Abstract
Candida albicans maintains both commensal and pathogenic states in humans. Here, we have defined the genomic response to osmotic stress mediated by transcription factor Sko1. We performed microarray analysis of a sko1Δ/Δ mutant strain subjected to osmotic stress, and we utilized gene sequence enrichment analysis and enrichment mapping to identify Sko1-dependent osmotic stress-response genes. We found that Sko1 regulates distinct gene classes with functions in ribosomal synthesis, mitochondrial function, and vacuolar transport. Our in silico analysis suggests that Sko1 may recognize two unique DNA binding motifs. Our C. albicans genomic analyses and complementation studies in Saccharomyces cerevisiae showed that Sko1 is conserved as a regulator of carbohydrate metabolism, redox metabolism, and glycerol synthesis. Further, our real time-qPCR results showed that osmotic stress-response genes that are dependent on the kinase Hog1 also require Sko1 for full expression. Our findings reveal divergent and conserved aspects of Sko1-dependent osmotic stress signaling.
Keywords: Yeast, Transcription factor, SKO1, Osmotic stress, Enrichment mapping
1. Introduction
Candida albicans is the most common fungal pathogen in humans, causing both superficial and fatal invasive infections. C. albicans occupies numerous niches within its human host including the urogenital and gastrointestinal tracts, skin, and abiotic surfaces such as indwelling catheters [1]. Also, C. albicans can infect the bloodstream and internal organs of immunocompromised patients [1]. As a consequence, C. albicans frequently encounters challenges from host defenses, resident microflora, antifungal drugs, and fluctuations in environmental pH and osmolarity. Signal transduction pathways allow C. albicans to detect and adapt to such varying microenvironments.
The mitogen activated kinase (MAPK) signaling cascade known as the High Osmolarity Glycerol (HOG) pathway mainly controls the response to osmotic, oxidative, and heavy metal stresses and is critical for processes such as cell wall stability, filamentation, and pathogenesis [2–8]. HOG pathway components are widely conserved in a variety of fungal organisms, and the cornerstone of HOG pathway signaling is the MAPK Hog1. The molecular mechanisms governing HOG pathway signaling were elucidated in the yeast Saccharomyces cerevisiae [9]. Genomic analyses determined that S. cerevisiae mounts a HOG pathway-dependent transcriptional response when exposed to hyperosmotic stress [9–11]. Membrane bound sensors initiate a phosphorylation cascade to activate Hog1 [12]. Subsequently, Hog1 activates transcription factors Sko1, Hot1, and Msn2/4 culminating in the expression/repression of target genes [10,13,14].
The Hog1–Sko1 signaling relationship has been extensively characterized in S. cerevisiae. Following oxidative or osmotic shock, activated Hog1 phosphorylates Sko1 [15]. Phosphorylated Sko1 forms a transcriptional complex with the coregulators Cyc8, Tup1, SAGA and Swi/Snf at target gene promoters, where it plays the role of both a gene activator and a repressor [16,17].
There are functionally divergent as well as conserved roles of Sko1 in C. albicans. Sko1 was identified as a regulator of the oxidative stress response similar to its counterpart in S. cerevisiae [18]. We previously found both HOG-independent and HOG-dependent roles for Sko1 in the responses to cell wall damage and osmotic stress, respectively [19]. The role of Sko1 in the response to cell wall damage by the anti-fungal agent caspofungin is controlled in part by the protein kinase Psk1 and is specific to C. albicans [19]. Interestingly, we discovered that following osmotic stress Hog1 phosphorylates Sko1, implicating Sko1 as a transcriptional regulator of osmotic stress-response genes [19]. However, the Sko1-dependent transcriptional output and the physiological role played by Sko1 in the osmotic stress response remain unknown in C. albicans.
Here, we utilized genetic profiling analyses coupled with the GSEA/Enrichment Map applications [20] to define the role of Sko1 in the osmotic stress response. We provide evidence of both divergent and conserved roles of Sko1 in the osmotic stress response.
2. Results
2.1. Global role of Sko1 in the osmotic stress response
The genomic response of C. albicans cells subjected to cationic osmotic stress was previously determined [3], but the identity of Sko1-dependent osmotic stress-response genes remains unknown. We performed microarray comparisons of a wild-type (wt) strain and sko1Δ/Δ mutant strain treated with 1.0 M NaCl. We identified Sko1-dependent targets as genes whose transcript abundance was consistently altered two-fold or greater (P-value < 0.05) in the sko1Δ/Δ mutant strain compared to the wt strain in the presence of salt. We found that Sko1 regulates 275 genes, 189 of which require Sko1 for full expression and 87 are repressed by Sko1 (Supplementary dataset worksheet 2). Thus, Sko1 acts as a repressor and activator in the C. albicans osmotic stress response. We also identified 21 genes with elevated expression in the sko1Δ/Δ mutant strain in the absence of salt stress and 3 genes that were downregulated (our unpublished data, [18]).
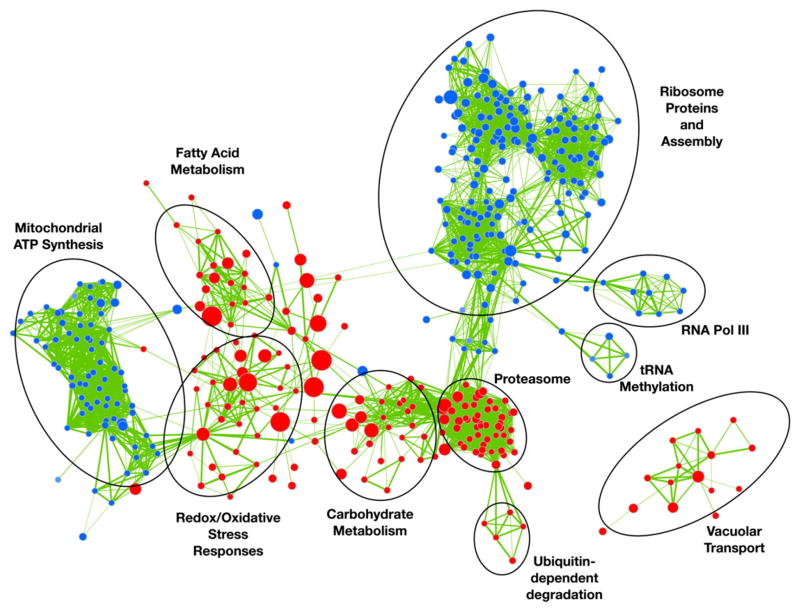
Gene Set Enrichment Analysis (GSEA) is used to identify statistically over-represented gene sets within a given dataset [20]. We used GSEA to identify gene sets (such as a metabolic pathway) that are statistically over-represented within a ranked list of Sko1-modulated genes. In order to clearly present gene sets that are enriched in the sko1Δ/Δ mutant under osmotic stress, we utilized the Cytoscape/Enrichment Map network plugin to group and organize the large number of correlated gene sets [20]. Surprisingly, our enrichment analysis revealed novel roles of Sko1 as an activator of ribosomal biogenesis and mitochondrial ATP synthesis genes. Also, we found that Sko1 acts as a repressor of vacuolar transport genes (Fig. 1). These gene sets were not identified in transcriptional profiling experiments with S. cerevisiae sko1Δ mutant strains [10,21,22]. In addition, we did not identify these gene sets after GSEA analysis using Capaldi et al.’s S. cerevisiae sko1Δ mutant transcriptional profiling dataset (our unpublished data, [10]). We also found a conserved role of Sko1 as a repressor of genes involved in redox metabolism and carbohydrate metabolism [10,15].
Fig. 1.
Enrichment Map of Sko1-dependent osmotic stress-responsive genes. Enrichment Map based on transcript profiles comparing the sko1Δ/Δ mutant strain (JMR104) against the wt reference strain (DAY185) following 1.0 M NaCl treatment. Nodes (circles) depict gene sets statistically overrepresented in the sko1Δ/Δ mutant strain, and node size represents the number of genes in a set. Red node color represents enrichment in the sko1Δ/Δ mutant strain (genes repressed by Sko1 under osmotic stress). Blue node color represents enrichment in the wt strain (genes activated by Sko1 under osmotic stress). Color intensity is proportional to the enrichment significance. Lines (edges) connecting nodes indicate the relatedness between nodes. Edge thickness represents the degree of overlap between gene sets.
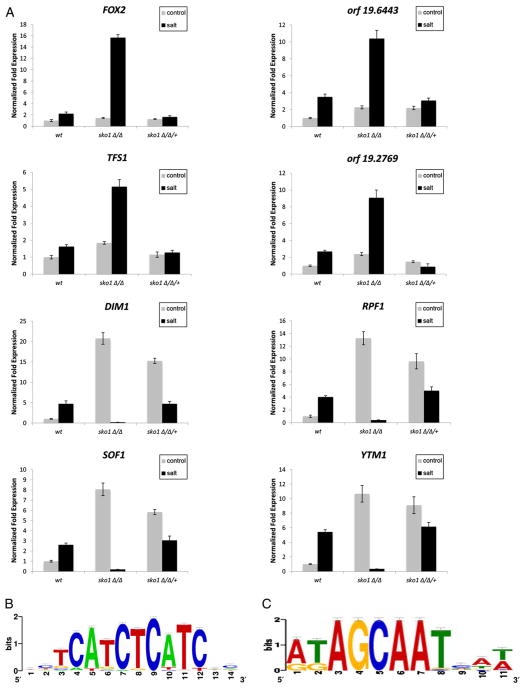
We measured transcript levels by real time (RT)-qPCR to confirm our microarray and enrichment map findings and chose a number of genes based on the presence of two putative Sko1 DNA binding motifs (see Section 2.2). Our results show that the fatty acid oxidase genes FOX2 and open reading frame (ORF) 19.6443 and the putative vacuolar protease inhibitor genes TFS1 and ORF19.2769 were induced in the wt strain and significantly overexpressed in the sko1Δ/Δ mutant following osmotic stress. Gene expression levels were restored to wt in the sko1Δ/Δ/+ complemented strain (Fig. 2A). In addition, the transcript levels of the ribosomal modification genes SOF1, DIM1, YTM1, and RPF1 were elevated in the wt strain under osmotic stress, but were almost completely absent in the sko1Δ/Δ mutant strain under similar conditions. Interestingly, in the absence of osmotic stress, these genes were overexpressed in the sko1Δ/Δ mutant and sko1Δ/Δ/+ complemented strain (Fig. 2A). Our observations in the sko1Δ/Δ/+ complemented strain suggest that both functional copies of Sko1 are required for full repression. These observations demonstrate a dual role of Sko1 for certain ribosomal genes: Sko1 is an activator during salt stress, but during vegetative growth, Sko1 functions as a repressor. Taken together, our genomic analyses and enrichment mapping highlight distinct features of Sko1 osmotic stress signaling in C. albicans.
Fig. 2.
Divergent aspects of Sko1 gene regulation in the osmotic stress response. (A) RT-qPCR analysis of osmotic stress-responsive genes identified from GSEA/enrichment mapping and containing a putative Sko1 binding site in the wt reference strain DAY185, sko1Δ/Δ mutant strain (JMR104), and sko1Δ/Δ/+ complemented strain (JMR109). Transcript levels were normalized to TDH3 expression, and fold changes between strains were normalized to the untreated wt adjusted to the value of 1.0. (B) Sequence logo for the putative DNA binding site of Sko1-repressed genes. (C) Sequence logo for the putative DNA binding site of Sko1-activated genes.
2.2. Identification of a putative Sko1 binding site
In S. cerevisiae, the ATF/CREB DNA motif (T(G/T)ACGT(C/A)A) is required for Sko1 binding to gene promoter regions [10,21]. In addition, S. cerevisiae Sko1 was shown to bind to promoter sequences that are different from the ATF/CREB motif [22]; however, the Sko1 DNA binding sequence is unknown in C. albicans. We used the RSAT, Align Ace, and MEME web-based applications to scan the promoter regions of the Sko1-dependent osmotic stress-response genes for a DNA-binding consensus sequence. We discovered two over-represented promoter motifs in genes that are repressed or activated by Sko1, respectively. These motifs were approximately 200 bp–300 bp upstream of the start codon. The sequence (A/T)ATAGCAAT(T/C)A was mainly associated with genes repressed by Sko1 and found 48 times in 34 genes with a P-value 1.8e–9 (Fig. 2B, Supplementary dataset worksheet 3). This sequence was present approximately 10 times more often in Sko1-dependent genes (10.55%) compared to the frequency found in all promoters of the C. albicans genome (1.77%). We compared this sequence to known transcription motifs and found a similar motif (P-value 0.0036) used by the S. cerevisiae transcription factor Rfx1.
The sequence (T/C)TCATCTCATC(G/T)CA(A/T) was found 131 times in 97 genes (P-value 1.0e–15), 85 of which were Sko1-activated genes (Fig. 2C, Supplementary dataset worksheet 3). This sequence was present approximately 2.8 times more often in Sko1-dependent genes (47%) compared to the genome (17%). Our similarity searches identified the binding motif (P-value 2.4e–4) of the S. cerevisiae transcription factors Tod6 and Dot6. We did not detect significant enrichment of the ATF/CREB motif in our Sko1-dependent gene set; however, this motif was present in several Sko1 osmotic stress-response genes (Supplementary dataset worksheet 3), so we cannot rule out the possibility that Sko1 may recognize this motif in C. albicans. Collectively, these findings indicate that Sko1 may regulate gene expression through two novel binding sequences in C. albicans.
2.3. Role of Sko1 in cell growth under osmotic stress
C. albicans wt strains exposed to 1.0 M NaCl show a slight increase in their doubling time compared to unstressed cells, and cell growth is drastically reduced in 2.0 M NaCl [23]. Because Sko1 activates genes involved in protein translation and mitochondrial function (Fig. 1), we reasoned that Sko1 may be required for cell growth. We monitored growth of the sko1Δ/Δ mutant strain in liquid nutrient medium and solid nutrient medium supplemented with 1.0 M and 1.5 M NaCl. Our results did not show a growth defect on 1.0 M growth medium (Supplementary Fig. 1). This finding was in concordance with a previous study using a different sko1Δ/Δ mutant strain [18]. However, we did observe a modest, but significant growth defect in solid and liquid media supplemented with 1.5 M NaCl (Supplementary Fig. 1). This phenotype was not due to cell death, since the yeast viability dyes methylene blue and propidium iodine did not stain the sko1Δ/ Δ mutant strain (our unpublished results). Growth was restored to wt levels in the sko1Δ/Δ/+ complemented strain. Therefore, Sko1 is required for optimal growth under moderate and high levels of osmotic stress.
2.4. Role of Sko1 in glycerol synthesis
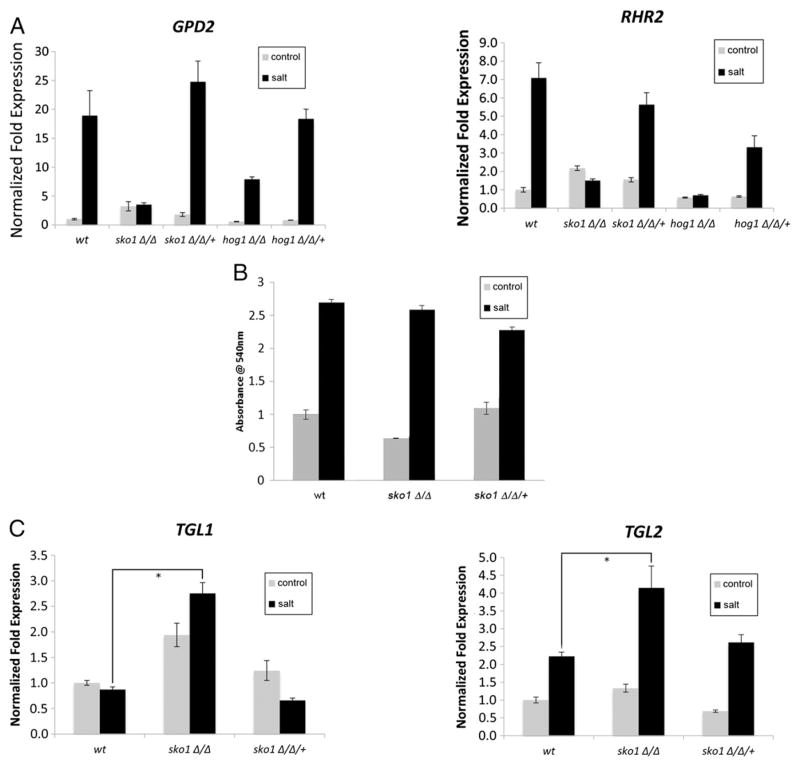
Glycerol accumulation is critical for osmoregulation, serving as an intracellular solute and molecular protectant [9]. Glycerol synthesis is catalyzed by Gpd1, Gpd2 and Rhr2, and C. albicans rhr2Δ/Δ mutants contain approximately 50% less glycerol than wt and grow poorly on hyperosmotic medium [24]. We hypothesized that a defect in RHR2 and GPD1/2 expression would lead to a defect in glycerol accumulation under hyperosmotic conditions. We found an expression defect of RHR2 and GPD2 (4.7-fold for RHR2 and 5.4-fold for GPD2) in the sko1Δ/Δ mutant strain under osmotic stress (Fig. 3A). However, glycerol accumulation in sko1Δ/Δ mutant strain was similar to the wt strain after 30 min of 1.0 M NaCl exposure (Fig. 3B). These findings suggest that Sko1 is involved in the activation of glycerol synthesis genes under osmotic stress, but that glycerol synthesis may also occur independently of Gpd1/2 and Rhr2 activity. Indeed, glycerol can be produced via triacylglycerol metabolism [25]. Our enrichment map shows that fatty acid metabolic genes are elevated in sko1Δ/Δ mutant strain (Fig. 1). We monitored expression of the triacylglycerol lipases TGL1 and TGL2 which catalyze the conversion of triacyglycerol to glycerol [25,26]. TGL1 expression was not induced in the wt strain following osmotic stress, but was elevated in the sko1Δ/Δ strain in the presence and absence of 1.0 M NaCl. TGL2 expression was induced in the wt strain following osmotic stress and significantly elevated in our sko1Δ/Δ mutant strain (Fig. 3C). Therefore, increased lipid metabolism could augment glycerol production in the sko1Δ/Δ mutant.
Fig. 3.
Requirement of Sko1 in glycerol production. (A) RT-qPCR analysis of the glycerol metabolic genes GPD2 and RHR2 in the wt reference strain DAY185, hog1Δ/Δ mutant strain (JMR121), hog1Δ/Δ/+ complemented strain (JMR123), sko1Δ/Δ mutant strain (JMR104), and sko1Δ/Δ/+ complemented strain (JMR109) following 1.0 M NaCl treatment. All genes were analyzed in triplicate. Transcript levels were normalized to TDH3, and fold changes between strains were normalized to the untreated wt strain adjusted to a value of 1.0. (B) Measurements of intracellular glycerol levels following growth in 1.0 M NaCl for the wt, sko1Δ/Δ mutant strain, and the sko1Δ/Δ/+ complemented strain. (C) RTq-PCR analysis of the triacylglycerol lipase genes TGL1 and TGL2. An asterisk indicates a P-value < 0.05 compared to the wt strain in salt.
2.5. Sko1 regulates expression of Hog1-dependent osmotic stress response genes
In C. albicans, transcriptional profiling studies showed that the majority of osmotic stress-response genes require Hog1 for their full expression or repression [3]. Further, Hog1–Sko1 signaling has been extensively characterized in S. cerevisiae, so we expected that Hog1 exerts control of osmotic stress-response genes partly by acting through Sko1 in C. albicans. To test this hypothesis, we measured transcript levels for 12 Hog1-dependent, osmotic stress-response genes by RT-qPCR following 1.0 M NaCl exposure in wt, hog1Δ/Δ, and sko1Δ/Δ mutant strains.
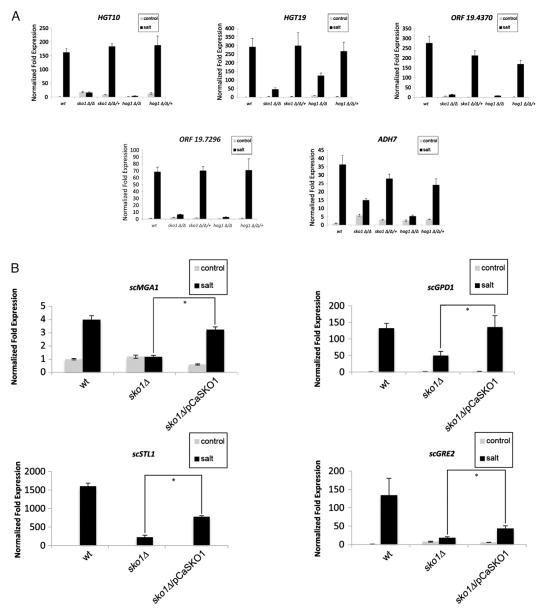
As in previous transcript profiling studies, 1.0 M NaCl exposure induced expression of osmotic stress response genes in the wt strain [27], and their expression was significantly reduced or absent in the hog1Δ/Δ mutant strain (Supplementary dataset worksheet 4 and Fig. 4A). Moreover, the 12 Hog1-dependent genes required Sko1 for full expression (Supplementary dataset worksheet 4, Fig. 4A). Notably, gene expression was reduced in the sko1Δ/Δ mutant strain for the glycerol permease HGT10, the alcohol dehydrogenase ADH7, the carbohydrate transporter HGT19, and two of the most highly induced osmotic stress response genes ORF19.4370 and ORF19.7296, which do not have orthologs in S. cerevisiae (Fig. 4A). In addition, the expression defect of GPD1 and RHR2 in the sko1Δ/Δ mutant strain was observed in the hog1Δ/Δ mutant strain (Fig. 3A). Gene expression levels were restored to wt in the sko1Δ/Δ/+ and hog1Δ/Δ/+ complemented strains. Taken together, these findings show that Sko1 is required for full expression of certain Hog1-dependent osmotic stress response genes.
Fig. 4.
Conserved aspects of Sko1 function in the osmotic stress response. (A) RT-qPCR analysis of osmotic stress-responsive genes in the C. albicans wt reference strain DAY185, hog1Δ/Δ mutant strain (JMR121), hog1Δ/Δ/+ complemented strain (JMR123), sko1Δ/Δ mutant strain (JMR104) and sko1Δ/Δ/+ complemented strain (JMR109) following exposure to 1.0 M NaCl. All genes were analyzed in triplicate. Transcript levels were normalized to TDH3, and fold changes between strains were normalized to the untreated wt reference strain adjusted to a value of 1.0. (B) RT-qPCR expression analysis of S. cerevisiae osmotic stress-response in the S. cerevisiae wt reference strain (LS07), sko1Δ mutant strain (LS04), and the sko1Δ/C. albicans SKO1 complemented strain (LS05). Transcript levels were normalized to S. cerevisiae TDH3 expression, and fold changes between strains were normalized to the untreated wt reference strain adjusted to the value of 1.0. An asterisk indicates a P-value < 0.05 compared to the reference strain in salt.
2.6. Conservation of C. albicans Sko1 function in S. cerevisiae
The basic leucine zipper (bZIP) DNA binding domain and Hog1 phosphorylation region are conserved between C. albicans Sko1 and S. cerevisiae Sko1 [28]. Therefore, we considered the possibility that C. albicans Sko1 would functionally complement an S. cerevisiae sko1Δ mutant strain in the osmotic stress response. We transformed a sko1Δ mutant strain with a multicopy plasmid containing C. albicans SKO1 and measured the expression of 10 S. cerevisiae Sko1-dependent osmotic stress response genes by RT-qPCR in the wt strain, sko1Δ mutant strain, and sko1Δ/pCaSKO1 complemented strain. We found that C. albicans Sko1 completely restored the gene expression defect of glycerol dehydrogenase GPD1 and the transcriptional regulator MGA1. Also, C. albicans Sko1 partially restored the gene expression defect of glycerol permease STL1 and the oxidoreductase GRE2 (Fig. 4B). Thus, these findings suggest that Sko1 function is partially conserved in the S. cerevisiae and C. albicans osmotic stress responses.
3. Discussion
Among the human niches for C. albicans, the kidneys have osmolarity levels over 1.0 M in the renal medulla [29]. C. albicans can survive in hyperosmotic environments with salt concentrations as high as 2.0 M [30]. We used genome-wide transcriptional analysis and enrichment mapping to define a novel signaling circuit for transcription factor Sko1 in the osmotic stress response. Further, we demonstrated conserved attributes of Sko1 as a downstream effector of HOG pathway signaling following osmotic stress based on similar transcriptional responses in hog1Δ/Δ and sko1Δ/Δ mutant strains. In C. albicans, as in S. cerevisiae, Sko1 has a conserved role as an activator of genes involved in glycerol and redox metabolism. Our findings reveal both divergent and conserved roles of Sko1 (Supplementary Fig. 2).
3.1. Functional divergence of Sko1 in the C. albicans osmotic stress response
Our transcriptional profiling, GSEA, and enrichment analysis uncovered distinct gene classes that are regulated by Sko1 in response to osmotic stress such as mitochondrial function, vacuolar transport, and ribosome synthesis. Although the physiological consequences of the C. albicans osmotic stress response have not been well-characterized, similar genomic responses were observed in S. cerevisiae, but Sko1 was not implicated in the transcriptional response. In S. cerevisiae, mitochondrial function is essential to counteract the toxic effects of reactive oxygen species (ROS) generated by cationic stress, and mutations to genes involved in mitochondrial biogenesis and energy transformation (citric acid cycle, electron transport chain, and ATP synthase subunits) causes severe growth defects on hyperosmotic medium [31,32]. In both C. albicans and S. cerevisiae, Sko1 function in the oxidative stress response is conserved. Our transcript profiling data shows that Sko1 is required to activate genes involved in general mitochondrial function such as MTO1 (mitochondrial tRNA modification), and MIS12 (folate synthesis, Supplementary datasheet worksheet 2). These findings suggest that Sko1 plays an expanded role in C. albicans to counter osmotic stress-induced ROS damage by mitochondrial nucleotide synthesis.
Elevated vacuolar fragmentation is observed following osmotic stress [33]. Also, vacuolar proton ATPase activity contributes in salt detoxification by sequestering Na+ in vacuolar vesicles [34]. Although, the expression of several vacuolar-associated genes was elevated in the sko1Δ/Δ mutant strain treated with 1.0 M NaCl, we did not detect any aberrant vacuolar phenotypes (our unpublished data), nor did we observe a significant change of gene expression for the vacuolar ATPases VMA2, VMA4, VMA10 and VMA13 [33,35]. Therefore, the phenotypic consequences of elevated vacuolar gene expression in the sko1Δ/Δ mutant strain remain unclear.
In S. cerevisiae, translation is transiently inhibited following 1.0 M salt stress — presumably to arrest cell division [11,36]. Transcriptional profiles in both C. albicans and S. cerevisiae show that ribosomal gene expression is mainly downregulated as a group following osmotic stress, even if the expression of certain genes is slightly induced [3,27,36]. Our microarray findings show that ribosomal gene expression is significantly less in the sko1Δ/Δ mutant strain than the wt strain following osmotic stress. Thus, Sko1 function may be to maintain basal levels of ribosomal gene expression following osmotic stress until translational activity is restored. Interestingly, the expression of the elongation factor gene ELF1 is significantly reduced in the sko1Δ/Δ mutant strain (Supplementary datasheet worksheet 1), and elf1Δ/Δ mutants have a growth defect [37]. Our growth assays showed that sko1Δ/Δ mutant strain grows slower that the wt strain under moderate (1.5 M NaCl) osmotic stress. Thus, we argue that reduced protein translational activity may contribute to the sko1Δ/Δ mutant growth defect.
Our RT-qPCR results show that the ribosomal genes SOF1, YTM1, RPF1, and DIM1 transcript levels are elevated in the sko1Δ/Δ mutant strain under vegetative growth, but absent during osmotic stress. This activator/repressor activity on the same gene was observed in S. cerevisiae Sko1 regulation of oxidative stress response genes and is dependent on the Tup1–Cyc8 complex [16,38]. Tup1–Cyc8 is recruited to promoters through direct interactions with transcriptional regulators to control gene expression [38]. We propose that Sko1 may associate with Tup1–Cyc8 in C. albicans to control ribosomal gene expression.
Our in silico gene promoter analysis provides insight of the cis-acting elements required for Sko1 function and the transcriptional regulatory components governing the C. albicans osmotic stress response. We found that the sequence (A/T)ATAGCAAT(T/C)A was significantly enriched in genes repressed by Sko1. This sequence is similar to the S. cerevisiae transcription factor Rfx1 which represses DNA damage response genes [39]. Moreover, similar to Sko1, Rfx1 recruits Tup1 and Cyc8 to gene promoters [39]. The C. albicans ortholog Rfx1 is functionally conserved in the DNA damage response and has not been associated with the osmotic stress response [40]. We also identified the consensus sequence (T/C)TCATCTCATC(G/T)CA(A/T) for genes activated by Sko1. This sequence is recognized by the S. cerevisiae transcriptional factors Tod6 and Dot6, which regulate ribosomal biogenesis [41]. C. albicans Dot6 has not been functionally characterized, and a TOD6 ortholog is not present in the C. albicans’ genome. Although we cannot completely rule out the possibility that Sko1 may indirectly regulate target genes by modulating RFX1 and DOT6 expression, our microarray findings show that DOT6 and RFX1 expression is at wt levels in the sko1Δ/Δ mutant strain (Supplementary dataset worksheet 1). An attractive hypothesis is that Sko1 may work synergistically with Rfx1 and Dot6 in the osmotic stress response in C. albicans. Nevertheless, our observations suggest that Sko1 may interact with two distinct DNA binding motifs in C. albicans.
3.2. Hog1–Sko1 signaling is conserved in the C. albicans osmotic stress response
Our findings establish Sko1 as a transcriptional regulator of the HOG pathway mediated osmotic stress response in C. albicans. This role was anticipated, as microarray and ChIP-chip analyses in S. cerevisiae showed that Sko1 regulates 18% of Hog1-dependent osmotic stress response genes [10]. There are three lines of evidence that the role is similar in C. albicans. First, a comparison of our microarray results against a previously published microarray study [3] indicates that Sko1 is required for full activation of 15% of Hog1-dependent genes. We have supported this analysis with RT-qPCR, which demonstrated that Hog1 and Sko1 are needed for activation of 12 genes following osmotic stress. Second, SKO1 expression and post-translational modification are dependent on Hog1 following osmotic stress (Supplementary dataset worksheet 4, [19]). Third, in C. albicans, Hog1 plays a role in mitochondrial function, and in S. cerevisiae, Hog1 controls vacuolar H+-ATPase function [34,42]. Taken together, these findings show that the HOG pathway acts, at least in part, through Sko1 in the C. albicans osmotic stress response.
Our enrichment map findings and complementation analysis in S. cerevisiae show that Sko1 is functionally conserved as a repressor of carbohydrate metabolism and transport, fatty acid oxidation, and redox metabolism. Carbohydrate metabolism is important for physiological processes, such as cell wall remodeling and production of the osmolyte glycerol. Our RT-qPCR results show that the glycerol metabolic genes GPD2 and RHR2 and the glycerol permease HGT10 require Hog1 and Sko1 for full expression. Furthermore, hog1Δ/Δ mutants fail to accumulate intracellular glycerol following osmotic stress. However, glycerol levels are similar to wt in our sko1Δ/Δ mutant strain, and our results suggest that increased fatty acid metabolism augments glycerol synthesis. It seems likely that in C. albicans, Hog1-dependent phosphorylation of Sko1 alleviates redox and carbohydrate metabolic gene repression that was found in S. cerevisiae Hog1–Sko1 signaling [13,15,16].
4. Conclusion
Although stress response components are conserved between S. cerevisiae and C. albicans, the evolutionary distance between these two yeasts predicts that the genetic response to environmental stress will be divergent. Indeed, transcriptional rewiring has been observed in the cell wall damage and oxidative stress signaling pathways [30]. Our findings demonstrate the utility of GSEA and enrichment mapping to uncover transcriptional networks. We established a divergent role of Sko1 in the C. albicans osmotic stress response as a regulator of translation, mitochondrial function and vacuolar transport. Further, we show that Sko1 is required for critical adaptive processes such as glycerol synthesis and carbohydrate and redox metabolism. This aspect of Sko1 signaling is evolutionarily conserved in yeasts.
5. Materials and methods
5.1. Media and growth conditions
C. albicans cultures were grown in Yeast Peptone Dextrose plus uridine media (YPD+uri: 2% dextrose, 2% bacto peptone, 1% yeast extract, and 80 mg/l uridine) at 30 °C with shaking. S. cerevisiae cultures were grown in YPD or complete synthetic medium (2% dextrose, 6.7% yeast nitrogen base [YNB] plus ammonium sulfate without uracil and the necessary auxotrophic supplements [csm−ura]). C. albicans cells were grown in YPD+uri and S. cerevisiae cells were grown in YPD or csm−ura in genomic, biochemical, and growth assay monitoring osmotic stress responses. Experimental cultures were supplemented with 1.0 M sodium chloride (NaCl) to induce osmotic stress.
5.2. Yeast strains and transformation procedures
C. albicans and S. cerevisiae strains used in this study are listed in Supplementary Table 1. All C. albicans strains were derived from the laboratory wt strain BWP17 (genotype: ura3Δ:: λimm434/ura3Δ:: λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG). S. cerevisiae strains were derived from the laboratory wt strain BY4741 (genotype: MATa, his3, leu2, met15, ura3).
All primers used in this study are listed on Supplementary Table 2. The S. cerevisiae sko1Δ mutant strain (LS03) was constructed as follows: Primers ScSKO1delfwd and ScSKO1delrev were used to amplify approximately a 1.7 kb DNA fragment consisting of the ORF for Kanamycin resistance flanked by 100 bp of S. cerevisiae SKO1 5′ and 3′ untranslated (UTR) sequences. The pFA6a-5FLAG-KanMX6 plasmid (Addgene) was used as a template, and yeast transformations were performed using the lithium chloride protocol [43]. Transformants were selected on YPD plates supplemented with 200 μg/ml G418 (Sigma) and verified by colony PCR. LS03 was brought to Ura prototrophy through transformation with the pYES2.1/V5-His-TOPO vector (Invitrogen) to create strain LS04. The S. cerevisiae sko1Δ/CaSKO1 complemented strain (LS05) was created by transforming strain LS03 with pYES-CaSKO1-V5 [19]. The wt BY4741 strain was brought to Ura prototrophy through transformation with the pYES2.1/ V5-His-TOPO vector to create strain LS07.
5.3. RNA isolation and real time-PCR analysis
Overnight cultures of designated C. albicans strains were diluted to a starting OD600 nm of 0.2 in 100 ml YPD+uri. The cultures were incubated with shaking at 30 °C to an OD600 nm of 1.0. A volume of 40 ml of this culture was transferred to a new flask containing 10 ml of 5.0 M NaCl (Cf = 1.0 M) to subject the cells to osmotic stress. As a control, an equivalent dilution was made of the remaining culture volume using water. The cultures were incubated at 30 °C for an additional 30 min. After salt (or water) treatment, OD600 of each culture was measured to approximate cell number, and an appropriate volume was removed to obtain 3 × 108 cells. Cells were collected by gentle centrifugation (2880 ×g), and total RNA was extracted using the RiboPure™ — Yeast Kit (Ambion). Manufacturer’s protocol was followed with the following minor adjustment: DNase treatment was for 1 h at 37 °C. RNA yield and quality were checked by UV absorbance at 260nm and 280nm in a BioRad Smart Spec spectrophotometer and by electrophoresis on a standard 0.8% agarose gel.
cDNA was synthesized using the AffinityScript Multiple Temperature cDNA Synthesis Kit (Agilent Technologies) following the manufacturer’s protocol. As a control for DNA contamination, parallel reactions were performed without reverse transcriptase. Primers were designed using Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). The reaction tubes were incubated at 42 °C for 90 min followed by 5 min at 90 °C to terminate the reaction.
PCR efficiency (E) was determined for all primers through amplification of C. albicans genomic DNA. Primer pairs yielding E-values between 99 and 103% were used in subsequent RT-qPCR experiments. RT reactions were prepared in triplicate using iQ SYBR Supermix (BioRad). RT-qPCR experiments and data analysis were performed using the BioRad MyiQ™2 thermocycler equipped with an iQ5 multicolor optical unit (BioRad) as previously described [19]. Transcript levels were normalized against C. albicans TDH3 or S. cerevisiae TDH3 expression (both of which encode glyceraldehyde-3-phosphate dehydrogenase), and gene expression changes were calculated by the ΔΔCT method [44,45]. Target-gene fold changes for treated or untreated cells were determined by comparison to the untreated wt strain. Significant differences between groups were determined in unpaired t-tests with a P-value of <0.05 being statistically significant.
5.4. Microarray analysis
Cultures of designated C. albicans strains were prepared, and RNA extraction was performed as described for RT-qPCR analysis. We performed four hybridizations that compared transcripts from a salt-treated sko1Δ/Δ mutant strain against a salt-treated wt strain. All RNA samples were produced from independent cultures. Transcriptional profiling was performed as previously described [46], and the resulting data was normalized and analyzed in GeneSpring GX version 7.3 (Agilent Technologies). The results of this analysis are listed in the Supplementary dataset worksheet 1, which includes significantly-modulated genes that exhibited a statistically-significant (t-test P-values < 0.05) change in transcript abundance of at least 2.0-fold. Functional analysis in GSEA, Cytoscape and Enrichment Map [20] software were conducted as described by Uwamahoro et al. [47].
5.5. In silico gene promoter analysis
All Sko1 gene targets with a 2-fold or greater change in transcription were considered. Upstream sequences were retrieved from the Regulatory Sequence Analysis Tools (RSAT, http://rsat.ulb.ac.be/ [48]) web-based application. The oligo-analysis program was chosen for matrix determination; parameters were left at default, and the C. albicans genome was chosen for the background model. In addition, the Multiple Em for Motif Elicitation (MEME, http://meme.sdsc.edu), and W-AlignACE programs were used to search for putative binding sites. Due to the query size restrictions for MEME (maximum of 60,000 characters/inquiry) and W-AlignACE programs, gene promoter sequences were run in successive groups with parameters set to default. To avoid bias, genome sequences were randomly selected for input; this was repeated three times to confirm results. The above procedure was then repeated with isolated data sets of genes with only increased expression (>2.0 fold) or only decreased expression (>2.0 fold). All returned position-specific scoring matrices (PSSMs) were compared to known matrices using TomTom, a component of the MEME suite. TomTom searches Jaspar Core, Transfac, and Uniprobe Mouse databases for known binding sites.
5.6. Intracellular glycerol assays
Yeast cultures were prepared as described for RNA analysis. Following 30 min of 1.0 M NaCl treatment, intracellular glycerol levels were measured using the free glycerol reagents kit (Sigma) following manufacturer’s protocol.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ygeno.2013.06.002.
Supplementary Material
Acknowledgments
This work was supported by NIH SCORE grant SC2 GM089556-01 and PSC-CUNY grant 65562-0043. LS and JRT were supported by the Program for Research Initiatives for Science Majors (PRISM) at John Jay College which is funded by the U.S. Department of Education, National Science Foundation, and the New York State’s Graduate Research and Technology Initiative. Candida research by the AN lab is supported by the Canadian Institutes of Health Research.
Footnotes
Author contributions
Conceived and designed the experiments: JMR, LS and DHM. Performed the experiments and analyzed the data: JMR, DHM, LS, AN, and JRT. Wrote the paper: DHM, JRT, AN, and JMR.
References
- 1.Richard CJC, Calderone A. Candida and Candidiasis. ASM Press; Washington, DC: 2012. p. 524. [Google Scholar]
- 2.Munro CA, Selvaggini S, de Bruijn I, Walker L, Lenardon MD, Gerssen B, Milne S, Brown AJ, Gow NA. The PKC, HOG and Ca2+ signalling pathways co-ordinately regulate chitin synthesis in Candida albicans. Mol Microbiol. 2007;63:1399–1413. doi: 10.1111/j.1365-2958.2007.05588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Enjalbert B, Smith DA, Cornell MJ, Alam I, Nicholls S, Brown AJ, Quinn J. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol Biol Cell. 2006;17:1018–1032. doi: 10.1091/mbc.E05-06-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith DA, Nicholls S, Morgan BA, Brown AJ, Quinn J. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol Biol Cell. 2004;15:4179–4190. doi: 10.1091/mbc.E04-03-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arana DM, Alonso-Monge R, Du C, Calderone R, Pla J. Differential susceptibility of mitogen-activated protein kinase pathway mutants to oxidative-mediated killing by phagocytes in the fungal pathogen Candida albicans. Cell Microbiol. 2007;9:1647–1659. doi: 10.1111/j.1462-5822.2007.00898.x. [DOI] [PubMed] [Google Scholar]
- 6.Alonso-Monge R, Navarro-Garcia F, Molero G, Diez-Orejas R, Gustin M, Pla J, Sanchez M, Nombela C. Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J Bacteriol. 1999;181:3058–3068. doi: 10.1128/jb.181.10.3058-3068.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eisman B, Alonso-Monge R, Roman E, Arana D, Nombela C, Pla J. The Cek1 and Hog1 mitogen-activated protein kinases play complementary roles in cell wall biogenesis and chlamydospore formation in the fungal pathogen Candida albicans. Eukaryot Cell. 2006;5:347–358. doi: 10.1128/EC.5.2.347-358.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Argimon S, Fanning S, Blankenship JR, Mitchell AP. Interaction between the Candida albicans high-osmolarity glycerol (HOG) pathway and the response to human beta-defensins 2 and 3. Eukaryot Cell. 2011;10:272–275. doi: 10.1128/EC.00133-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Capaldi AP, Kaplan T, Liu Y, Habib N, Regev A, Friedman N, O’Shea EK. Structure and function of a transcriptional network activated by the MAPK Hog1. Nat Genet. 2008;40:1300–1306. doi: 10.1038/ng.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez-Montanes F, Pascual-Ahuir A, Proft M. Toward a genomic view of the gene expression program regulated by osmostress in yeast. OMICS. 2010;14:619–627. doi: 10.1089/omi.2010.0046. [DOI] [PubMed] [Google Scholar]
- 12.Tatebayashi K, Tanaka K, Yang HY, Yamamoto K, Matsushita Y, Tomida T, Imai M, Saito H. Transmembrane mucins Hkr1 and Msb2 are putative osmosensors in the SHO1 branch of yeast HOG pathway. EMBO J. 2007;26:3521–3533. doi: 10.1038/sj.emboj.7601796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Proft M, Pascual-Ahuir A, de Nadal E, Arino J, Serrano R, Posas F. Regulation of the Sko1 transcriptional repressor by the Hog1 MAP kinase in response to osmotic stress. EMBO J. 2001;20:1123–1133. doi: 10.1093/emboj/20.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rep M, Krantz M, Thevelein JM, Hohmann S. The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J Biol Chem. 2000;275:8290–8300. doi: 10.1074/jbc.275.12.8290. [DOI] [PubMed] [Google Scholar]
- 15.Rep M, Proft M, Remize F, Tamas M, Serrano R, Thevelein JM, Hohmann S. The Saccharomyces cerevisiae Sko1p transcription factor mediates HOG pathway-dependent osmotic regulation of a set of genes encoding enzymes implicated in protection from oxidative damage. Mol Microbiol. 2001;40:1067–1083. doi: 10.1046/j.1365-2958.2001.02384.x. [DOI] [PubMed] [Google Scholar]
- 16.Proft M, Struhl K. Hog1 kinase converts the Sko1–Cyc8–Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol Cell. 2002;9:1307–1317. doi: 10.1016/s1097-2765(02)00557-9. [DOI] [PubMed] [Google Scholar]
- 17.Zapater M, Sohrmann M, Peter M, Posas F, de Nadal E. Selective requirement for SAGA in Hog1-mediated gene expression depending on the severity of the external osmostress conditions. Mol Cell Biol. 2007;27:3900–3910. doi: 10.1128/MCB.00089-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alonso-Monge R, Roman E, Arana DM, Prieto D, Urrialde V, Nombela C, Pla J. The Sko1 protein represses the yeast-to-hypha transition and regulates the oxidative stress response in Candida albicans. Fungal Genet Biol. 2010;47:587–601. doi: 10.1016/j.fgb.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 19.Rauceo JM, Blankenship JR, Fanning S, Hamaker JJ, Deneault JS, Smith FJ, Nantel A, Mitchell AP. Regulation of the Candida albicans cell wall damage response by transcription factor Sko1 and PAS kinase Psk1. Mol Biol Cell. 2008;19:2741–2751. doi: 10.1091/mbc.E08-02-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ni L, Bruce C, Hart C, Leigh-Bell J, Gelperin D, Umansky L, Gerstein MB, Snyder M. Dynamic and complex transcription factor binding during an inducible response in yeast. Genes Dev. 2009;23:1351–1363. doi: 10.1101/gad.1781909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Proft M, Gibbons FD, Copeland M, Roth FP, Struhl K. Genomewide identification of Sko1 target promoters reveals a regulatory network that operates in response to osmotic stress in Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:1343–1352. doi: 10.1128/EC.4.8.1343-1352.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaloriti D, Tillmann A, Cook E, Jacobsen M, You T, Lenardon M, Ames L, Barahona M, Chandrasekaran K, Coghill G, Goodman D, Gow NA, Grebogi C, Ho HL, Ingram P, McDonagh A, de Moura AP, Pang W, Puttnam M, Radmaneshfar E, Romano MC, Silk D, Stark J, Stumpf M, Thiel M, Thorne T, Usher J, Yin Z, Haynes K, Brown AJ. Combinatorial stresses kill pathogenic Candida species. Med Mycol. 2012;50:699–709. doi: 10.3109/13693786.2012.672770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan J, Whiteway M, Shen SH. Disruption of a gene encoding glycerol 3-phosphatase from Candida albicans impairs intracellular glycerol accumulation-mediated salt-tolerance. FEMS Microbiol Lett. 2005;245:107–116. doi: 10.1016/j.femsle.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 25.Wei M, Fabrizio P, Madia F, Hu J, Ge H, Li LM, Longo VD. Tor1/Sch9-regulated carbon source substitution is as effective as calorie restriction in life span extension. PLoS Genet. 2009;5:e1000467. doi: 10.1371/journal.pgen.1000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jandrositz A, Petschnigg J, Zimmermann R, Natter K, Scholze H, Hermetter A, Kohlwein SD, Leber R. The lipid droplet enzyme Tgl1p hydrolyzes both steryl esters and triglycerides in the yeast, Saccharomyces cerevisiae. Biochim Biophys Acta. 2005;1735:50–58. doi: 10.1016/j.bbalip.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 27.Enjalbert B, Moran GP, Vaughan C, Yeomans T, Maccallum DM, Quinn J, Coleman DC, Brown AJ, Sullivan DJ. Genome-wide gene expression profiling and a forward genetic screen show that differential expression of the sodium ion transporter Ena21 contributes to the differential tolerance of Candida albicans and Candida dubliniensis to osmotic stress. Mol Microbiol. 2009;72:216–228. doi: 10.1111/j.1365-2958.2009.06640.x. [DOI] [PubMed] [Google Scholar]
- 28.Krantz M, Becit E, Hohmann S. Comparative analysis of HOG pathway proteins to generate hypotheses for functional analysis. Curr Genet. 2006;49:152–165. doi: 10.1007/s00294-005-0039-9. [DOI] [PubMed] [Google Scholar]
- 29.Kwon MS, Lim SW, Kwon HM. Hypertonic stress in the kidney: a necessary evil. Physiology (Bethesda) 2009;24:186–191. doi: 10.1152/physiol.00005.2009. [DOI] [PubMed] [Google Scholar]
- 30.Nikolaou E, Agrafioti I, Stumpf M, Quinn J, Stansfield I, Brown AJ. Phylogenetic diversity of stress signalling pathways in fungi. BMC Evol Biol. 2009;9:44. doi: 10.1186/1471-2148-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez-Pastor M, Proft M, Pascual-Ahuir A. Adaptive changes of the yeast mitochondrial proteome in response to salt stress. Omics. 2010;14:541–552. doi: 10.1089/omi.2010.0020. [DOI] [PubMed] [Google Scholar]
- 32.Pastor MM, Proft M, Pascual-Ahuir A. Mitochondrial function is an inducible determinant of osmotic stress adaptation in yeast. J Biol Chem. 2009;284:30307–30317. doi: 10.1074/jbc.M109.050682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Izawa S, Ikeda K, Miki T, Wakai Y, Inoue Y. Vacuolar morphology of Saccharomyces cerevisiae during the process of wine making and Japanese sake brewing. Appl Microbiol Biotechnol. 2010;88:277–282. doi: 10.1007/s00253-010-2758-1. [DOI] [PubMed] [Google Scholar]
- 34.Li SC, Diakov TT, Rizzo JM, Kane PM. Vacuolar H+-ATPase works in parallel with the HOG pathway to adapt Saccharomyces cerevisiae cells to osmotic stress. Eukaryot Cell. 2012;11:282–291. doi: 10.1128/EC.05198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nett JE, Lepak AJ, Marchillo K, Andes DR. Time course global gene expression analysis of an in vivo Candida biofilm. J Infect Dis. 2009;200:307–313. doi: 10.1086/599838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melamed D, Pnueli L, Arava Y. Yeast translational response to high salinity: global analysis reveals regulation at multiple levels. RNA. 2008;14:1337–1351. doi: 10.1261/rna.864908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sturtevant J, Cihlar R, Calderone R. Disruption studies of a Candida albicans gene, ELF1: a member of the ATP-binding cassette family. Microbiology. 1998;144(Pt 8):2311–2321. doi: 10.1099/00221287-144-8-2311. [DOI] [PubMed] [Google Scholar]
- 38.Wong KH, Struhl K. The Cyc8–Tup1 complex inhibits transcription primarily by masking the activation domain of the recruiting protein. Genes Dev. 2011;25:2525–2539. doi: 10.1101/gad.179275.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang M, Zhou Z, Elledge SJ. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell. 1998;94:595–605. doi: 10.1016/s0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- 40.Hao B, Clancy CJ, Cheng S, Raman SB, Iczkowski KA, Nguyen MH. Candida albicans RFX2 encodes a DNA binding protein involved in DNA damage responses, morphogenesis, and virulence. Eukaryot Cell. 2009;8:627–639. doi: 10.1128/EC.00246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu C, Byers KJ, McCord RP, Shi Z, Berger MF, Newburger DE, Saulrieta K, Smith Z, Shah MV, Radhakrishnan M, Philippakis AA, Hu Y, De Masi F, Pacek M, Rolfs A, Murthy T, Labaer J, Bulyk ML. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alonso-Monge R, Carvaihlo S, Nombela C, Rial E, Pla J. The Hog1 MAP kinase controls respiratory metabolism in the fungal pathogen Candida albicans. Microbiology. 2009;155:413–423. doi: 10.1099/mic.0.023309-0. [DOI] [PubMed] [Google Scholar]
- 43.David DJB, Amberg C, Jefferey N. Strathern Methods in Yeast Genetics. 2005. John Inglis; New York: 2005. [Google Scholar]
- 44.Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonak J, Lind K, Sindelka R, Sjoback R, Sjogreen B, Strombom L, Stahlberg A, Zoric N. The real-time polymerase chain reaction. Mol Aspects Med. 2006;27:95–125. doi: 10.1016/j.mam.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 45.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffi MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 46.Nantel A, Rigby T, Hogues H, Whiteway M. Microarrays for Studying Pathology in Candida albicans. Wiley Press; Hoboken, NJ: 2006. [Google Scholar]
- 47.Uwamahoro N, Qu Y, Jelicic B, Lo TL, Beaurepaire C, Bantun F, Quenault T, Boag PR, Ramm G, Callaghan J, Beilharz TH, Nantel A, Peleg AY, Traven A. The functions of mediator in Candida albicans support a role in shaping species-specific gene expression. PLoS Genet. 2012;8:e1002613. doi: 10.1371/journal.pgen.1002613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turatsinze JV, Thomas-Chollier M, Defrance M, van Helden J. Using RSAT to scan genome sequences for transcription factor binding sites and cis-regulatory modules. Nat Protoc. 2008;3:1578–1588. doi: 10.1038/nprot.2008.97. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.