Abstract
The zebrafish Danio rerio has emerged as a powerful vertebrate model system that lends itself particularly well to quantitative investigations with live imaging approaches, owing to its exceptionally high optical clarity in embryonic and larval stages. Recent advances in light microscopy technology enable comprehensive analyses of cellular dynamics during zebrafish embryonic development, systematic mapping of gene expression dynamics, quantitative reconstruction of mutant phenotypes and the system-level biophysical study of morphogenesis.
Despite these technical breakthroughs, it remains challenging to design and implement experiments for in vivo long-term imaging at high spatio-temporal resolution. This article discusses the fundamental challenges in zebrafish long-term live imaging, provides experimental protocols and highlights key prop1erties and capabilities of advanced fluorescence microscopes. The article focuses in particular on experimental assays based on light sheet-based fluorescence microscopy, an emerging imaging technology that achieves exceptionally high imaging speeds and excellent signal-to-noise ratios, while minimizing light-induced damage to the specimen. This unique combination of capabilities makes light sheet microscopy an indispensable tool for the in vivo long-term imaging of large developing organisms.
Keywords: Zebrafish, Fluorescence microscopy, Light sheet microscopy, In vivo imaging, Quantitative developmental biology, Image processing
1. Introduction
The ability to acquire and quantitatively analyze light microscopy data of living biological organisms is crucial in many areas of the life sciences. However, live imaging of large multicellular specimens is fundamentally constrained by the limited optical penetration depth of light microscopy. In many model systems, light scattering and light absorption obscure the view of the inside of the specimen and preclude functional imaging or the live observation of developmental processes. Zebrafish embryos and larvae, which are highly transparent, are a remarkable exception. They offer the exciting opportunity to study development and function in a highly complex vertebrate model system by live imaging.
By using standard light microscopy techniques, detailed morphogenetic information can be obtained on the cellular and subcellular levels in a completely non-invasive manner [1]. By using advanced imaging approaches optimized for high-speed long-term imaging, it is even possible to study highly dynamic morphological processes over long periods of time and from the whole-embryo perspective, to obtain comprehensive gene expression atlases with high spatio-temporal resolution, to follow the formation of morphological defects in mutants, to reconstruct the dynamic building plans of entire organs and tissues, and to assess the level of morphogenetic variability between different embryos [2,3].
2. Challenges in long-term live imaging experiments
Several conflicting requirements need to be balanced in in vivo imaging applications. It is often desirable to achieve high imaging speed, low photo-bleaching and photo-toxicity, good three-dimensional resolution, high signal-to-noise ratio and excellent physical coverage at the same time. The latter aspect is particularly difficult to realize for large biological specimens, since limitations in optical penetration depth usually make it impossible to extract comprehensive information of the sample from a single view.
Some forms of light microscopy provide good overall performance, whereas others excel in one area by trading off performance in other areas. Conventional wide-field fluorescence microscopes provide high imaging speed and a good signal-to-noise ratio, but lack optical sectioning capability and are therefore unsuited for three-dimensional imaging. Point-scanning confocal fluorescence microscopes provide good spatial resolution and the intrinsic capability to eliminate scattered light [4], but perform poorly with respect to imaging speed, signal-to-noise ratio, photo-bleaching/toxicity and penetration depth. Pointscanning two-photon microscopes improve over confocal fluorescence microscopes with respect to penetration depth and usually also photo-bleaching/toxicity [5,6], but further reduce signal-to-noise ratio and imaging speed. Spinning disk confocal microscopes provide higher imaging speed than (single) point-scanning confocal fluorescence microscopes [7], but reduce spatial resolution.
To address the limitations arising from the intrinsic performance trade-offs encountered in conventional light microscopy techniques, new light sheet-based fluorescence microscopy techniques have been developed over the course of the past decade. Light sheet-based fluorescence microscopy, which will be discussed in Section 3, provides a particularly powerful approach to biological live imaging. Such microscopes combine intrinsic optical sectioning capability with high imaging speeds, good signal-to-noise ratio and low light exposure of the specimen [8-11].
A particular strength of this unique combination of capabilities is the possibility to study embryonic development on the system level while preserving the ability to follow cellular processes at the high spatio-temporal resolution required for global cell tracking [3]. For example, the reconstruction of zebrafish embryogenesis during the first 24 h of development requires the simultaneous tracking of several tens of thousands of cells, e.g. by fast long-term imaging of a nuclear fluorescent marker expressed in all cells [2]. High spatio-temporal resolution, low photo-bleaching rates, and an excellent signal-to-noise ratio are crucial. A volume of ~1 mm3 must be recorded approximately once per minute to keep track of cell identities, since cells move in average several micrometer per minute. In order to reliably detect all nuclei, good spatial resolution and axial sampling are necessary, which requires acquiring several hundred images at each time point to achieve full coverage of the embryo. Overall, the observation must thus be performed at a continuous imaging speed of at least 10 million volume elements (voxels) per second, which is approximately one order of magnitude faster than the typical acquisition speed of conventional point-scanning confocal or two-photon microscopes [2]. At the same time, a high dynamic range is required to address the varying expression levels of genetically encoded markers and signal heterogeneity due to signal degradation with increasing imaging depth into the embryo. The embryo’s central yolk cell is opaque at visible wavelengths and imaging along multiple directions (providing a set of complementary views of the embryo) is needed to capture the development of the entire embryo. Finally, photobleaching and photo-toxicity [12] must be minimized to ensure the physiological development of the embryo [2].
Point-scanning confocal fluorescence microscopy is still the standard imaging technology used in most labs, and advanced light sheet fluorescence microscopy technology has become commercially available only very recently. However, the latter method provides a number of key advantages, which are of particular importance in long-term live imaging experiments. A comparison of both techniques is therefore provided in Section 4, which summarizes and expands on a previous discussion of these methods [34]. Two-photon fluorescence microscopy is not discussed as a separate method in this comparison, since two-photon excitation can effectively be implemented in both imaging techniques mentioned above [13-15].
3. Light sheet microscopy
3.1. Introduction to light sheet microscopy
Conventional and cofocal epi-fluorescence microscopes employ the same lens for fluorescence excitation and detection. In contrast, light sheet microscopes rely on the principle of sample illumination with a planar light sheet perpendicular to the axis of fluorescence detection (Fig. 1A) [16-18]. The light sheet is coplanar with the focal plane of the fluorescence detection system. This approach directly provides optical sectioning: Fluorophores are only excited in the illuminated plane and thus photo-bleaching and other types of photo-induced damage are avoided outside the thin volume of interest. This circumstance gives light sheet microscopes a decisive advantage in the fast imaging of sensitive biological specimens as well as in in vivo imaging over long periods of time [2,15,18-21].
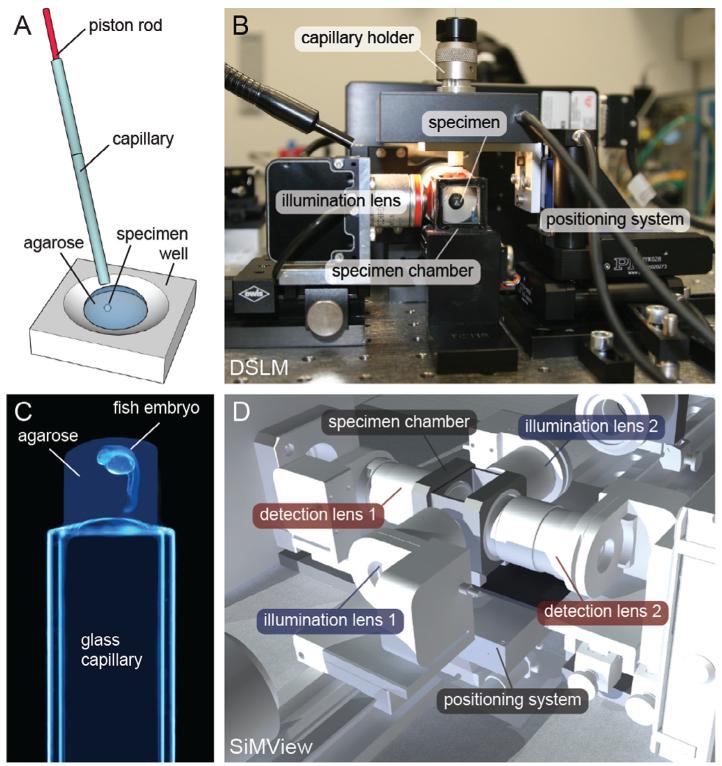
Fig. 1.
Scanned light sheet fluorescence microscopy. (A) The illustration shows the principle behind Digital Scanned Laser Light Sheet Fluorescence Microscopy (DSLM) [2]. The f-theta lens converts the tilting movement of the scan mirror into a vertical displacement of the laser beam. The tube lens and the illumination objective focus the laser beam into the specimen, which is positioned in front of the detection lens. The laser beam thus illuminates the specimen from the side and excites fluorophores along a single line. Rapid scanning of a thin volume and fluorescence detection at a right angle to the illumination axis provides an optically sectioned image. (B) Photograph of a basic DSLM implementation, using a single CCD camera for fluorescence detection. The optical axis in the DSLM illumination sub-system is shown as a dashed cyan line, whereas the optical axis in the DSLM illumination detection sub-system is shown as a dashed green line. (C) Computer model of the opto-mechanical implementation of a light sheet microscope for simultaneous multi-view imaging. The opto-mechanical modules of the instrument consist of two illumination arms for fluorescence excitation with scanned light sheets (blue), two fluorescence detection arms equipped with sCMOS cameras (red) as well as beam-coupling modules, specimen chamber and specimen positioning system (grey). Credits: Panel (A) was reprinted from Science, vol. 322, Keller et al., “Reconstruction of Zebrafish Early Embryonic Development by Scanned Light Sheet Microscopy”, 1065-1069, Copyright (2008), with permission from AAAS. Panel (B) was reprinted from Cold Spring Harbor Protocols, vol. 2011 No. 10, Keller et al., “Digital scanned laser light-sheet fluorescence microscopy (DSLM) of zebrafish and Drosophila embryonic development”, 1235-1243, Copyright (2011), with permission from Cold Spring Harbor Laboratory Press. Panel (C) was reprinted from Nature Methods, vol. 9 No. 7, Tomer et al., “Quantitative High-Speed Imaging of Entire Developing Embryos with Simultaneous Multiview Light-Sheet Microscopy”, 753-763, Copyright (2012), with permission from Macmillan Publishers Ltd.
Light sheet microscopes are furthermore particularly well suited for “multi-view imaging”, which refers to the strategy of observing the same specimen along multiple different directions. Thereby, parts of the specimen become visible that would otherwise be hidden or obscured in the observation along a single direction [22]. The next two sections discuss the building blocks of a basic light sheet-based microscope [2], which is particularly costefficient yet very effective, as well as an advanced light sheet microscope for simultaneous multi-view imaging [15], which provides excellent physical coverage even of very large biological specimens.
3.2. Basic implementation of scanned light sheet microscopy
Different approaches have been developed to facilitate light sheet illumination, including the use of cylindrical optics to focus a Gaussian laser beam into a sheet of light [17,18] and fast laserscanning of a laser beam that enters the specimen chamber from the side [2]. In scanned light sheet microscopy (DSLM), a “light sheet” is generated with a laser scanner that rapidly moves a micrometer-thin beam of laser light vertically through the specimen (Fig. 1A) [2]. The specimen is positioned in front of a second lens, the lens for fluorescence detection, and typically embedded in an aqueous gel [18] or in a transparent plastic compartment[19,23].
The basic DSLM implementation is shown in Fig. 1B. The DSLM illumination system comprises a multi-line argon-krypton laser (Melles Griot, 35 LTL 835-230), an acousto-optical tunable filter for laser wavelength selection and intensity control (AA Opto-Electronic, AA.AOTF.nC-400-650 nm-PV-TN), a two-axis high-speed scan head (GSI Lumonics, VM500+), an f-theta lens (Sill Optics, S4LFT0061/065/) and a low-NA illumination objective lens (Carl Zeiss, Plan-Apochromat 5 ×/0.16) operated with a regular tube lens. The illumination/excitation objective lens is mounted on a piezo nanofocus (Physik Instrumente, P-725.CLQ), which can move the lens by up to 400 μm along its optical axis [2].
The DSLM detection system can take advantage of several detection objective lenses (in particular Carl Zeiss C-Apochromat 10 ×/0.45 W and Plan-Apochromat 20 ×/1.0 W for zebrafish whole-embryo imaging) mounted on a second independently operated piezo nano-focus, a filter wheel (Ludl) with exchangeable long-pass filters (Semrock, RazorEdge RU 488 LP, RU 568 LP and RU 647 LP) and a detection tube, equipped with a tube lens and a CCD camera (PCO, pco.2000) [2].
3.3. Advanced implementation of light sheet microscopy for simultaneous multi-view imaging
The SiMView light sheet microscopy platform provides exceptionally high imaging speeds and allows simultaneous acquisition of four complementary views of the specimen for optimal physical coverage [15]. In order to realize simultaneous multi-view imaging, SiMView uses an orthogonal arrangement of four independently operated optical arms (Fig. 1C, Video 1). One pair of opposite arms is used for bi-directional light sheet illumination with two long working distance air objectives, similar to the illumination arrangement used in other light sheet microscopes[14,24,25]. The other pair, arranged at a right angle to the first, is used for bi-directional fluorescence detection with high numerical aperture water-dipping objectives and fast sCMOS cameras [15].
In addition to the illumination and detection arms, the SiMView microscope platform includes a laser light source, a four-view specimen chamber and a four-axis specimen positioning system (Fig. 1C, Video 1).
For imaging with one-photon excitation, the laser beams of two multi-laser units (Omicron Laserage, SOLE-3 and SOLE-6) are combined by fiber optics and split into two illumination arms for fluorescence excitation from near-UV to near-IR, providing the laser excitation wavelengths 405, 445, 488, 515, 561, 594, 642 and 685 nm [15].
Each illumination arm consists of two custom collimator modules, a beam coupling-unit, a high-speed laser shutter with controller (Uniblitz, VS14S2ZM1-100 with AlMgF2 coating, VMM-D3 three-channel driver), a compact illumination filter wheel with controller (Ludl, 96A351, MAC6000 DC servo controller), a miniature piezo tip/tilt mirror with amplifier (Physik Instrumente, S-334, E-503.00S amplifier, E-509.S3 servo controller, E-500 chassis), an f-theta lens (Sill Optics, S4LFT4375), a tube lens module (Olympus, U-TLU-1-2 camera tube) and a piezo-positioned illumination objective with controller (Physik Instrumente, P-725 piezo, E-665 piezo amplifier and servo controller; equipped with Olympus XLFLUOR 4 ×/340/0.28 objective) [15].
Each detection arm consists of a piezo-positioned detection objective with controller (Physik Instrumente, P-725 piezo, E-665 piezo amplifier and servo controller; equipped with long working distance water-dipping objectives from Nikon or Carl Zeiss), a detection filter wheel with controller (Ludl, 96A354, MAC6000 DC servo controller), a custom tube module with a tube lens (Nikon, CFI second lens unit) and a water-cooled sCMOS camera (Hamamatsu, Orca Flash 4.0; or Andor, Neo, with Koolance ExosII re-circulator; or PCO, pco.edge) [15].
The four-view specimen chamber comprises a custom specimen holder, a custom mechanical scaffold manufactured from black Delrin, a multi-stage adapter module for connecting the specimen holder to the specimen positioning system and a custom perfusion system. Using the positioning system, the medical-grade stainless steel specimen holder can be translated in three dimensions and rotated around its main axis without breaking the water seal. The chamber is open to two opposite sides to accommodate the water-dipping detection objectives and contains two windows with coverslips on the remaining two sides for laser light sheet illumination. The top of the chamber is open for mechanical or optical access and allows specimen illumination with a cold light source [15].
The four-axis (X−Y−Z−Theta) specimen positioning system consists of three customized translation stages (Physik Instrumente, M-111K046) and one rotary stage (Physik Instrumente, M-116), a motion I/O interface and amplifier (Physik Instrumente, C-809.40 4-channel servo-amplifier) and a motion controller (National Instruments, PXI-7354 4-axis stepper/servo motion controller).
Video 1 shows the complete mechano-optical assembly of the one-photon SiMView implementation, with the exception of the multi-laser units.
For imaging with two-photon excitation, additional optical modules are required. The laser light source is a pulsed Ti:Sapphire laser (Coherent, Chameleon Ultra II). Laser intensity modulation and IR beam splitting are performed with a module positioned between light source and illumination arms, which consists of an (optional) beam attenuation sub-module (Thorlabs, AHWP05M-980 mounted achromatic half-wave plate and GL10-B Glan-laser polarizer), a Pockels cell with driver (Conoptics, Model 350-80-LA-02 KD/P series electro-optic modulator and Model 302RM driver) and an IR beam splitter (Melles Griot, PBSH-450-2000-100 broadband polarizing cube beam splitter, and Casix, WPA1312-2-700-1000 achromatic 1/2 wave plate). Customized f-theta lenses (Special Optics, 66-S80-30T-488-1100 nm custom lens) and different illumination objectives (Olympus, LMPLN10XIR 10/0.30) are used in the illumination arms. The illumination and detection × objectives are mounted on different piezo-positioners (Physik Instrumente, P-622.1CD PIHera piezo linear stage and E-665 piezo amplifier and servo controller). Finally, different filters are used in the detection arms (Semrock, BrightLine short-pass and band-pass filters) [15].
4. Comparison of light sheet microscopy and confocal microscopy
This section summarizes and expands on a previous comparison of light sheet microscopy and confocal microscopy [34].
4.1. Light exposure, laser power and photo-bleaching/toxicity
A distinct advantage of light sheet microscopy is that only the observed plane is actually illuminated. In contrast, point-scanning confocal fluorescence microscopes perform optical sectioning by discriminating against out-of-focus light with the detection pinhole. The detection of the signal from a single plane requires the entire sample to be illuminated, and fluorescence light that is emitted from out-of-focus regions (the majority of the signal) is simply blocked by the detection pinhole.
The corresponding reduction of light exposure in light sheet microscopes compared to confocal microscopes can be approximated as a factor n equal to the total thickness of the recorded image stack divided by the light sheet thickness. Photo-bleaching rates in confocal fluorescence microscopes are thus at least by a factor of n higher than in light sheet microscopy [34]. In practice, this factor is usually even larger for point-scanning confocal fluorescence microscopes, since these implementations often operate in non-linear photo-bleaching regimes, owing to the high laser power (see below).
Since scanned light sheet microscopes perform one-dimensional laser scanning, the time available for light exposure of a single voxel in the recorded image is several orders of magnitude longer than in point-scanning confocal fluorescence microscopes, which employ two-dimensional laser scanning. In a typical highspeed imaging experiment with DSLM (~15 frames per second for an image size of 2048 × 2048 pixels), fluorescence signal is collected for approximately 500 μs for each camera pixel [2]. In contrast, point-scanning confocal and two-photon fluorescence microscopes use pixel dwell times of typically no more than 5 μs when operating in a regime of reasonably high imaging speeds (at least 1 frame per second for an image size of 512 512 pixels). In scanned light sheet microscopes, the laser power required to collect a certain number of signal photons per time unit is therefore typically reduced by at least a factor of 100 compared to point-scanning confocal fluorescence microscopes [34].
4.2. Spatial resolution
The lateral and axial resolution of light sheet microscopes and confocal fluorescence microscopes is given by the wavelength of the fluorescence light, the numerical apertures of the lenses and by the signal-to-noise ratio in the image. In theory, point-scanning confocal fluorescence microscopy provides a lateral resolution that is by a factor of square-root of 2 better than in light sheet microscopy. In practice, however, the limited signal-to-noise ratio of confocal fluorescence microscopes often makes it difficult to take advantage of this improved performance [4,26,34].
The situation is different for the axial resolution. Light sheet microscopy performs optical sectioning with a diffraction-limited illumination profile and provides multi-view capability, i.e. the option to record multiple data sets of a specimen along different directions. In single-view experiments with lenses of low numerical aperture, the axial resolution is mainly determined by the thickness of the light sheet. In multi-view experiments, the axial resolution can be improved by fusing the information obtained along different recording angles. For highly transparent specimens, the fusion of a few angles already leads to an almost isotropic three-dimensional resolution, i.e. the usually low axial resolution becomes comparable to the considerably better lateral resolution[22,34].
It has been shown that light sheet microscopy outperforms confocal fluorescence microscopes for the entire range of detection NAs in multi-view experiments [27]. In single-view experiments and for numerical apertures up to about 0.6-0.8 (depending on the field-of-view of the light sheet microscope’s camera), light sheet microscopy provides an at least 50% better axial resolution. Confocal fluorescence microscopes perform better than light sheet microscopes with numerical apertures above 0.6-0.8 (depending on the field-of-view of the light sheet microscope’s camera) [27].
The use of Bessel beams provides another option for improving axial resolution in the light sheet microscope [28,29]. Bessel beams have a thinner central intensity peak compared to Gaussian beams produced with the same optical configuration. However, unlike the Gaussian beams used in conventional light sheet microscopy, Bessel beams also comprise a system of concentric rings surrounding this central peak. If left unaccounted for, this ring system will contribute to the generation of fluorescence signal and thereby degrade image quality. In order to improve axial resolution over that obtained in Gaussian beam light sheet microscopes, the contribution from the ring system can be suppressed by using structured illumination (i.e. patterned light sheets [30]) or two-photon excitation.
Finally, it is important to note that the high signal-to-noise ratio in light sheet microscopy (see below) renders the images particularly amenable to image deconvolution, providing a further increase in lateral and axial resolution [31].
4.3. Signal-to-noise ratio and image quality
At reasonably high imaging speeds (at least 1 million voxels per second) and reasonably low laser power settings compatible with long-term live imaging, confocal fluorescence microscopes provide images with a signal-to-noise ratio of typically not more than 100:1, owing to physical limitations arising from short pixel dwell times (1 μs or less). In high-speed imaging experiments with light sheet microscopy, pixel dwell times are considerably longer (typically on the order of 500 μs) as a direct result of the signal detection from many volume elements simultaneously. Using CCD or sCMOS cameras with high dynamic range, light sheet microscopes provide images with a signal of up to ~14 bit above the background and a standard deviation of ~2 bit in the background. The signal-to-noise ratio is typically in the range 100:1 to 10,000:1 [34].
As indicated in the previous section, the possibility to increase resolution by image deconvolution is one of the advantages of the high dynamic range in light sheet microscopy. More importantly, however, the good signal-to-noise ratio is crucial to ensure robust performance of automated computational approaches to the analysis of large image data sets.
4.4. Imaging speed
Light sheet microscopy accommodates exceptionally high imaging speeds through the use of camera-based fluorescence detection. For example, in a typical DSLM detection system, intensity data is collected in parallel for ~6 to 12 entire lines on the camera chip, constituting ~12,000-24,000 pixels of the final image [34]. The SiMView imaging platform, which performs simultaneous image acquisition from multiple views, is capable of recording data at a continuous rate of 175 million voxels per second. This level of parallelization in light sheet microscopes leads to a dramatic speed advantage over point-scanning confocal fluorescence microscopes, which employ two-dimensional scanning of the specimen, i.e. record fluorescence data voxel per voxel at maximum rates of about 1–10 million voxels per second.
Line-scanning implementations of confocal fluorescence microscopes suffer from a substantial decrease in image quality and contrast. In this context, it is important to note that each sample provides a fixed photon “budget”, i.e. faster scanning will also inevitably lead to an accordingly increased photo-bleaching rate. Thus, line-scanning or resonant scanner confocal microscopy implementations perform poorly in long-term high-speed imaging experiments due to the high photo-bleaching rates [34].
Similar to light sheet microscopy, spinning-disk confocal fluorescence microscopes are able to take advantage of the high recording speed and high quantum efficiency provided by CCD and sCMOS cameras. However, image quality in spinning-disk confocal microscopes is reduced compared to point-scanning implementations, owing to the simultaneous use of multiple pinholes and associated issues arising from light scattering and loss of incoherence [34].
5. Live imaging of zebrafish embryos with light sheet microscopy
In the following three sections, the basic steps involved in setting up a zebrafish live imaging experiment with light sheet microscopy are discussed, starting with the specimen preparation (Fig. 2) and including a list of required materials. Please see [32] for a detailed description of specimen embedding and trouble-shooting strategies in light sheet microscopy.
Fig. 2.
Specimen preparation. (A) The specimen is suspended in a droplet of liquid low-melting temperature agarose in a plastic well. A few dozen microliters of agarose, followed by the specimen, are drawn into a thin glass capillary using a piston rod. The agarose gel forms within a few minutes at room temperature. (B) Photograph of central imaging chamber in the DSLM after loading the capillary holder with the glass capillary. The detection lens is located directly behind the imaging chamber and faces the specimen. (C) For conventional DSLM imaging, the zebrafish embryos are embedded in 1% low-melting temperature agarose in a thin glass capillary. In contrast, the upright specimen holder geometry in the SiMView microscope allows using agarose concentrations as low as 0.3–0.4% without compromising specimen stability during imaging. After gelling, the section of the agarose cylinder containing the embryo is gently pushed out of the capillary. The agarose section containing the embryo must be sufficiently exposed such that neither the light sheet nor the fluorescence light pass through the glass wall of the capillary during imaging. (D) View of the central part of the SiMView microscope, where the four optical arms for simultaneous multi-view imaging meet at the specimen chamber. The specimen positioning system is located underneath the specimen chamber, which provides three essential advantages: full access to the specimen chamber from the top, sufficient space for opto-mechanical components in the four-arm arrangement surrounding the sample chamber, and mechanical specimen support by an upright specimen holder. The latter feature enables specimen embedding in ultra-low concentration agarose gels. Credits: Panels (A–C) were reprinted from Cold Spring Harbor Protocols, vol. 2011 No. 10, Keller et al., “Digital scanned laser light-sheet fluorescence microscopy (DSLM) of zebrafish and Drosophila embryonic development”, 1235–1243, Copyright (2011), with permission from Cold Spring Harbor Laboratory Press. Panel (D) was reprinted from Nature Methods, vol. 9 No. 7, Tomer et al., “Quantitative High-Speed Imaging of Entire Developing Embryos with Simultaneous Multiview Light-Sheet Microscopy”, 753–763, Copyright (2012), with permission from Macmillan Publishers Ltd.
5.1. Materials for sample preparation
Transgenic or mRNA-injected zebrafish eggs.
Sharp forceps and an agarose-coated Petri dish for dechorionation.
Stereo binocular for sample mounting.
0.4% (SiMView light sheet microscope) or 1.0% (DSLM light sheet microscope) low-melting temperature agarose (Cambrex, SeaPlaque GTG low-melting temperature agarose) in zebrafish medium, several aliquots in 1.5 ml tubes placed in heating block at 37 °C.
Glass capillaries and plunger (Brand, 701910 and 701938).
Glass Pasteur pipette, glass beaker.
1.5 ml Eppendorf tube or inert plastic well (Fig. 2A).
Dissecting needle or cut-off microloader pipette tip (Eppendorf, catalog number 930001007).
5.2. Dechorionation and agarose embedding of zebrafish embryos
Carefully dechorionate zebrafish embryos in an agarose-coated petri dish using sharp forceps. The agarose coating prevents the embryos from adhering to the plastic bottom.
Transfer the dechorionated zebrafish embryos directly into a tube or well with liquid agarose (Fig. 2A) using a glass Pasteur pipette. The temperature of the agarose should be at least about 32 °C, but not more than 37 °C to prevent damage to the embryos.
Insert the glass capillary with the plunger into the agarose-filled tube and draw up agarose to the 2 cm mark, followed by one egg together with its surrounding agarose.
Position the egg in the preferred orientation along the axis of the glass capillary under the stereo-microscope using a dissecting needle or a cut-off microloader pipette tip. It is vital not to damage the zebrafish embryo in this process. Avoid touching the embryo itself and instead try to achieve the optimal orientation only by swirling the agarose around the embryo and taking advantage of gravitational forces by turning the capillary accordingly.
Once the agarose is fully gelled, place the sample-loaded glass capillaries in a beaker filled with zebrafish mium to prevent drying out of agarose and embryo. Keep the beaker at 28 °C until the start of the imaging experiment.
Imaging is not possible through the glass capillary, owing to the substantial difference in the refractive indices of water and glass. The illuminating light sheet thus has to be able to reach the embryo directly. Thus, before attaching the glass capillary to the light sheet microscope’s specimen holder, gently push out the short section (~2 mm) of the agarose cylinder containing the embryo using the plunger. Excess agarose above the embryo can be sliced off with a razor blade to maximize the mechanical stability of the exposed section of the agarose cylinder.
It should be noted that, in the SiMView light sheet microscope, the gel-embedded specimen is extruded at the top of the vertically-oriented glass capillary (Fig. 2C and D). This upright positioning of the specimen allows using exceptionally low concentrations of agarose for specimen embedding (as low as 0.3-0.4%) [15], since additional mechanical support is provided by the glass capillary just below the gel section containing the specimen (Fig. 2C). This approach is particularly well-suited for long-term imaging experiments with minimal mechanical constraints arising from specimen embedding. In contrast, conventional SPIM and DSLM designs use an inverted geometrical arrangement (Fig. 2B), i.e. the gel-embedded specimen is extruded at the bottom of the vertically-oriented glass capillary (imagine a horizontally flipped version of Fig. 2C). This approach requires using higher agarose concentrations to achieve sufficiently high specimen stability, since mechanical support is provided only by the agarose gel [2,18].
5.3. Single-view and multiple-view imaging of zebrafish embryos
The basic DSLM set-up uses a single light sheet and a single detection arm, oriented at a right angle to the light sheet. Using this simple single-view imaging arrangement, a relatively large fraction of a zebrafish embryo can be recorded (Fig. 3), but coverage is insufficient for whole-embryo imaging [2]. This limitation arises primarily from the highly light-scattering and light-absorbing central yolk cell, which makes it impossible to acquire high-quality images from the far side (with respect to the detection axis) of the embryo, but also from the decrease in signal strength and image quality with increasing imaging depth into the embryo proper. In order to improve physical coverage for whole-embryo imaging experiments, multiple views can be recorded sequentially (Fig. 4), by simply turning the specimen using a rotary stage. In this sequential multi-view imaging mode, several complementary three-dimensional image stacks are acquired and subsequently computationally registered and combined [2].
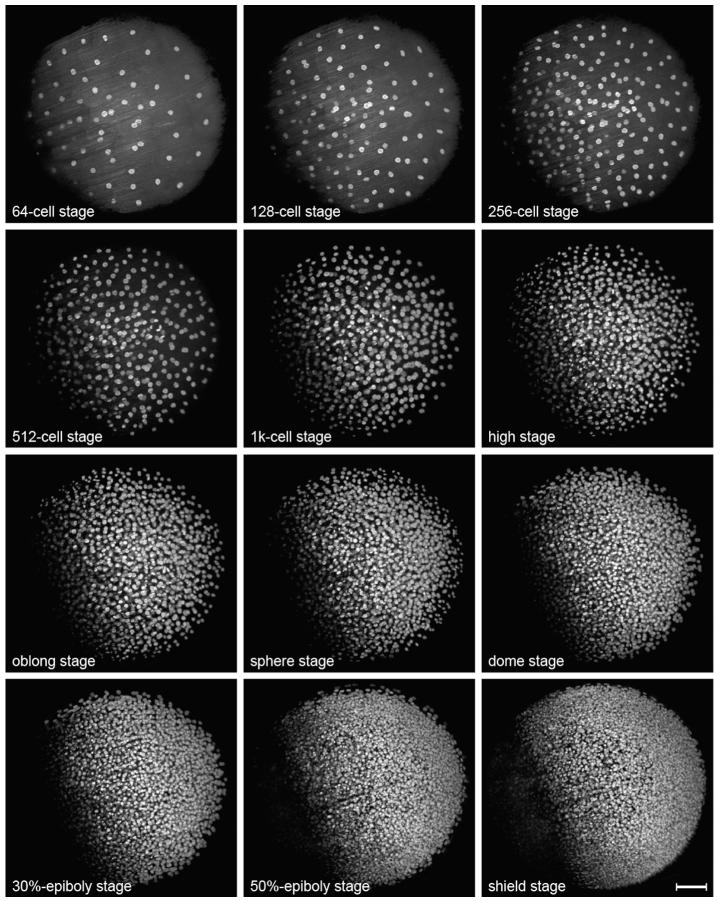
Fig. 3.
Imaging zebrafish embryonic development with scanned light sheet microscopy. Maximum-intensity projections of a DSLM time-lapse recoding of zebrafish embryonic development, providing a view of the animal hemisphere. The wild-type zebrafish embryo was injected with H2B-eGFP mRNA in the one cell stage. Imaging was started in the transition from the 32-to the 64-cell stage. The entire data set consists of 1470 time points (301,350 images, 2.3 terabytes), which were recorded in 60 s intervals, and is available as Supplementary Movie 7 in [2] and on the website www.digital-embryo.org. At each time point, 205 images with 2048 × 2048 pixels each were recorded with a z-spacing of 2.96 μm. Since the illumination laser beam has to travel through the yolk in order to excite fluorophores on the far left side of the animal hemisphere, the average nuclear brightness is lower in this part of the embryo. This limitation is addressed in the SiMView advanced light sheet microscopy implementation [15]. The images were deconvolved with 20 iterations of the Lucy–Richardson algorithm and intensity-normalized to compensate for the increase in intensity over time due to GFP expression. Fluorescence was detected with a Carl Zeiss C-Apochromat 10 ×/0.45 W detection objective. Scale-bar: 100 μm.
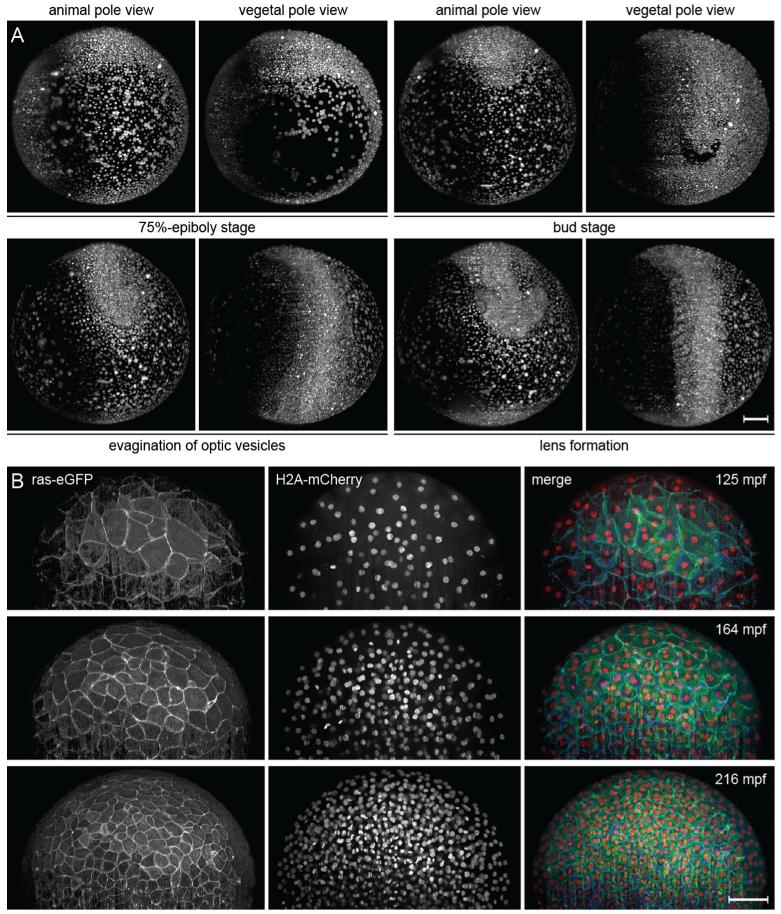
Fig. 4.
Multi-view and multi-color time-lapse imaging of zebrafish embryogenesis. (A) Maximum-intensity projections of a DSLM time-lapse multi-view recoding of zebrafish embryonic development, providing views of the animal and vegetal hemispheres. The wild-type zebrafish embryo was injected with H2B-eGFP mRNA in the one cell stage. Imaging was started in the 64-cell stage. The entire data set consists of 1226 time points (453,620 images, 3.5 terabytes), which were recorded in 90 s intervals, and is available as Supplementary Movie 2 in [2] and on the website www.digital-embryo.org. At each time point, 370 images with 2048 × 2048 pixels each were recorded with a z-spacing of 2.96 μm. The images were deconvolved with 10 iterations of the Lucy–Richardson algorithm and intensity-normalized to compensate for the increase in intensity over time due to GFP expression. Fluorescence was detected with a Carl Zeiss C-Apochromat 10 ×/0.45 W detection objective. (B) Fast multi-color time-lapse imaging of a membrane-and nuclei-labeled zebrafish embryo with light sheet-based structured illumination microscopy (DSLM-SI). The embryo was injected with ras-eGFP mRNA and H2A-mCherry mRNA in the one-cell stage. Membranes were imaged using structured illumination and nuclei using standard light sheet illumination. In total, 125,280 images were recorded at 261 time points from 2 to 8.5 hpf. The complete data set is available as Supplementary Movie 2 in [30] and on the website www.digital-embryo.org. Images were deconvolved with 10 iterations of the Lucy-Richardson algorithm. Fluorescence was detected with a Carl Zeiss C-Apochromat 10 ×/0.45 W detection objective. Scalebars: 100 μm (A,B). Credits: Panel (A) was reprinted from Science, vol. 322, Keller et al., “Reconstruction of Zebrafish Early Embryonic Development by Scanned Light Sheet Microscopy”, 1065–1069, Copyright (2008), with permission from AAAS. Panel (B) was reprinted from Nature Methods, vol. 7, No. 8, Keller et al., “Fast, high-contrast imaging of animal development with scanned light sheet-based structured-illumination microscopy”, 637–642, Copyright (2010), with permission from Macmillan Publishers Ltd.
It should be noted, however, that sequential multi-view imaging only partially addresses the challenge of imaging a large living biological specimen. Since each sequential acquisition and rotation step takes a certain amount of time – during which development of the specimen continues –, the resulting multi-view image content can usually not be correctly registered. Migrating or dividing cells are captured in different locations in the respective overlap regions of the recorded stacks, and automated segmentation and tracking approaches make mistakes in the subsequent data analysis. These limitations are overcome by simultaneous multi-view imaging: SiMView, the light sheet-based imaging platform for simultaneous multi-view imaging described in Section 3.3, is particularly well-suited for the fast live imaging of large developing specimens [15], such as entire zebrafish embryos. In SiMView experiments, the time delay of multi-view image acquisition is zero when using multi-photon excitation and on the order of a few tens of milliseconds when using one-photon excitation. If single-view imaging is insufficient to record a living specimen in its entirety, the SiMView microscopy platform is thus the optimal choice for obtaining quantitative image data.
Irrespective of the type of light sheet microscopy implementation used for a given experiment, it is important to select appropriate illumination and detection objectives, determine the optimal light sheet thickness and assign a corresponding z-step size for three-dimensional image acquisition. Generally, long working distance water-dipping detection objectives are an excellent choice for light sheet microscopy. Sample size and camera chip size define the optimal magnification of the detection system as well as the optimal light sheet thickness [27]. The z-step size should be defined to yield sufficient axial overlap between images, while preventing unnecessary oversampling. A z-step size equivalent to approximately 50-70% of the central light sheet thickness is a conservative choice in this respect.
For example, when imaging a GFP-expressing early zebrafish embryo (corresponding to a field-of-view of approximately 700 μm) with a basic DSLM-like imaging arrangement, a central light sheet FWHM (full width at half maximum) thickness of 5.6 μm provides the minimum average light sheet thickness across the field-of-view. A z-step size of 3–4 μm is recommended to obtain a well-sampled image stack. When using a light sheet microscopy implementation with bidirectional light sheet illumination, each of the two light sheets can be dedicated to the illumination of one half of the field-of-view [24], thus improving axial resolution without compromising light sheet uniformity across the field-of-view. In this case, the central light sheet thickness can be reduced by a factor of square-root of two (4.0 μm FWHM). Even thinner Gaussian beam light sheets are straight-forward to generate but require a restriction of the size of the field-of-view. When aligning the microscope based on the Rayleigh criterion (the ratio of light sheet thickness at the edge and in the center of the field-of-view equals square-root of two), the size of the field-of-view is proportional to the square of the light sheet thickness. As discussed in Section 4.2, the axial resolution in the light sheet microscope can alternatively also be improved by using Bessel beams instead of Gaussian beams [28].
The choice of time-lapse recording interval and total recording duration depends largely on the biological process of interest. For example, in order to follow cell migration using ubiquitously-expressed nuclear markers in early zebrafish embryos, a temporal resolution of 60–90 s ensures proper temporal sampling. Shorter time intervals simplify automated computational cell tracking, but increase exposure of the embryo to laser light and lead to a larger amount of image data collected in the experiment. In contrast, longer time intervals may lead to temporal under-sampling and compromise the ability to follow cells over long periods of time.
It is important to monitor fluorescence levels and specimen position over the course of a long-term imaging experiment. When imaging very early embryos using an mRNA-injection-based fluorescent marker strategy, fluorescence levels can increase by up to 1–2 orders of magnitude during the first 10 h of development [2]. Moreover, embryos embedded in a soft agarose gel for light sheet-based imaging can easily drift by several tens of micrometers at the onset of the imaging experiment, if imaging is started very shortly after embedding. For long-term imaging experiments, an automated software module that tracks and corrects changes in fluorescence intensity levels and center-of-mass positions can thus be very helpful.
6. Image processing and computational data analysis
It is important to keep in mind that data sets resulting from long-term imaging experiments are typically very large, in particular when using high-speed light sheet-based imaging methods. Unless a qualitative inspection is sufficient to evaluate the data set, it is thus often not possible to use manual approaches to the data analysis. An adequate computational infrastructure and robust automated approaches to image processing and data analysis are crucial.
In order to minimize computational bottlenecks in the routine use of light-sheet microscopy, the acquisition of a file server with adequate storage capacity (on the order of at least several tens of terabytes) and a dedicated image processing workstation with adequate memory capacity (on the order of at least several tens of gigabytes) is highly recommended. Fast connections between image acquisition workstation, image processing workstation and data storage server, ideally via glass fiber, are crucial to efficiently transfer large data sets. Detailed configurations of the systems used for the SiMView platform are provided in the supplement of [15].
For simple image processing tasks and small data sets, off-the-shelf commercial and non-commercial software suites for multipurpose image processing, such as Imaris (Bitplane), Amira (Visage Imaging) or ImageJ/FIJI (http://rsbweb.nih.gov/ij and http://fiji.sc/wiki/index.php/Fiji), may already provide the required functionality. Another popular approach is the implementation of custom processing scripts using MATLAB (The Mathworks), a computer language that enables fast prototyping and provides decent performance at the same time. For bead-based or content-based multiview image registration, free software packages are publicly available [15,33].
For complex tasks, such as robust automated image segmentation and cell tracking (Fig. 5), no standardized approaches exist. The design of the most appropriate algorithmic solution simply depends on too many factors, including marker strategies in the imaging experiment, image quality, accuracy and speed requirements as well as data set size. For example, a software solution optimized for confocal time-lapse microscopy recordings with data volumes on the order of a few hundred gigabytes may require filtering and processing steps that are not suitable for light sheet microscopy data. Such software will also likely be too slow to efficiently process e.g. the tens of terabytes of data recorded in a typical SiMView light sheet microscopy experiment. A fast approach to nuclear segmentation and cell lineage tracking in this latter scenario, providing close to real-time performance while achieving high accuracy, is described in [15].
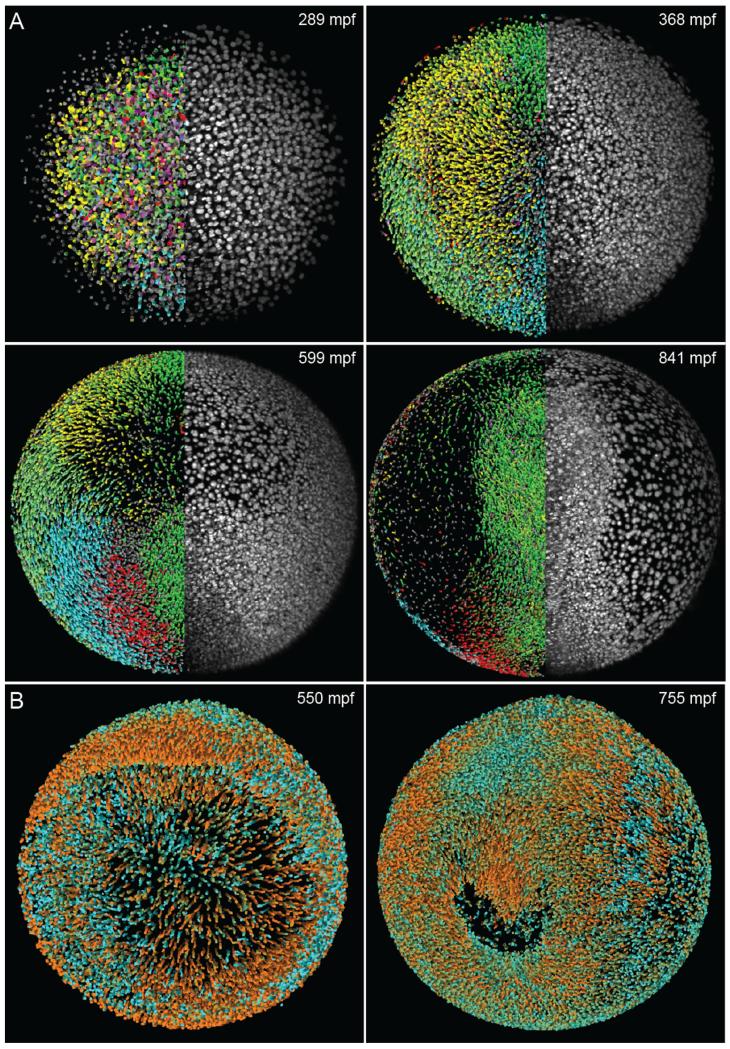
Fig. 5.
Computational analysis of zebrafish time-lapse microscopy recordings. (A) Cell tracking in the digital zebrafish embryo (www.digital-embryo.org). Microscopy data are shown for the right half of the embryo (animal view, maximum intensity projections) and digital embryo reconstructions are shown for the left half, using color-encoded migration directions (see Supplementary Movie 9 in [2]). Color code: dorsal migration (cyan), ventral migration (green), toward or away from body axis (red or yellow), toward yolk (pink). (B) Orthographic rendering of the digital zebrafish embryo obtained from the microscopy data set shown in Fig. 4A. The color code indicates the current movement speed of each nucleus. Bright orange corresponds to an average movement speed of 1.2 μm/min or more, whereas bright cyan highlights effectively non-migratory nuclei. Intermediate speeds are shown by linear interpolation between these two colors. The complete data set is available as Supplementary Movie 3 in [2] and on the website www.digital-embryo.org. Credits: Panel (A) and the left image in panel (B) were reprinted from Science, vol. 322, Keller et al., “Reconstruction of Zebrafish Early Embryonic Development by Scanned Light Sheet Microscopy”, 1065–1069, Copyright (2008), with permission from AAAS.
7. Conclusions
The successful implementation of long-term live imaging experiments with advanced fluorescence microscopy requires thoughtful design of fluorescent marker strategy, specimen preparation assay, imaging method and image processing strategy. Importantly, a complete set of compatible methods is required. The optimal approach to specimen preparation depends on the imaging technique as well as the biological process of interest. Similarly, the optimal approach to image processing and computational data analysis is intrinsically linked to key characteristics of the image data, which are substantially influenced by the imaging method and marker strategy. Depending on the complexity of the imaging experiment, developing this complete set of methods can be very challenging, and often, commercial/standard techniques, if at all available, have to be customized for this purpose.
Long-term imaging strategies based on emerging light sheet microscopy techniques have proven to be particularly powerful in enabling new live imaging experiments. Light sheet microscopy pushes the limit in live imaging by providing high imaging speeds, high signal-to-noise ratios and low rates of photo-bleaching/toxicity. With these key capabilities, it is now possible to comprehensively follow and quantify cellular dynamics in entire embryos throughout their development, reconstruct cell lineage information, correlate changes in gene expression and morphogenetic processes on the system level, and perform biophysical analyses of cell shape changes and cellular forces even for very complex organisms.
Supplementary Material
Acknowledgement
This work was supported by the Howard Hughes Medical Institute.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.ymeth.2013.03.015.
References
- [1].Vermot J, Fraser SE, Liebling M. HFSP J. 2008;2:143–155. doi: 10.2976/1.2907579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EHK. Science. 2008;322:1065–1069. doi: 10.1126/science.1162493. [DOI] [PubMed] [Google Scholar]
- [3].Khairy K, Keller PJ. Genesis. 2011;49:488–513. doi: 10.1002/dvg.20698. [DOI] [PubMed] [Google Scholar]
- [4].Pawley JB. Handbook of Biological Confocal Microscopy. Springer; New York: 2006. [Google Scholar]
- [5].Diaspro A, Chirico G, Collini M. Q. Rev. Biophys. 2005;38:97–166. doi: 10.1017/S0033583505004129. [DOI] [PubMed] [Google Scholar]
- [6].Helmchen F, Denk W. Nat. Methods. 2005;2:932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- [7].Graf R, Rietdorf J, Zimmermann T. Adv. Biochem. Eng. Biotechnol. 2005;95:57–75. doi: 10.1007/b102210. [DOI] [PubMed] [Google Scholar]
- [8].Tomer R, Khairy K, Keller PJ. Curr. Opin. Genet. Dev. 2011;21:558–565. doi: 10.1016/j.gde.2011.07.003. [DOI] [PubMed] [Google Scholar]
- [9].Keller PJ, Dodt HU. Curr. Opin. Neurobiol. 2011;22:138–143. doi: 10.1016/j.conb.2011.08.003. [DOI] [PubMed] [Google Scholar]
- [10].Weber M, Huisken J. Curr. Opin. Genet. Dev. 2011;21:566–572. doi: 10.1016/j.gde.2011.09.009. [DOI] [PubMed] [Google Scholar]
- [11].Mertz J. Nat. Methods. 2011;8:811–819. doi: 10.1038/nmeth.1709. [DOI] [PubMed] [Google Scholar]
- [12].Tsien RY, Ernst L, Waggoner A. In: Handbook of Biological Confocal Microscopy. Pawley JB, editor. Springer; New York: 2006. [Google Scholar]
- [13].Palero J, Santos SI, Artigas D, Loza-Alvarez P. Opt. Express. 2010;18:8491–8498. doi: 10.1364/OE.18.008491. [DOI] [PubMed] [Google Scholar]
- [14].Truong TV, Supatto W, Koos DS, Choi JM, Fraser SE. Nat. Methods. 2011;8:757–760. doi: 10.1038/nmeth.1652. [DOI] [PubMed] [Google Scholar]
- [15].Tomer R, Khairy K, Amat F, Keller PJ. Nat. Methods. 2012;9:755–763. doi: 10.1038/nmeth.2062. [DOI] [PubMed] [Google Scholar]
- [16].Siedentopf H, Zsigmondy R. Ann. Phys. 1903;315:1–39. [Google Scholar]
- [17].Voie AH, Burns DH, Spelman FA. J. Microsc. 1993;170:229–236. doi: 10.1111/j.1365-2818.1993.tb03346.x. [DOI] [PubMed] [Google Scholar]
- [18].Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EHK. Science. 2004;305:1007–1009. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- [19].Keller PJ, Pampaloni F, Stelzer EH. Nat. Methods. 2007;4:843–846. doi: 10.1038/nmeth1087. [DOI] [PubMed] [Google Scholar]
- [20].Arnaout R, Ferrer T, Huisken J, Spitzer K, Stainier DY, Tristani-Firouzi M, Chi NC. Proc. Natl. Acad. Sci. USA. 2007;104:11316–11321. doi: 10.1073/pnas.0702724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Scherz PJ, Huisken J, Sahai-Hernandez P, Stainier DY. Development. 2008;135:1179–1187. doi: 10.1242/dev.010694. [DOI] [PubMed] [Google Scholar]
- [22].Swoger J, Verveer P, Greger K, Huisken J, Stelzer EH. Opt. Express. 2007;15:8029–8042. doi: 10.1364/oe.15.008029. [DOI] [PubMed] [Google Scholar]
- [23].Keller PJ, Pampaloni F, Stelzer EHK. Curr. Opin. Cell Biol. 2006;18:117–124. doi: 10.1016/j.ceb.2005.12.012. [DOI] [PubMed] [Google Scholar]
- [24].Huisken J, Stainier DY. Opt. Lett. 2007;32:2608–2610. doi: 10.1364/ol.32.002608. [DOI] [PubMed] [Google Scholar]
- [25].Dodt HU, Leischner U, Schierloh A, Jahrling N, Mauch CP, Deininger K, Deussing JM, Eder M, Zieglgansberger W, Becker K. Nat. Methods. 2007;4:331–336. doi: 10.1038/nmeth1036. [DOI] [PubMed] [Google Scholar]
- [26].Stelzer EHK. J. Microsc. 1998;1:15–24. [Google Scholar]
- [27].Engelbrecht CJ, Stelzer EHK. Opt. Lett. 2006;31:1477–1479. doi: 10.1364/ol.31.001477. [DOI] [PubMed] [Google Scholar]
- [28].Planchon TA, Gao L, Milkie DE, Davidson MW, Galbraith JA, Galbraith CG, Betzig E. Nat. Methods. 2011;8:417–423. doi: 10.1038/nmeth.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gao L, Shao L, Higgins CD, Poulton JS, Peifer M, Davidson MW, Wu X, Goldstein B, Betzig E. Cell. 2012;151:1370–1385. doi: 10.1016/j.cell.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Keller PJ, Schmidt AD, Santella A, Khairy K, Bao Z, Wittbrodt J, Stelzer EH. Nat. Methods. 2010;7:637–642. doi: 10.1038/nmeth.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Verveer PJ, Swoger J, Pampaloni F, Greger K, Marcello M, Stelzer EHK. Nat. Methods. 2007;4:311–313. doi: 10.1038/nmeth1017. [DOI] [PubMed] [Google Scholar]
- [32].Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EH. Cold Spring Harbor Protoc. 2011;2011:1235–1243. doi: 10.1101/pdb.prot065839. [DOI] [PubMed] [Google Scholar]
- [33].Preibisch S, Saalfeld S, Schindelin J, Tomancak P. Nat. Methods. 2010;7:418–419. doi: 10.1038/nmeth0610-418. [DOI] [PubMed] [Google Scholar]
- [34].Keller PJ, Stelzer EHK. Curr. Opin. Neurobiol. 2008;18:624–632. doi: 10.1016/j.conb.2009.03.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.