36920 in two allotetraploids (Figure 3c), Aa-3 allele of At45g60120 in Allo738 (Figure 3d) and Aa-3 allele of At2g628850 in both allotetraploids (Figure 3e) were associated with the corresponding genomic fragments that were present in A. arenosa but absent in one or both allotetraploids. This suggests a role for genomic changes and/or allelic polymorphisms of outcrossing A. arenosa in the expression difference in newly formed allotetraploids. In addition, expression levels of At-1 allele of At5g60120 (Figure 3d) and At-1 and At-2 alleles of At2g628850 (Figure 3e) were slightly lower in the allotetraploids than that in A. thaliana. The data may suggest that in addition to translational repression of mRNA by miR172 as shown in A. thaliana (Chen, 2004), miR172 also cleaves target transcripts of A. thaliana alleles in allotetraploids (Ha et al., 2009). Note there is a caveat that, in spite of the advantage for allelic discrimination by RT-PCR and SSCP, this assay is semi-quantitative.
Expression origins of miR172 in resynthesized allotetraploids
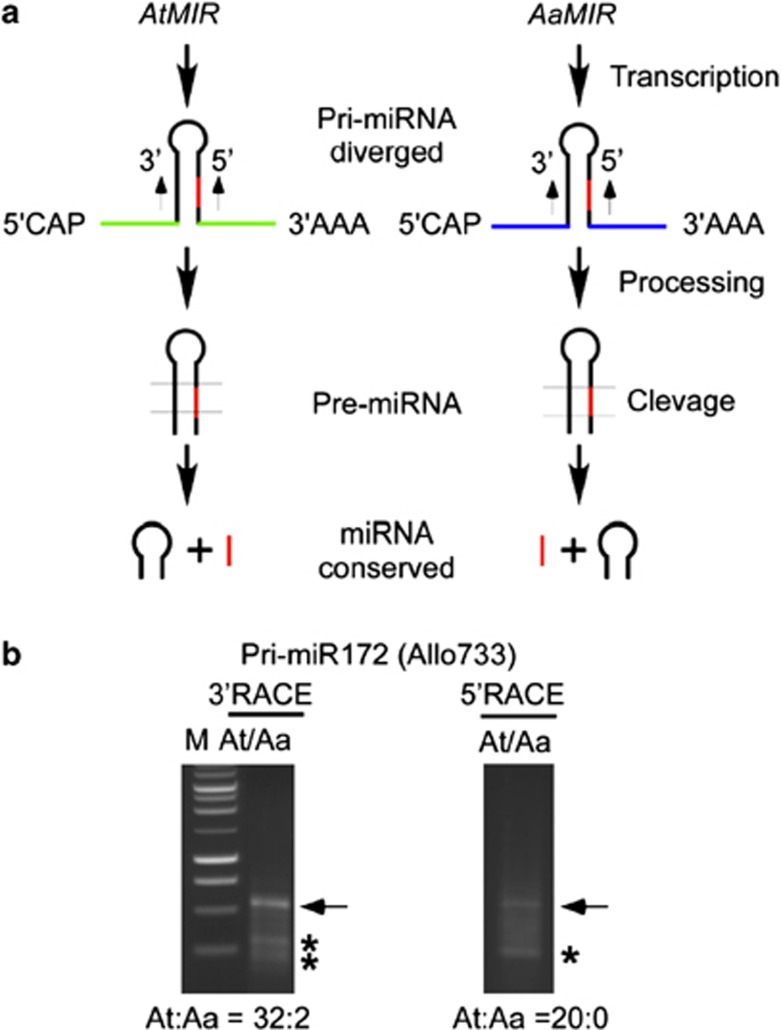
We now consider the origins of miRNAs in allotetraploids because sequence divergence and expression diversity in A. thaliana and A. arenosa may contribute to differential accumulation of miRNAs in the allotetraploids. Using RNA ligase-mediated and 5′ or 3′ rapid amplification of complementary DNA ends (RACE) assays (Kurihara and Watanabe, 2004), we analyzed miR172b pre-miRNAs in the allotetraploids (Figure 4a). We amplified and cloned A. thaliana and A. arenosa pre-miR172b in each parent and in the allotetraploid (Allo733) using the same primer pair (Figure 4b and Supplementary Table S2). In the allotetraploid, of 34 insert sequences recovered in 3′ RACE assays, 32 matched A. thaliana MIR172b, whereas only 2 were derived from A. arenosa. In 5′ RACE assays, all 20 pre-miR172b sequences were derived from A. thaliana. As expected, A. thaliana and A. arenosa generated similar 5′ and 3′ RACE patterns relative to those of allotetraploid, and sequencing confirmed that all fragments were derived from respective species in each assay. The data suggest that MIR172b precursors in the allotetraploids primarily originate from the A. thaliana parent. This is consistent with relatively low expression levels of some A. thaliana target transcripts in allotetraploids (Figures 3d and e). Sequence analysis indicated that additional fragments (with asterisks) detected in RACE assays were alternative splicing products of pri-miR172 transcripts as reported in A. thaliana (Aukerman and Sakai, 2003), suggesting a similar mechanism for miRNA processing in the allotetraploids.
Figure 4.
Detection of pre-miR172 in resynthesized allotetraploids. (a) Diagram of miRNA processing from pri-miRNA, pre-miRNA to miRNA in A. thaliana and A. arenosa. Green and blue colors in the stem-loop structure indicate sequence divergence between related species. Mature miRNAs are in red color. Species-specific primer pairs were designed for MIR172b and used for 5′ and 3′ RACE-PCR amplification. (b) Preferential accumulation of A. thaliana pri-miR172 in Allo733. The ratios of A. thaliana vs A. arenosa transcripts were determined by cloning and sequencing the amplified PCR products (indicated by arrows). Asterisks indicate alternatively spliced products in the 5′ and 3′ ends of AtMIR172.
Progenitor-dependent repression of MIR172b and neighboring genes in allotetraploids
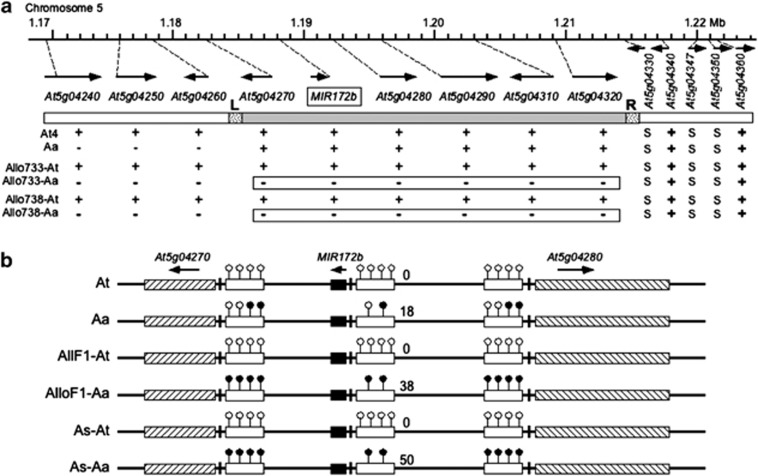
To test whether this repression of A. arenosa miR172 also occurs in neighboring genes in the vicinity of the chromosomal domain in the allotetraploids, we examined expression patterns of 13 genes flanking MIR172b in a 55-kb genomic region using RT-PCR and SSCP assays that can distinguish the transcripts between homoeologous loci (Figure 5a and Supplementary Table S3). Like MIR172b, eight loci in this genomic region were highly expressed in A. thaliana and allotetraploids, suggesting that the expression states of A. thaliana loci are maintained after allopolyploid formation. In A. arenosa, these loci displayed expression diversity: three loci in the 3′ of MIR172 were expressed at very low levels, whereas one locus (At5g04270) in the 3′ of MIR172 and four loci in the 5′ were expressed at high levels similar to those of A. thaliana. Interestingly, six loci spanning from At5g04270 to At5g04320 that were expressed in A. arenosa were repressed in two independently derived allotetraploids. At5g04360 was expressed from both A. thaliana and A. arenosa loci. The expression differences are not caused by genomic sequence changes as both A. thaliana and A. arenosa genomic DNA in all loci studied were equally amplified. The data suggest that the A. arenosa genomic region in the vicinity of MIR172b is subjected to repression after allotetraploid formation. The repressive region spread over 30-kb from the left border (L) near MIR172b to a few silenced genes (S) in the right border (R). Therefore, in contrast to many protein-coding genes that show expression dominance of A. arenosa over A. thaliana loci (Wang et al., 2006b), a miRNA-containing chromosomal domain originating from A. arenosa is subjected to repression in the allopolyploids.
Figure 5.
Repression of A. arenosa MIR172b and its neighboring genes in Arabidopsis allotetraploids. (a) Diagram of an ∼55-kb genomic region containing MIR172b and 13 neighboring genes. Arrows indicate the orientation of gene transcription in each locus. Plus (+), minus (−) and ‘S' indicate expression, low expression and silencing or undetectable in RT-PCR and SSCP assays. The expression states suggest putative left (L) and right (R) borders of the repression domain surrounding A. arenosa-derived MIR172b. The domain containing silenced A. arenosa loci in two allotetraploids is boxed. (b) Repression of A. arenosa MIR172b is associated with DNA methylation in the promoter regions. The genomic DNA located at ∼400-bp upstream of the transcription start site (Xie et al., 2005) was treated with bisulfite and amplified by PCR. The PCR products were purified and cloned, and individually cloned fragments were sequenced. The percentage of CG and CHG (H=A, T or C) methylation is calculated by the number of fragments containing methylated cytosines divided by the total number of fragments analyzed, as shown by the numbers near the promoters (Supplementary Table S5). Open and filled lollipops indicate unmethylated and methylated cytosines, respectively. Open, filled and slashed boxes indicate promoter regions, MIR172b sequences and coding regions of the two neighboring loci, respectively. Arrows indicate transcriptional orientation. AlloF1-At, AlloF1-Aa, As-At and As-Aa indicate A. thaliana (At) and A. arenosa (Aa) alleles in F1 resynthesized allotetraploids (AlloF1) and natural A. suecica (As), respectively.
Hypermethylation of a genomic region containing MIR172b locus
We then tested if the repressed A. arenosa MIR172b locus is associated with DNA methylation in the allotetraploids. We analyzed DNA methylation patterns in 400-bp promoter regions upstream of the pri-mi172b transcription start site in A. thaliana (Xie et al., 2005), A. arenosa and resynthesized allotetraploids (F1) (Figure 5b and Supplementary Table S4). The promoter regions were AT-rich contained four adjacent CG sites and one CHG (H=A, T or C) site (−92 to −400) in A. thaliana and A. arenosa. We estimated relative ratios of methylated and unmethylated sequences by sequencing 50 individual fragments (Supplementary Table S5). A sequence is considered being methylated if methyl-cytosine is present in one or more sites. The promoter regions of A. thaliana MIR172b and its neighboring genes were free of cytosine methylation, whereas ∼18% of A. arenosa MIR172b promoter fragments were methylated. The methylation patterns originating in the progenitors were inherited and enhanced in the first generation of allotetraploid: that is, cytosine methylation in the promoter regions of AtMIR172b and two genes flanking AtMIR172b was undetectable, and ∼38% of A. arenosa MIR172b promoter fragments were methylated. In A. suecica, the A. thaliana fragment was unmethylated but ∼50% of A. arenosa promoter fragments were methylated. The data suggest that hypermethylation of this genomic region is inherited from the A. arenosa parent and correlated with repression of MIR172b and its neighboring genes in the allotetraploids.
Discussion
Polyploidy induces extensive genomic changes including chromosomal rearrangements and transpositions, as observed in allopolyploids in Brassica (Xiong et al., 2011), Tragopogon (Chester et al., 2012) and wheat (Feldman et al., 1997). Some of these changes are associated with variation in flowering time (Pires et al., 2004) and epigenetic changes in DNA methylation (Gaeta et al., 2007) in Brassica napus, with transposon activation and altered expression of neighboring genes in wheat (Kashkush et al., 2003) and with gene expression changes in Tragopogon (Tate et al., 2006). In contrast, genomic changes are not commonly found in allopolyploids of Arabidopsis (Comai et al., 2000) and cotton (Liu et al., 2001). Gene expression changes observed in allopolyploids of cotton (Adams et al., 2003; Flagel et al., 2008) and Arabidopsis (Wang et al., 2004a, 2006b) are likely caused by epigenetic changes (Chen, 2007). A. arenosa is an outcrossing tetraploid, which displays high levels of sequence divergence between alleles of the same miR172 locus relative to A. thaliana ones, and in some cases loss of target alleles, which is related to allelic expression differences in allotetraploids. This level of diversity may be underestimated in previous studies and should be revisited.
miRNA loci in plants originated by inverted duplication of target genes (Allen et al., 2004; Cuperus et al., 2011). Although mature miRNA sequences are conserved, the flanking sequences including the foldback are often less conserved compared with protein sequences. Among four homoeologous loci in the miR172 family, the foldback sequences contain many changes including small deletions and insertions. The level of nucleotide sequence divergence is very high (15–25%) in miRNA loci compared with protein-coding genes (∼5%) (Hanfstingl et al., 1994; Henikoff and Comai, 1998; Lee and Chen, 2001; Wang et al., 2006a). Indeed, miRNA loci evolve rapidly between A. thaliana and A. lyrata (Fahlgren et al., 2010). This does not preclude a wide range of gene evolutionary rates that exist among various ecotypes within A. thaliana (Clark et al., 2007). Plant miRNA foldback sequences are highly variable and may contribute to the specificity of miRNA targeting (Ha et al., 2008). Although the cause for this is unknown, the preference for degrading homoeologous transcripts of miR163 (Ng et al., 2011) and miR164 and miR168 (Ha et al., 2008) is also found in allotetraploids.
Many miRNAs in the A. thaliana genome affect multiple mRNA targets encoding transcription factors and other proteins important to plant development (Reinhart et al., 2002; Wang et al., 2004b; Xie et al., 2005; Cuperus et al., 2011). miR172 affects flowering time and floral organ identity (Aukerman and Sakai, 2003), regulates stem cell fate and defines the inner boundary of APETALA3 and PISTILLATA expression domain in Arabidopsis (Zhao et al., 2007). Differential accumulation of miR172 in allotetraploids and their progenitors may suggest a role of miR172 in developmental differences between the closely related species. Like A. arenosa, allotetraploids flower late, whereas A. thaliana flowers early. Although late flowering is associated with complementation of A. arenosa FRIGIDIA with nonfunctional A. thaliana FRI in the allotetraploids (Wang et al., 2006a), it does not rule out a possibility that miR172 may also affect flowering time in the allotetraploids that display a wide range of flowering time variation. Differential accumulation of miR172 is correlated with expression changes in two of the four target genes, suggesting that miR172 affects target gene regulation by both mRNA cleavage and translational repression (Aukerman and Sakai, 2003; Chen, 2004; Schwab et al., 2005).
Abundance levels of miR172 also change in A. thaliana diploids and autotetraploids, suggesting that both genome doubling and hybridization affect miRNA accumulation. Expression levels of miRNAs and their target genes are also different between the progenitors and subjected to changes in the allotetraploids. These quantitative and qualitative changes in miRNA accumulation correlate inversely with the expression levels of target genes, leading to gene expression changes in the allotetraploids. Species- and locus-specific changes in miRNA accumulation may account for some aspects of developmental and phenotypic variation in different allopolyploid lineages and their progenitors, which may facilitate adaptive evolution of allopolyploids and their progenitors.
Data archiving
Sequences of Arabidopsis arenosa MIR172 loci have been deposited in GenBank: Accession numbers KF543109-KF543110, KF543112, KF543113, KF543116-KF543117, KF543119-KF543120.
Acknowledgments
The work is supported by grants (IOS0733857 and IOS1025947) from the NSF Plant Genome Research Program (ZJC).
The authors declare no coflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Adams KL, Cronn R, Percifield R, Wendel JF. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc Natl Acad Sci USA. 2003;100:4649–4654. doi: 10.1073/pnas.0630618100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Sung GH, Spatafora JW, Carrington JC. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet. 2004;36:1282–1290. doi: 10.1038/ng1478. [DOI] [PubMed] [Google Scholar]
- Arteaga-Vazquez M, Caballero-Perez J, Vielle-Calzada JP. A family of microRNAs present in plants and animals. Plant Cell. 2006;18:3355–3369. doi: 10.1105/tpc.106.044420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- Buggs RJ, Zhang L, Miles N, Tate JA, Gao L, Wei W, et al. Transcriptomic shock generates evolutionary novelty in a newly formed, natural allopolyploid plant. Curr Biol. 2011;21:551–556. doi: 10.1016/j.cub.2011.02.016. [DOI] [PubMed] [Google Scholar]
- Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. Small RNAs and their roles in plant development. Ann Rev Cell Dev Biol. 2009;25:21–44. doi: 10.1146/annurev.cellbio.042308.113417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol. 2007;58:377–406. doi: 10.1146/annurev.arplant.58.032806.103835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester M, Gallagher JP, Symonds VV, Cruz da Silva AV, Mavrodiev EV, Leitch AR, et al. Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae) Proc Natl Acad Sci USA. 2012;109:1176–1181. doi: 10.1073/pnas.1112041109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RM, Schweikert G, Toomajian C, Ossowski S, Zeller G, Shinn P, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- Comai L. The advantages and disadvantages of being polyploid. Nat Rev Genet. 2005;6:836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- Comai L, Tyagi AP, Winter K, Holmes-Davis R, Reynolds SH, Stevens Y, et al. Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell. 2000;12:1551–1568. doi: 10.1105/tpc.12.9.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus JT, Fahlgren N, Carrington JC. Evolution and functional diversification of miRNA genes. Plant Cell. 2011;23:431–442. doi: 10.1105/tpc.110.082784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Jogdeo S, Kasschau KD, Sullivan CM, Chapman EJ, Laubinger S, et al. MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana. Plant Cell. 2010;22:1074–1089. doi: 10.1105/tpc.110.073999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M, Liu B, Segal G, Abbo S, Levy AA, Vega JM. Rapid elimination of low-copy DNA sequences in polyploid wheat: a possible mechanism for differentiation of homoeologous chromosomes. Genetics. 1997;147:1381–1387. doi: 10.1093/genetics/147.3.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel L, Udall J, Nettleton D, Wendel J. Duplicate gene expression in allopolyploid Gossypium reveals two temporally distinct phases of expression evolution. BMC Biol. 2008;6:16. doi: 10.1186/1741-7007-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaeta RT, Pires JC, Iniguez-Luy F, Leon E, Osborn TC. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell. 2007;19:3403–3417. doi: 10.1105/tpc.107.054346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Lu J, Tian L, Ramachandran V, Kasschau KD, Chapman EJ, et al. Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc Natl Acad Sci USA. 2009;106:17835–17840. doi: 10.1073/pnas.0907003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Pang M, Agarwal V, Chen ZJ. Interspecies regulation of microRNAs and their targets. Biochim Biophys Acta. 2008;1779:735–742. doi: 10.1016/j.bbagrm.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanfstingl U, Berry A, Kellogg EA, Costa JT, 3rd, Rudiger W, Ausubel FM. Haplotypic divergence coupled with lack of diversity at the Arabidopsis thaliana alcohol dehydrogenase locus: Roles for both balancing and directional selection. Genetics. 1994;138:811–828. doi: 10.1093/genetics/138.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K. PCR-SSCP: a simple and sensitive method for detection of mutations in the genomic DNA. PCR Methods Appl. 1991;1:34–38. doi: 10.1101/gr.1.1.34. [DOI] [PubMed] [Google Scholar]
- Hegarty MJ, Barker GL, Wilson ID, Abbott RJ, Edwards KJ, Hiscock SJ. Transcriptome shock after interspecific hybridization in senecio is ameliorated by genome duplication. Curr Biol. 2006;16:1652–1659. doi: 10.1016/j.cub.2006.06.071. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Comai L. A DNA methyltransferase homolog with a chromodomain exists in multiple polymorphic forms in Arabidopsis. Genetics. 1998;149:307–318. doi: 10.1093/genetics/149.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S, Chen ZJ. Genomic and expression plasticity of polyploidy. Curr Opin Plant Biol. 2010;13:153–159. doi: 10.1016/j.pbi.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Sakai H, Finnegan EJ, Cao X, Meyerowitz EM. Ectopic hypermethylation of flower-specific genes in Arabidopsis. Curr Biol. 2000;10:179–186. doi: 10.1016/s0960-9822(00)00324-9. [DOI] [PubMed] [Google Scholar]
- Kashkush K, Feldman M, Levy AA. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat Genet. 2003;33:102–106. doi: 10.1038/ng1063. [DOI] [PubMed] [Google Scholar]
- Kenan-Eichler M, Leshkowitz D, Tal L, Noor E, Melamed-Bessudo C, Feldman M, et al. Wheat hybridization and polyploidization results in deregulation of small RNAs. Genetics. 2011;188:263–272. doi: 10.1534/genetics.111.128348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch MA, Haubold B, Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae) Mol Biol Evol. 2000;17:1483–1498. doi: 10.1093/oxfordjournals.molbev.a026248. [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci USA. 2004;101:12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Chen ZJ. Protein-coding genes are epigenetically regulated in Arabidopsis polyploids. Proc Natl Acad Sci USA. 2001;98:6753–6758. doi: 10.1073/pnas.121064698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch AR, Leitch IJ. Genomic plasticity and the diversity of polyploid plants. Science. 2008;320:481–483. doi: 10.1126/science.1153585. [DOI] [PubMed] [Google Scholar]
- Levy AA, Feldman M. The impact of polyploidy on grass genome evolution. Plant Physiol. 2002;130:1587–1593. doi: 10.1104/pp.015727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Brubaker G, Cronn RC, Wendel JF. Polyploid formation in cotton is not accompanied by rapid genomic changes. Genome. 2001;44:321–330. [PubMed] [Google Scholar]
- Llave C, Kasschau KD, Rector MA, Carrington JC. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002;14:1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Tej SS, Luo S, Haudenschild CD, Meyers BC, Green PJ. Elucidation of the small RNA component of the transcriptome. Science. 2005;309:1567–1569. doi: 10.1126/science.1114112. [DOI] [PubMed] [Google Scholar]
- Masterson J. Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science. 1994;264:421–424. doi: 10.1126/science.264.5157.421. [DOI] [PubMed] [Google Scholar]
- Ng DW, Lu J, Chen ZJ. Big roles for small RNAs in polyploidy, hybrid vigor, and hybrid incompatibility. Curr Opin Plant Biol. 2012;15:154–161. doi: 10.1016/j.pbi.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Ng DW, Zhang C, Miller M, Palmer G, Whiteley M, Tholl D, et al. cis- and trans-Regulation of miR163 and target genes confers natural variation of secondary metabolites in two Arabidopsis species and their allopolyploids. Plant Cell. 2011;23:1729–1740. doi: 10.1105/tpc.111.083915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP. The evolutionary consequences of polyploidy. Cell. 2007;131:452–462. doi: 10.1016/j.cell.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Pires JC, Zhao JW, Schranz ME, Leon EJ, Quijada PA, Lukens LN, et al. Flowering time divergence and genomic rearrangements in resynthesized Brassica polyploids (Brassicaceae) Biol J Linnean Soc. 2004;82:675–688. [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Tate JA, Ni Z, Scheen AC, Koh J, Gilbert CA, Lefkowitz D, et al. Evolution and expression of homeologous loci in Tragopogon miscellus (Asteraceae), a recent and reciprocally formed allopolyploid. Genetics. 2006;173:1599–1611. doi: 10.1534/genetics.106.057646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Chen ZJ. Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc Natl Acad Sci USA. 2001;98:200–205. doi: 10.1073/pnas.011347998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Tian L, Lee HS, Chen ZJ. Nonadditive regulation of FRI and FLC loci mediates flowering-time variation in Arabidopsis allopolyploids. Genetics. 2006;173:965–974. doi: 10.1534/genetics.106.056580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Tian L, Lee HS, Wei NE, Jiang H, Watson B, et al. Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics. 2006;172:507–517. doi: 10.1534/genetics.105.047894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Tian L, Madlung A, Lee HS, Chen M, Lee JJ, et al. Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics. 2004;167:1961–1973. doi: 10.1534/genetics.104.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Reyes JL, Chua NH, Gaasterland T. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004;5:R65. doi: 10.1186/gb-2004-5-9-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC. Expression of Arabidopsis miRNA genes. Plant Physiol. 2005;138:2145–2154. doi: 10.1104/pp.105.062943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z, Gaeta RT, Pires JC. Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc Natl Acad Sci USA. 2011;108:7908–7913. doi: 10.1073/pnas.1014138108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Kim Y, Dinh TT, Chen X. miR172 regulates stem cell fate and defines the inner boundary of APETALA3 and PISTILLATA expression domain in Arabidopsis floral meristems. Plant J. 2007;51:840–849. doi: 10.1111/j.1365-313X.2007.03181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.