Summary
STRA6 is a multitransmembrane domain protein that was recently identified as the cell-surface receptor for plasma retinol binding protein (RBP), the vitamin A carrier protein in the blood. STRA6 binds to RBP with high affinity and mediates cellular uptake of vitamin A from RBP. It is not homologous to any known receptors, transporters, and channels, and it represents a new class of membrane transport protein. Consistent with the diverse physiological functions of vitamin A, STRA6 is widely expressed in diverse adult organs and throughout embryonic development. Mutations in human STRA6 that abolish its vitamin A uptake activity cause severe pathological phenotypes in many human organs including the eye, brain, lung and heart. This chapter describes functional assays for STRA6 in live cells and on cellular membranes. These assays can be employed to study the mechanism of this new membrane transport mechanism and its roles in the physiology and pathology of many organs.
Keywords: STRA6, retinol binding protein receptor, retinol, retinyl ester, vitamin A, retinoid, vitamin A uptake, HPLC
1. Introduction
Vitamin A has diverse biological functions (1–3). It exists in diverse forms, including alcohol, aldehyde, acid, and ester. Except for the ester, which is the storage form, all other forms of vitamin A are known to have biological activities. The acid form of vitamin A (retinoic acid) has the most diverse functions. Nuclear retinoic acid receptors regulate the transcription of a large number of genes (4, 5). In addition to its essential roles in embryonic developmental (6, 7), retinoic acid is also important in the function of many adult organs such as the nervous system (8), the immune system (9, 10), the male and female reproductive systems (11, 12), the respiratory system (12, 13), and the skin (14). Retinoic acid was recently discovered to regulate protein translation in neurons (15, 16). The aldehyde form of vitamin A functions as the chromophore for visual pigments in the eye (17). It was also recently discovered to inhibit adipogenesis (18). The alcohol form of vitamin A serves as the substrate for retinol binding protein (RBP) for delivery in the blood. In addition, the alcohol derivatives of vitamin A have distinct biological activities. For example, they control the growth of B lymphocytes (19) and function as survival factors in serum for fibroblasts (20). Retinol, but not retinoic acid, regulates BMP4 expression in male germ line cells (21) and maintains the pluripotency of embryonic stem cells (22).
Plasma retinol binding protein (RBP) is the principal carrier of vitamin A in the blood and is essential in mobilizing the hepatic vitamin A store (23–26). It was first proposed in the 1970s that there exists a cell surface receptor that mediates vitamin A uptake from RBP on the retinal pigment epithelium and small intestine cells (27–32). During the past three decades, there has been mounting evidence for the existence of RBP receptors on diverse types of tissues including the placenta (33–35), choroid plexus (34, 36), Sertoli cells and peritubular cells of the testis (34, 37–40), macrophages (41), and skin (34, 42). There are also indirect pieces of evidence for the existence of an RBP receptor. For example, in an unbiased search for a serum factor that stimulates the growth of B cells, it was found that vitamin A/RBP complex (holo-RBP) is this factor (43). Using an unbiased strategy combining ligand-specific photo-crosslinking, high affinity purification, and mass spectrometry, the RBP receptor was identified as STRA6, a multitransmembrane protein of previously unknown function (44) (Figure 1). STRA6 binds to RBP with high affinity and specificity and mediates the uptake of vitamin A into the cell. STRA6 was originally identified as a retinoic acid-stimulated gene in cancer cell lines (45, 46). STRA6 is widely expressed in embryonic development and adult organ systems (44, 45). Consistent with the diverse functions of vitamin A in human, STRA6 mutations cause severe pathological phenotypes including the absence of eyes (anophthalmia), mental retardation, congenital heart defects, and lung hyperplasia (47, 48). The point mutations identified in the human patients have been shown to abolish vitamin A uptake activity of STRA6 (49).
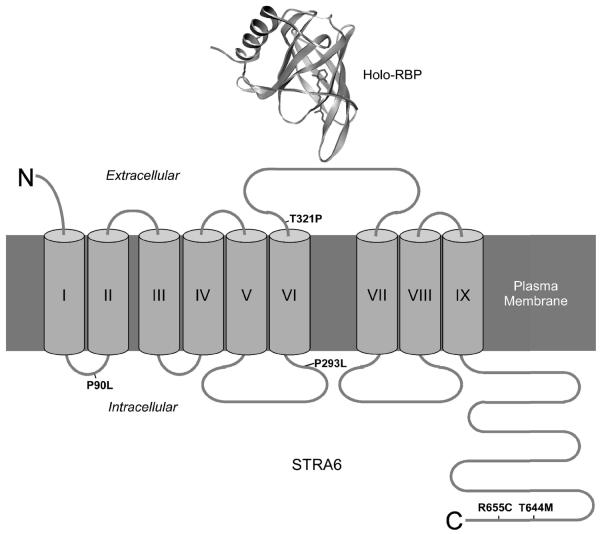
Figure 1.
Transmembrane topology of STRA6, the high-affinity cell-surface receptor for RBP (50). Transmembrane domains are indicated with Roman numerals. Missense mutations in human STRA6 associated with severe birth defects are indicated (47). Crystal structure of holo-RBP is based on structure 1HBP in Protein Data Bank.
In this chapter, we described techniques to study RBP receptor-mediated vitamin A uptake: a radioactive retinoid uptake assay, an HPLC-based retinol uptake assay, a cell-free retinoid uptake assay, and a method to quantitate of RBP binding to live cells. The advantage of the radioactive retinoid uptake assay is its high sensitivity. It does not distinguish between retinoids, but measures total retinoid uptake. Although the HPLC-based assay is less sensitive, it can distinguish between retinoids. In addition, the HPLC-based assay can use serum as the source of holo-RBP and most closely mimics the in vivo uptake process due to the presence of transthyretin/RBP complex in the serum. The cell-free assay is based on the radioactive assay but is performed using cellular membranes. This assay makes it possible to acutely change the composition of the reaction. Quantitation of RBP binding to live cells is achieved using alkaline phosphatase-tagged RBP (AP-RBP). These assays have been successfully used for structural and functional analysis of STRA6's role as the RBP receptor in mediating vitamin A uptake (44, 49, 50).
2. Materials
2.1. RBP production
-
1.
Human retinol binding protein with a 6XHis tag on the N-terminus (His-RBP) cloned into the pET3a vector (New England Biolabs).
-
2.
Competent BL-21 bacteria
-
3.
Luria-Bertani (LB) media with 0.1 mg/ml carbenicillin
-
4.
0.1M IPTG (sterilized by filtration)
-
5.
Complete protease inhibitors (Roche)
-
6.
Phosphate buffered saline (PBS)
-
7.
20% Triton X-100
-
8.
Sonicator
2.2. RBP refolding
-
1.
7.5 M guanidine hydrochloride
-
2.
Sonicator
-
3.
25 mM Tris-HCl, pH 9.0
-
4.
30 mM cystine (prepared fresh): Dissolve 10.8 mg cystine in 50 μl of 1N NaOH. Heating at 37°C helps solubilization. Once cystine is completely dissolved, add water to 1.5ml.
-
5.
300 mM cysteine (prepared fresh)
-
6.
Refolding Buffer (prepared fresh): 25 mM Tris, pH9.0, 0.3 mM cystine, 3.0 mM cysteine, 1 mM EDTA
-
7.
Amicon Ultra 15 concentrator, MWCO 10 K (Millipore)
-
8.
87 mM all-trans-retinol (ACROS): Dissolve 25 mg all-trans-retinol in 1 ml of 10 mM Butylated hydroxytoluene (BHT) in ethanol. Keep the solution in dark. Store at −80°C.
-
9.
2 M dithiothreitol (DTT) (stored at −20°C in aliquots)
2.3. RBP purification
-
1.
Ni-NTA resin (Qiagen)
-
2.
10 mM imidazole in PBS
-
3.
100 mM imidazole in PBS
-
4.
Dialysis tubing with MWCO of 10 kD
-
5.
25 mM Tris, pH 8.4 and 120 mM NaCl
-
6.
HPLC system with a photo-diode array detector (Agilent Technologies)
-
7.
Weak anion exchange column AX-300 (Eprogen)
-
8.
25 mM Tris, pH 8.4
-
9.
25 mM Tris, pH 8.4 and 2 N NaCl
-
10.
Nanodrop spectrophotometer
2.4. Apo-RBP production and loading of 3H-retinol into apo-RBP
-
1.
Heptane
-
2.
Syringe with 22 gauge needle (Hamilton)
-
3.
Nanodrop spectrophotometer
-
4.
[11,12-3H] all-trans retinol (PerkinElmer) (stored at −20°C)
-
5.
Ni-NTA resin (Qiagen)
-
6.
100 mM imidazole in PBS
2.5. Radioactive retinoid uptake assay
-
1.
Transfected COS cells (24 hours after transfection)
-
2.
3H-retinol/RBP
-
3.
Hank's buffered salt solution (HBSS)
-
4.
Serum free medium (SFM)
-
5.
1% Triton-X100 in PBS
-
6.
Scintillation fluid (PerkinElmer)
-
7.
Scintillation counter
2.6. Cell-free retinoid uptake assay
-
1.
PBS with protease inhibitors
-
2.
3H-retinol/RBP
-
3.
2X reaction buffer: 3H-retinol/RBP (40,000–50,000 CPM/reaction), 2.5 mg/ml BSA, 10 mM DTT, 500 mM sucrose, 1X PBS and cØmplete protease inhibitors
-
4.
Mechanical grinder (KONTES)
-
5.
Syringe with 22 gauge needle (Hamilton)
-
6.
Flask that connects to a vacuum source for radioactive waste
-
7.
Multiscreen vacuum manifold (Millipore)
-
8.
MultiScreenHTS low protein binding 96-well filter plates (Millipore)
-
9.
Scintillation fluid (PerkinElmer)
-
10.
Scintillation counter
2.7. HPLC-based retinoid uptake assays to detect retinyl esters and retinol
-
1.
HPLC system with a photo-diode array detector (Agilent Technologies)
-
2.
ZORBAX Eclipse XDB-C18 column, 5 μm, 4.6×150 mm (Agilent Technologies)
-
3.
Purified holo-RBP
-
4.
Pooled human sera (Innovative Research)
-
5.
Serum free medium (SFM)
-
6.
Hank's buffered salt solution (HBSS)
-
7.
PBS with 5 mM EDTA
-
8.
Mechanical grinder (KONTES)
-
9.
10 mM butylated hydroxytoluene (BHT) in ethanol
-
10.
Hexane
-
11.
Nitrogen tank
-
12.
Ethylacetate (HPLC grade)
-
13.
Methanol (HPLC grade)
-
14.
Dark room with red lights
2.8. AP-RBP production and binding
-
1.
AP-RBP fusion protein in SFM
-
3.
Hank's buffered salt solution (HBSS)
-
4.
Serum free medium (SFM)
-
5.
2% formaldehyde in PBS (freshly made)
-
6.
65°C incubator
-
7.
AP buffer: 0.1 M Tris, pH 9.5, 0.1 M NaCl and 50 mM MgCl2
-
8.
BCIP (Roche)
-
9.
NBT (Roche)
-
10.
pNPP (Sigma)
-
11.
3 N NaOH
-
12.
Microplate spectrophotometer
3. Methods
3.1. Production of His tagged-RBP (His-RBP) in bacteria
-
1.
Tag human cDNA for RBP on the N-terminus with 6X His tag and clone it into the pET3a vector (see Note 1). Transform the construct into competent BL-21 bacteria.
-
2.
Grow transformed BL-21 in 50 ml LB Medium containing 0.1 mg/ml carbenicillin at 37 °C with vigorous shaking.
-
3.
Add 0.5 ml of 0.1M IPTG to the culture medium when the O.D. at 660 nm reaches 0.1. Grow further for 5 hours (see Note 2).
-
4.
Pellet the cells by centrifugation at 10,000 g for 20 min.
-
5.
Wash the cells with 50 ml PBS, pellet again, and resuspend in cold 20 ml PBS with complete protease inhibitors.
-
6.
Sonicate the cells on ice for 1 min followed by 1 min cooling. This process is repeated 4 times.
-
7.
Add Triton X-100 to a final concentration of 0.1%.
-
8.
Freeze and thaw the cell lysate twice.
-
9.
Pellet down the inclusion bodies at 20,000g for 30 min at 4°C. RBP expressed in E.coli is mainly found in inclusion bodies (see Note 3).
3.2. Solubilization and refolding of RBP (see Note 3)
-
1.
Solubilize the inclusion body pellet in 20 ml 7.5 M guanidine hydrochloride. Brief sonication helps in solubilization.
-
2.
After adding half volume (10 ml) of 25 mM Tris buffer, pH 9.0, add DTT to a final concentration of 10 mM (see Note 4). Incubate overnight at room temperature to fully reduce and denature proteins.
-
3.
Centrifuge the protein solution to remove guanidine-insoluble material at 16,000 g for 30 min. Split the solution into two batches of 15 ml. Freeze one batch at −20°C (see Note 5).
-
4.
Bring all-trans-retinol stock to room temperature, keeping it in the dark (see Note 6). Prepare 4 volumes (60 ml) of refolding buffer (25 mM Tris, pH9.0, 0.3 mM cystine, 3.0 mM cysteine, 1 mM EDTA) (see Note 7).
-
5.
Degas the refolding buffer for 15 min and chill on ice. At the same time, cool both the protein solution and refolding buffer on ice since high temperature can lead to protein aggregation.
-
6.
Add 690 μl of 87 mM all-trans retinol to the refolding buffer (to a final concentration of 1 mM) right before the refolding reaction.
-
7.
Add refolding buffer to the protein solution dropwise with vigorous stirring of the protein solution (see Note 8). Carry out the refolding in the dark on ice or at 4°C for 5 hours.
-
8.
Spin down the refolding reaction at 24,000 g for 30 min at 4°C to remove protein aggregates. Misfolded RBP tends to aggregate together with correctly folded RBP. Therefore, it is essential to remove protein aggregates as soon as refolding is done.
-
9.
Concentrate the supernatant solution using Amicon Ultra 15 concentrator (MWCO 10 K) to about 10 ml. Do not concentrate the refolded solution more than 10 fold. Too much concentrating causes protein aggregation and lowers the final yield of correctly folded His-RBP.
-
10.
Dilute the concentrated sample with PBS to 100 ml to reduce the DTT and EDTA concentration (see Note 9).
-
11.
Spin the solution one more time at 24,000 g for 20 min at 4°C, and transfer the supernatant to two 50-ml tubes. Add 1 ml of Ni-NTA resin to each tube and rotate the solution with the resin for 2 hours (see Note 10). Longer incubation may increase yield slightly. Adding protease inhibitors may help, however no major degradation has been observed without them.
-
12.
Wash the resin with at least 20 column volumes of 10 mM imidazole in PBS before eluting His-RBP with 100 mM imidazole in PBS (see Note 11).
3.3. HPLC purification of refolded holo-RBP
-
1.
Dialyze His-RBP purified from the Ni-NTA resin against 25 mM Tris, pH 8.4 and 120 mM NaCl.
-
2.
Set up HPLC with weak anion exchange column AX-300 (see Note 12).
-
3.
Clear samples for protein purification on HPLC by centrifugation at 16,000 g for 10 min at 4°C before each run.
-
4.
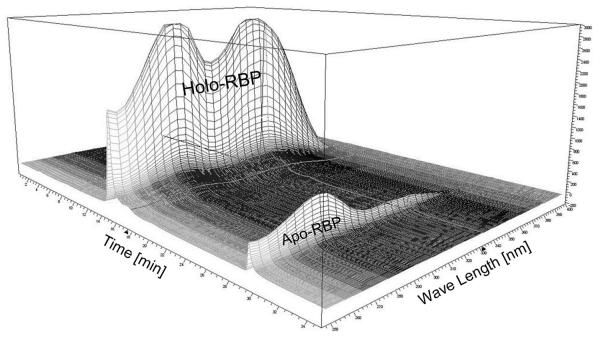
Separate proteins by a NaCl step gradient (220 mM 10 min, 360 mM 15 min, and 1000 mM 15 min) using 25 mM Tris, pH 8.4 as mobile phase at 1 ml/min. As NaCl concentration rises, holo-His-RBP is released whereas apo-His-RBP and misfolded RBP stay bound to the column. Monitor the full absorption spectrum of entire HPLC run (Figure 2) (see Note 13).
-
5.
Recover holo-RBP from the fractions at 360 mM NaCl. At 1000 mM NaCl, all His-RBP bound to the column is released.
-
6.
Fractions containing holo-RBP are pooled, concentrated, and dialyzed against PBS.
-
7.
Purified holo-RBP can be quantified by Nanodrop spectrophotometer (see Note 14).
-
8.
Store HPLC-purified RBP at 4°C (see Note 15).
Figure 2.
Monitoring of the full absorption spectrum of the entire HPLC run during the holo-RBP purification. The peaks for holo-RBP and apo-RBP are indicated.
3.4. Production of apo-RBP and loading of 3H-retinol onto apo-RBP
Apo-RBP is prepared by removing retinol from holo-RBP using organic solvents such as hexane or heptane.
-
1.
Add an equal volume of heptane to purified holo-RBP and mix gently by rotating overnight at 4°C. Harsh mixing will result in loss of the protein due to aggregation.
-
2.
Centrifuge at 16,000 g for 10 min at 4°C to separate the aqueous phase and organic phase. Use a 22-gauge needle to carefully transfer the aqueous phase (bottom) to a new tube. A protein precipitate may form at the interphase. Be careful to avoid the precipitate.
-
3.
Repeat this process 3 more times with a 3-hour heptane incubation for each.
-
4.
Centrifuge the aqueous phase at 16,000 g for 10 min at 4°C to remove any insoluble materials. Check retinol depletion from His-RBP using a Nanodrop spectrophotometer. Apo-RBP should not show an absorption peak at 330 nm. If the protein solution still has significant absorption at 330 nm, repeat the heptanes extraction until the absorption at 330 nm is the same as the baseline.
-
5.
Add 40 μl of 15-30 μM 3H-all-trans retinol stock to 10–20 μg of apo-His-RBP in 960 μl of PBS (see Note 16).
-
6.
Rotate overnight at 4°C and purify His-RBP with 15 μl of Ni-NTA resin.
-
7.
Wash the resin with 1ml PBS 4 times, and elute the 3H-retinol/RBP in 200-1000 μl of 100 mM imidazole in PBS. Measure the radioactivity of the eluted materials by scintillation.
3.5. Retinol uptake assay based on 3H-retinol/RBP
The following protocol is designed for COS cells grown on 24-well dishes (see Note 17). An example of this experiment is shown in Figure 3. The volume of each buffer can be adjusted proportionally for cells grown on wells or dishes of larger areas.
-
1.
At 24 four hours after transfection, media from each well is removed by vacuum suction. The cells in each well are washed with 500 μl HBSS.
-
2.
Remove HBSS and add 250 μl of SFM containing 3H-retinol/RBP (see Note 18).
-
3.
After incubating for desired length of time at 37°C (e.g., 1 hour), remove the supernatant and wash the cells in each well with 500 μl HBSS (see Note 19).
-
4.
Lyse the cells by adding 500 μl 1% Triton X-100 in PBS to each well.
-
5.
Transfer the cell lysate to a scintillation vial.
-
6.
Repeat steps 4 and 5 for each well before counting with a scintillation counter (see Note 20).
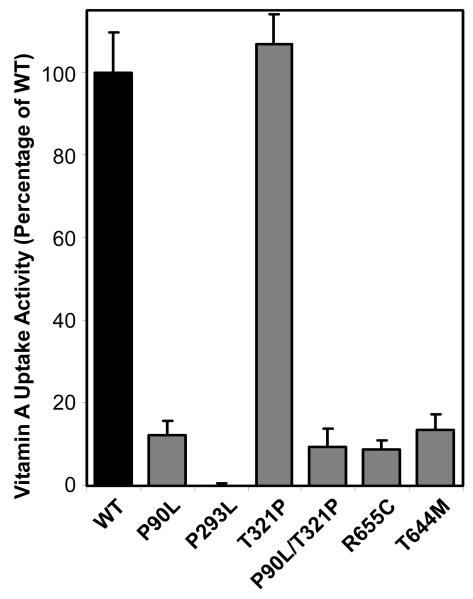
Figure 3.
Vitamin A uptake activity from holo-RBP for human STRA6 mutants associated with severe birth defects (47, 49). Locations of these mutations in the transmembrane topology model of STRA6 are shown in Figure 1. The activity of wild-type STRA6 is defined as 100%.
3.6. A cell-free assay for 3H-retinol uptake (see Note 21)
-
1.
To prepare crude membranes, break up cell pellets on ice with a mechanical grinder in PBS with protease inhibitors.
-
2.
Dilute the cell lysates in PBS with protease inhibitors. After centrifugation at 1,000 g for 3 min at 4°C to remove cell nuclei, centrifuge the lysate at 16,000 g for 30 min at 4°C to pellet the cell membranes.
-
3.
Resuspend the membrane pellets in PBS (>50 μl) and break up the pellets by repeated passage through a 22 gauge needle (see Note 22).
-
4.
Prepare 2X reaction buffer that contains 3H-retinol-RBP solution and add an equal volume to the membrane suspension. A typical final reaction volume is 100 μl.
-
5.
Incubate for a defined amount of time (e.g., 1 hour) at 37°C.
-
6.
During the incubation, assemble the 96-well filtration plate and multiscreen vacuum manifold according to the manufacturer's protocol. Open valve and test vacuum suction (see Note 23).
-
7.
Wet each well with 10 μl PBS. Seal unused wells.
-
8.
Mix samples by pipetting and add them to wells. Once the liquid is removed by suction, add 200 μl PBS. Repeat this wash to each well once more. Use of a multichannel pipettor is recommended when dealing with many samples.
-
9.
Let the membrane dry by keeping suction force for 10 min. Then remove the back support from the MultiscreenHTS 96-well plate. Place the multiscreen plate on a few layers of Kimwipes to remove liquid on the backside. Let the plate dry for another 15 min.
-
10.
Punch the membrane screen out of the plate and place it in a scintillation vial. Count radioactivity by scintillation.
3.7. Non-radioactive retinoid uptake assay
The following protocol is designed for transfected COS cells (see Note 24):
-
1.
At 6 hours after transfection, remove medium and quickly wash cells once with HBSS before culturing the cells in SFM at 37°C (see Note 25).
-
2.
At 18 hours after culturing in SFM, remove SFM and add holo-RBP freshly diluted in SFM. The source of holo-RBP can be either purified holo-RBP or normal human serum, which contains RBP in the form of holo-RBP/transthyretin complex (see Note 26).
-
3.
After incubation at 37°C for a defined period of time (e.g., 1 hour), remove the medium and wash the cells briefly with 10 ml PBS.
-
4.
Add 1.5 ml PBS containing 5 mM EDTA. Incubate for 2 min.
-
5.
Detach cells by pipetting and collect cells in microcentrifuge tubes.
-
6.
Pellet cells at 1,000 g for 3 min. Repeat steps 4 and 5 and combine cells.
-
7.
Store the cell pellets at −80°C before retinoid extraction.
3.8. HPLC analysis of retinyl ester and retinol contents in the cells
All steps in the following procedure are done under red light.
-
1.
Resuspend cell pellet in 50 μl PBS in the presence of protease inhibitors. The following protocol describes experiments at this scale.
-
2.
Add 50 μl of 10 mM BHT in ethanol to the cell lysate. Vortex for 30 sec.
-
3.
Add 500 μl of hexane. Vortex for 5 min.
-
4.
Pellet down cellular debris at 5,000 g for 5 min.
-
5.
Carefully remove and save the upper phase (hexane).
-
6.
Repeat steps 3–5 three times and combine all hexane extracts.
-
7.
Dry the hexane extractable material under nitrogen in the dark.
-
8.
Solublize the dried material in 450 μl of methanol by vortexing for 1 min.
-
9a.
For retinol analysis, add 50 μl water to the sample and filter the sample before HPLC (see Note 27). Separate samples using ZORBAX Eclipse XDB-C18 column (5 μm, 4.6×150 mm) at 1 ml/min using 90% methanol as the mobile phase. The retinol peak at 325 nm appears at around 7 min.
-
9b.
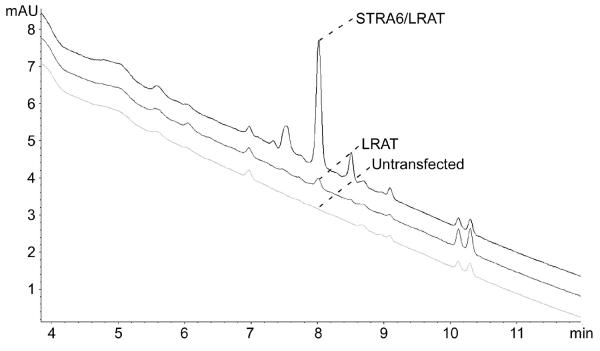
For retinyl ester analysis, separate filtered samples in 100% methanol using a linear gradient (100% methanol at time zero and 100% ethylacetate at 10 min). Flow rate is fixed at 1 ml/min. The same column for retinol analysis is used. The retinyl ester peak at 325 nm appears at around 8 min (Figure 4) (see Note 28).
-
10.
Peak areas for retinoid HPLC analysis are usually normalized by the protein peak by following the 280 nm absorbance. Using the conditions of this protocol, the protein peak appears at 2.3 min for retinyl ester or at 4.5 min for retinol analysis (see Note 29).
Figure 4.
HPLC assay for retinyl ester uptake using transfected or untransfected COS-1 cells that were incubated with 20% normal human serum for 4 hrs. Retinyl ester was extracted and analyzed by HPLC. Mobile phase was changed linearly from 100% methanol to 100% ethylacetate. Retinylpalmitate peaks are indicated.
3.9. Visualization and quantitation of the binding of AP-RBP to live cells
The following protocol is designed to visualize AP-RBP binding to transfected or untransfected COS cells (see Note 30).
-
1.
Wash COS cells grown on gelatin-coated coverslips once with PBS.
-
2.
Incubate with AP-RBP diluted in SFM for 1 hour at room temperature.
-
3.
After washing the cells twice with HBSS, fix in fresh 2% formaldehyde in PBS at room temperature for 10 min.
-
4.
After 3 more PBS washes, heat the cells in PBS for 1 hour at 65°C.
-
5.
After washing the cells once with AP buffer, perform the AP color reactions in the AP buffer containing 165 μg/ml of BCIP and 330 μg/ml of NBT for 1 hour at room temperature.
The following protocol is the liquid AP assay designed to quantitate AP-RBP binding to transfected or untransfected COS cells. An example of this experiment is shown in Figure 5.
-
1.
Wash COS cells grown on 12-well cell culture plates once with HBSS and incubated with AP-RBP diluted in SFM at 37°C for 1 hour.
-
2.
Stop the reactions by quickly washing cells three times with HBSS.
-
3.
Lyse the cells in 150 μl of cold PBS containing 1% Triton X-100 and protease inhibitors per well.
-
4.
Centrifuge the cell lysates at 3,000 g at 4°C for 5 min.
-
5.
Remove the supernatants and heat at 65°C for 1 hour.
-
6.
Mix 50 μl of the heated lysate with 200 μl of pNPP and incubate at 37°C for 1 hour for the AP color reaction. Use the lysate from untransfected cells as the negative control.
-
7.
Stop the reactions by adding 50 μl 3 M NaOH and transfer to a 96-well plate for reading in a microplate reader at 405 nm.
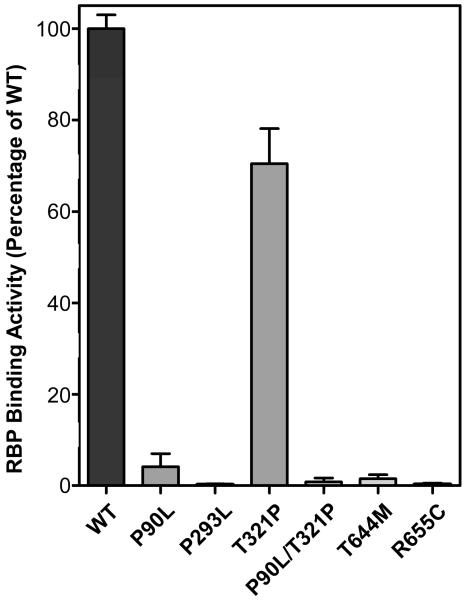
Figure 5.
Quantitation of RBP binding activities of human STRA6 mutants associated with severe birth defects (47, 49). Locations of these mutations in the transmembrane topology model of STRA6 are shown in Figure 1. The activity of wild-type STRA6 is defined as 100%.
4. Notes
-
1.
Tagging RBP on the N-terminus does not interfere with its interaction with STRA6, but a tag on the C-terminus may interfere with this interaction (44). Therefore, the 6XHis tag is engineered into the N-terminus of RBP.
-
2.
IPTG may or may not be necessary in RBP induction. Basal RBP expression is already high (Figure 2). LB is better than rich media for RBP production.
-
3.
As an extracellular protein, native RBP has three pairs of disulfide bonds. Bacteria express RBP intracellularly and cannot properly form the correct disulfide bonds. For this reason, RBP expressed in E.coli is mainly found in inclusion body as misfolded proteins that are mostly inactive and do not bind retinol. This misfolded RBP needs to be denatured and refolded. Good refolding is critical for producing high-quality holo-RBP. Without good refolding, even HPLC cannot purify holo-RBP 100% loaded with all-trans retinol.
-
4.
The concentration of DTT in the solublizing solution is critical. Excess DTT can inhibit the oxidation reaction during the refolding reaction and makes refolded--RBP unstable without disulfide bonds.
-
5.
Increasing the scale of the refolding reaction can decrease the yield of correctly folded RBP.
-
6.
The refolding solution includes all-trans-retinol, which helps proper refolding of RBP. Good quality retinol is essential during the refolding reaction. Without all-trans-retinol, refolding efficiency is very low (<10%). Once solubilized in ethanol, add BHT to 10 mM to prevent oxidation, seal the tube tightly and store the unused retinol at −80°C.
-
7.
Cystine and cysteine solutions need to be made fresh. Cystine is not easily solubilized without high pH and heat. Do not add water directly to solubilize cystine. To make the 30 mM cystine stock solution, dissolve 10.8 mg cystine in 50 μl of 1N NaOH. Heating at 37°C helps solubilization. Once cystine is completely dissolved add water to 1.5 ml.
-
8.
Refolding buffer containing cystine and cysteine has to be added slowly at the start of the refolding reaction when the solution is being stirred. The slow addition of refolding buffer prevents the dramatic reduction of local guanidine concentration. As guanidine concentration is gradually diluted, RBP refolding starts. Refolding buffer contains cystine and cysteine at a ratio of 1:10. Cystine and cysteine facilitate the shuffling reaction by reducing and oxidizing the disulfide bonds in RBP.
-
9.
High DTT and EDTA concentrations can reduce the efficiency of subsequent His-RBP purification using Ni-NTA resin. Dialysis to remove DTT and EDTA is not recommended because long buffer exchange tends to cause serious protein aggregation.
-
10.
Ni-NTA resin binding of refolded RBP should be done on the same day as the refolding reaction. Refolded RBP solution without dilution tends to aggregate over an extended time period.
-
11.
The typical yield at this step is >20 mg/100 ml of bacteria culture. In a typical preparation with successful refolding, 75– 80% of His-RBP should be bound with retinol based on the 280 nm and 330 nm readings.
-
12.
Since His-RBP has a pI of 6.47, a basic buffer is used to keep the protein in the negatively charged state. At physiological ionic strength, His-RBP binds to weak anion exchange column. It is important to calibrate the pH meter to prepare the mobile phase because the binding of RBP to the column is very sensitive to pH. High pH (pH> 8.4) can reduce the protein binding capacity of the column and low pH (between pH 6.47 and 8.4) can make proteins bind too strongly to elute.
-
13.
Holo-RBP can be monitored during purification either by its characteristic absorption at 330 nm or by the fluorescence intensity of the bound retinol.
-
14.
Accurate quantitation of holo-RBP is not possible by spectrophotometer if free retinol is present in the solution. Even for purified holo-RBP, retinol in holo-RBP consistently causes a problem in accurate quantitation because the presence of retinol in RBP lowers the absorbance baseline significantly. Therefore, the baseline needs to be adjusted to reflect the presence of retinol. On a Nanodrop spectrophotometer, use PBS as baseline if holo-RBP is in PBS. Then read the absorbance of holo-RBP solution at 280, 330 and 500 nm. Note that the 500 nm value is reduced due to the presence of retinol in the solution. Add the reduced absorbance value at 550 nm to the values 280 nm and 330 nm to get the correct absorbance values. Alternatively, purified holo-RBP can be quantified using retinol fluorescence using holo-RBP of known concentrations. Although fluorescence is more sensitive, it cannot reveal how well retinol is loaded into RBP.
-
15.
High quality holo-RBP is very stable and can be stored at 4°C in PBS for years without any loss or degradation of retinol. Freezing HPLC-purified RBP is not recommended, as it lowers the 330/280 ratio upon thawing. If RBP is frozen, centrifuge at 16,000g for 10 min to remove any protein aggregates.
-
16.
Apo-His-RBP tends to denature in the presence of a high concentration of ethanol. Therefore, the ethanol concentration should not exceed 4% in the loading reaction. The [11,12-3H] retinol purchased from PerkinElmer has a specific activity of 30–60 Ci/mmol and a concentration of 1 mCi/ml or about 15−30 μM (in ethanol). At this concentration, the maximum concentration of 3H-retinol is 0.6–1.2 μM in the loading reaction if 4% of the 3H-retinol stock in ethanol is added. To achieve higher 3H-retinol incorporation, the stock 3H-retinol solution can be concentrated before use. Alternatively, purify His-RBP and repeat the loading reaction with fresh 3H-retinol.
-
17.
Since retinol uptake assays on live cells involve multiple washes, COS cells are advantageous because they don't detach as easily as HEK293 cells. HEK293 cells are more easily transfected and can be used for membrane-based assays.
-
18.
For 24-well assays, stock 3H-retinol/RBP is diluted in SFM so that each well contains 250 μl of SFM and 40,000–50,000 CPM of 3H-retinol-RBP. The sensitivity of the radioactive assay allows the use of 3H-retinol-RBP at a much lower concentration (e.g., 3 nM) than the Kd of the RBP/STRA6 interaction.
-
19.
The presence of divalent ions in HBSS makes it a better solution for live cell washes than PBS because it can minimize cell loss due to the wash. It helps to visually monitor cell attachment during the wash process. If the experiment is designed to account for 3H-retinol/RBP bound to cell surface, it is important to perform the wash quickly since the RBP/STRA6 interaction is transient (e.g., wash 2 wells at a time).
-
20.
Compared with HPLC-based assays, radioactive assays use much lower concentrations of holo-RBP (3H-retinol/RBP) and have overall lower concentrations of proteins. Therefore, nonspecific sticking of retinol/RBP to plastic dishes is a more significant source of background “uptake” signal in radioactive retinol-based assays, while it is negligible for HPLC-based assays. For a typical 3H-retinol/RBP-based assay, this background signal is less than 10% of the real uptake signal. This background signal can be easily meausred by incubating 3H-retinol/RBP with empty wells that have been incubated with culture media but without cells. The equivalent amount of background in wells with cells can be calculated by taking into account of the confluence of the cells on the bottom of this dish and the area of the side wall in contact with the medium.
-
21.
The cell-free assay for retinol uptake has the advantage of using more concentrated STRA6 in a small volume and allowing for the addition of soluble factors during the reaction. However, cellular membrane normally needs to be spun down to wash off non-specific bound 3H-retinol-RBP. Repeated centrifugation can be time consuming. A filtering device can greatly reduce the washing time for cellular membrane-based retinol uptake.
-
22.
Resuspend the membrane in 50–100 μl of PBS per reaction. Avoid bubbles during needle passage. Since a 96-well filtration device is used in this assay during the wash step, it is important not to use membranes prepared from more than ¼ of a confluent 100-mm dish of cells per well (per reaction). Too much membrane can clog the filter and make it impossible to wash the membranes.
-
23.
To provide constant vacuum pressure for the multiscreen system, cap the top of each well that will not be used. Alternatively, once PBS is added to wash each well, place a cover over the multiscreen plate and sealed the side briefly.
-
24.
Radioactive assays and HPLC assays for retinoid uptake require different scales of experiments due to the difference in sensitivity of detection. Radioactive assays can be performed on 24-well plates due to their high sensitivity. An HPLC-based assay to detect retinol uptake needs to be done at a much larger scale than an HPCL-based assay to detect retinyl esters. HPLC to detect retinyl esters can be performed on 6-well plates. In contrast, a 100-mm dish is necessary for HPLC-based assay to detect retinol accumulation.
-
25.
It is ideal to perform the retinoid uptake assay 24 hours after transfection, since this is the time at which the transfected proteins just reach the peak of expression. However, fetal bovine serum used in cell culture does contain retinol binding protein. Prolonged incubation after trasnfection will result in significant background retinoid uptake. The background level of uptake before the assay can be easily detected in HPLC by measuring retinoid levels without adding exogenous retinol binding protein or human serum. Changing the culture media to SFM 6 hours after transfection can reduce background retinoid uptake to non-detetable levels.
-
26.
Normal human serum contains high concentrations of holo-RBP (~2 μM), which is much higher than the Kd of STRA6/RBP binding (~50 nM). Therefore, retinol uptake from blood samples usually uses serum diluted in SFM (e.g., 25% serum).
-
27.
Samples for retinoid analysis are filtered. Use of a guard column is preferred for good long-term performance of the column.
-
28.
Flush the entire HPLC system with water if you do not plan to run HPLC for more than a few days. Salt precipitation will damage the pump.
-
29.
An alternative way to normalize is to spike a fixed amount of retinyl acetate into the starting materials as an external control, and use the peak areas for retinyl acetate to normalize retinoid extraction efficiency across the samples. For absolute quantitation, a standard curve is made with stock retinyl palmitate or retinol solution.
-
30.
AP fusion is an established method to label secreted proteins and study their interactions with cell-surface receptors (51). In this system, the GPI anchor of human placental AP was removed to make it a secreted protein. AP tagged at the N-terminus of RBP (AP-RBP) does not interfere with its interaction with STRA6 (44). To produce AP-RBP, COS cells are transfected with AP-RBP cDNA cloned into a mammalian expression vector. COS cell is preferred due to its strong attachment to the culture dish. At 12–24 hours after transfection, the media is changed to SFM. AP-RBP fusion protein can be harvested from the supernatant of transfected cells in SFM 48 hours later. If purified AP-RBP is desired, a 6XHis tag can be inserted between AP and RBP. This tag allows convenient purification of AP-RBP from the SFM. Quantitation of AP-RBP concentration can be performed by comparing the AP signal in AP-RBP with AP proteins with known concentrations. Detection of AP signal after AP-RBP binding is an effective method to quantitative study of the interaction between RBP and STRA6. Since this AP is heat resistant, heating is an effective way to eliminate endogenous AP activity.
References
- 1.Blomhoff R. Overview of Vitamin A Metabolism and Function. In: Blomhoff R, editor. Vitamin A in Health and Disease. Marcel Dekker, Inc.; 1994. pp. 1–35. [Google Scholar]
- 2.Ross AC, Gardner EM. The function of vitamin A in cellular growth and differentiation, and its roles during pregnancy and lactation. Adv Exp Med Biol. 1994;352:187–200. doi: 10.1007/978-1-4899-2575-6_15. [DOI] [PubMed] [Google Scholar]
- 3.Napoli JL. Biochemical pathways of retinoid transport, metabolism, and signal transduction. Clin Immunol Immunopathol. 1996;80:S52–62. doi: 10.1006/clin.1996.0142. [DOI] [PubMed] [Google Scholar]
- 4.Evans RM. The molecular basis of signaling by vitamin A and its metabolites. Harvey Lect. 1994;90:105–117. [PubMed] [Google Scholar]
- 5.Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- 6.Maden M. Role of Retinoids in Embryonic Development. In: Blomhoff R, editor. Vitamin A in Health and Disease. Marcel Dekker, Inc.; 1994. [Google Scholar]
- 7.Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008;134:921–931. doi: 10.1016/j.cell.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drager UC. Retinoic acid signaling in the functioning brain. Sci STKE. 2006;2006:pe10. doi: 10.1126/stke.3242006pe10. [DOI] [PubMed] [Google Scholar]
- 9.Blomhoff HK, Smeland EB. Role of Retinoids in Normal Hemtopoiesis and the Immune System. In: Blomhoff R, editor. Vitamin A in Health and Disease. Marcel Dekker, Inc.; 1994. pp. 451–484. [Google Scholar]
- 10.Stephensen CB. Vitamin A, infection, and immune function. Annu Rev Nutr. 2001;21:167–192. doi: 10.1146/annurev.nutr.21.1.167. [DOI] [PubMed] [Google Scholar]
- 11.Chung SS, Wolgemuth DJ. Role of retinoid signaling in the regulation of spermatogenesis. Cytogenet Genome Res. 2004;105:189–202. doi: 10.1159/000078189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolbach SR, Howe PR. Tissue change following deprivation of fat-soluble A vitamin. J. Exp. Med. 1925;42:753–777. doi: 10.1084/jem.42.6.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biesalski HK. The significance of vitamin A for the development and function of the lung. Forum Nutr. 2003;56:37–40. [PubMed] [Google Scholar]
- 14.Vahlquist A. Role of Retinoids in Normal and Diseased Skin. In: Blomhoff R, editor. Vitamin A in Health and Disease. Marcel Dekker, Inc.; 1994. pp. 365–424. [Google Scholar]
- 15.Chen N, Onisko B, Napoli JL. The nuclear transcription factor RARalpha associates with neuronal RNA granules and suppresses translation. J Biol Chem. 2008;283:20841–20847. doi: 10.1074/jbc.M802314200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aoto J, Nam CI, Poon MM, Ting P, Chen L. Synaptic signaling by all-trans retinoic acid in homeostatic synaptic plasticity. Neuron. 2008;60:308–320. doi: 10.1016/j.neuron.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Travis GH, Golczak M, Moise AR, Palczewski K. Diseases caused by defects in the visual cycle: retinoids as potential therapeutic agents. Annu Rev Pharmacol Toxicol. 2007;47:469–512. doi: 10.1146/annurev.pharmtox.47.120505.105225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ziouzenkova O, Orasanu G, Sharlach M, Akiyama TE, Berger JP, Viereck J, Hamilton JA, Tang G, Dolnikowski GG, Vogel S, Duester G, Plutzky J. Retinaldehyde represses adipogenesis and diet-induced obesity. Nat Med. 2007;13:695–702. doi: 10.1038/nm1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buck J, Myc A, Garbe A, Cathomas G. Differences in the action and metabolism between retinol and retinoic acid in B lymphocytes. J Cell Biol. 1991;115:851–859. doi: 10.1083/jcb.115.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y, Derguini F, Buck J. Vitamin A in serum is a survival factor for fibroblasts. Proc Natl Acad Sci U S A. 1997;94:10205–10208. doi: 10.1073/pnas.94.19.10205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baleato RM, Aitken RJ, Roman SD. Vitamin A regulation of BMP4 expression in the male germ line. Dev Biol. 2005;286:78–90. doi: 10.1016/j.ydbio.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 22.Chen L, Khillan JS. Promotion of feeder-independent self-renewal of embryonic stem cells by retinol (vitamin A) Stem Cells. 2008;26:1858–1864. doi: 10.1634/stemcells.2008-0050. [DOI] [PubMed] [Google Scholar]
- 23.Blomhoff R, Green MH, Berg T, Norum KR. Transport and storage of vitamin A. Science. 1990;250:399–404. doi: 10.1126/science.2218545. [DOI] [PubMed] [Google Scholar]
- 24.Newcomer ME, Ong DE. Plasma retinol binding protein: structure and function of the prototypic lipocalin. Biochim Biophys Acta. 2000;1482:57–64. doi: 10.1016/s0167-4838(00)00150-3. [DOI] [PubMed] [Google Scholar]
- 25.Zanotti G, Berni R. Plasma retinol-binding protein: structure and interactions with retinol, retinoids, and transthyretin. Vitam Horm. 2004;69:271–295. doi: 10.1016/S0083-6729(04)69010-8. [DOI] [PubMed] [Google Scholar]
- 26.Quadro L, Hamberger L, Gottesman ME, Wang F, Colantuoni V, Blaner WS, Mendelsohn CL. Pathways of vitamin A delivery to the embryo: insights from a new tunable model of embryonic vitamin A deficiency. Endocrinology. 2005;146:4479–4490. doi: 10.1210/en.2005-0158. [DOI] [PubMed] [Google Scholar]
- 27.Bok D, Heller J. Transport of retinol from the blood to the retina: an autoradiographic study of the pigment epithelial cell surface receptor for plasma retinol-binding protein. Exp Eye Res. 1976;22:395–402. doi: 10.1016/0014-4835(76)90177-9. [DOI] [PubMed] [Google Scholar]
- 28.Heller J. Interactions of plasma retinol-binding protein with its receptor. Specific binding of bovine and human retinol-binding protein to pigment epithelium cells from bovine eyes. J Biol Chem. 1975;250:3613–3619. [PubMed] [Google Scholar]
- 29.Heller J, Bok D. Transport of retinol from the blood to the retina: involvement of high molecular weight lipoproteins as intracellular carriers. Exp Eye Res. 1976;22:403–410. doi: 10.1016/0014-4835(76)90178-0. [DOI] [PubMed] [Google Scholar]
- 30.Rask L, Peterson PA. In vitro uptake of vitamin A from the retinol-binding plasma protein to mucosal epithelial cells from the monkey's small intestine. J Biol Chem. 1976;251:6360–6366. [PubMed] [Google Scholar]
- 31.Maraini G, Gozzoli F. Binding of retinol to isolated retinal pigment epithelium in the presence and absence of retinol-binding protein. Invest Ophthalmol. 1975;14:785–787. [PubMed] [Google Scholar]
- 32.Chen CC, Heller J. Uptake of retinol and retinoic acid from serum retinol-binding protein by retinal pigment epithelial cells. J Biol Chem. 1977;252:5216–5221. [PubMed] [Google Scholar]
- 33.Sivaprasadarao A, Findlay JB. The interaction of retinol-binding protein with its plasma-membrane receptor. Biochem J. 1988;255:561–569. [PMC free article] [PubMed] [Google Scholar]
- 34.Smeland S, Bjerknes T, Malaba L, Eskild W, Norum KR, Blomhoff R. Tissue distribution of the receptor for plasma retinol-binding protein. Biochem J. 1995;305(Pt 2):419–424. doi: 10.1042/bj3050419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sivaprasadarao A, Boudjelal M, Findlay JB. Solubilization and purification of the retinol-binding protein receptor from human placental membranes. Biochem J. 1994;302(Pt 1):245–251. doi: 10.1042/bj3020245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacDonald PN, Bok D, Ong DE. Localization of cellular retinol-binding protein and retinol-binding protein in cells comprising the blood-brain barrier of rat and human. Proc Natl Acad Sci U S A. 1990;87:4265–4269. doi: 10.1073/pnas.87.11.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shingleton JL, Skinner MK, Ong DE. Characteristics of retinol accumulation from serum retinol-binding protein by cultured Sertoli cells. Biochemistry. 1989;28:9641–9647. doi: 10.1021/bi00451a015. [DOI] [PubMed] [Google Scholar]
- 38.Bishop PD, Griswold MD. Uptake and metabolism of retinol in cultured Sertoli cells: evidence for a kinetic model. Biochemistry. 1987;26:7511–7518. doi: 10.1021/bi00397a046. [DOI] [PubMed] [Google Scholar]
- 39.Bhat MK, Cama HR. Gonadal cell surface receptor for plasma retinol-binding protein. A method for its radioassay and studies on its level during spermatogenesis. Biochim Biophys Acta. 1979;587:273–281. doi: 10.1016/0304-4165(79)90360-x. [DOI] [PubMed] [Google Scholar]
- 40.Davis JT, Ong DE. Retinol processing by the peritubular cell from rat testis. Biol Reprod. 1995;52:356–364. doi: 10.1095/biolreprod52.2.356. [DOI] [PubMed] [Google Scholar]
- 41.Hagen E, Myhre AM, Smeland S, Halvorsen B, Norum KR, Blomhoff R. Uptake of vitamin A in macrophages from physiologic transport proteins: role of retinol-binding protein and chylomicron remnants. J Nutr Biochem. 1999;10:345–352. doi: 10.1016/s0955-2863(99)00013-3. [DOI] [PubMed] [Google Scholar]
- 42.Torma H, Vahlquist A. Vitamin A uptake by human skin in vitro. Arch Dermatol Res. 1984;276:390–395. doi: 10.1007/BF00413360. [DOI] [PubMed] [Google Scholar]
- 43.Buck J, Ritter G, Dannecker L, Katta V, Cohen SL, Chait BT, Hammerling U. Retinol is essential for growth of activated human B cells. J Exp Med. 1990;171:1613–1624. doi: 10.1084/jem.171.5.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawaguchi R, Yu J, Honda J, Hu J, Whitelegge J, Ping P, Wiita P, Bok D, Sun H. A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science. 2007;315:820–825. doi: 10.1126/science.1136244. [DOI] [PubMed] [Google Scholar]
- 45.Bouillet P, Sapin V, Chazaud C, Messaddeq N, Decimo D, Dolle P, Chambon P. Developmental expression pattern of Stra6, a retinoic acid-responsive gene encoding a new type of membrane protein. Mech Dev. 1997;63:173–186. doi: 10.1016/s0925-4773(97)00039-7. [DOI] [PubMed] [Google Scholar]
- 46.Szeto W, Jiang W, Tice DA, Rubinfeld B, Hollingshead PG, Fong SE, Dugger DL, Pham T, Yansura DG, Wong TA, Grimaldi JC, Corpuz RT, Singh JS, Frantz GD, Devaux B, Crowley CW, Schwall RH, Eberhard DA, Rastelli L, Polakis P, Pennica D. Overexpression of the retinoic acid-responsive gene Stra6 in human cancers and its synergistic induction by Wnt-1 and retinoic acid. Cancer Res. 2001;61:4197–4205. [PubMed] [Google Scholar]
- 47.Pasutto F, Sticht H, Hammersen G, Gillessen-Kaesbach G, Fitzpatrick DR, Nurnberg G, Brasch F, Schirmer-Zimmermann H, Tolmie JL, Chitayat D, Houge G, Fernandez-Martinez L, Keating S, Mortier G, Hennekam RC, von der Wense A, Slavotinek A, Meinecke P, Bitoun P, Becker C, Nurnberg P, Reis A, Rauch A. Mutations in STRA6 Cause a Broad Spectrum of Malformations Including Anophthalmia, Congenital Heart Defects, Diaphragmatic Hernia, Alveolar Capillary Dysplasia, Lung Hypoplasia, and Mental Retardation. Am J Hum Genet. 2007;80:550–560. doi: 10.1086/512203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Golzio C, Martinovic-Bouriel J, Thomas S, Mougou-Zrelli S, Grattagliano-Bessieres B, Bonniere M, Delahaye S, Munnich A, Encha-Razavi F, Lyonnet S, Vekemans M, Attie-Bitach T, Etchevers HC. Matthew-Wood Syndrome Is Caused by Truncating Mutations in the Retinol-Binding Protein Receptor Gene STRA6. Am J Hum Genet. 2007;80:1179–1187. doi: 10.1086/518177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawaguchi R, Yu J, Wiita P, Honda J, Sun H. An essential ligand-binding domain in the membrane receptor for retinol-binding protein revealed by large-scale mutagenesis and a human polymorphism. J Biol Chem. 2008;283:15160–15168. doi: 10.1074/jbc.M801060200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawaguchi R, Yu J, Wiita P, Ter-Stepanian M, Sun H. Mapping the membrane topology and extracellular ligand binding domains of the retinol binding protein receptor. Biochemistry. 2008;47:5387–5395. doi: 10.1021/bi8002082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Flanagan JG, Cheng HJ. Alkaline phosphatase fusion proteins for molecular characterization and cloning of receptors and their ligands. Methods Enzymol. 2000;327:198–210. doi: 10.1016/s0076-6879(00)27277-7. [DOI] [PubMed] [Google Scholar]