Abstract
The characterization of the structure of highly hierarchical biosamples such as collagen-based tissues at the scale of tens of nanometers is essential to correlate the tissue structure with its growth processes. Coherent x-ray Bragg ptychography is an innovative imaging technique that gives high resolution images of the ordered parts of such samples. Herein, we report how we used this method to image the collagen fibrillar ultrastructure of intact rat tail tendons. The images show ordered fibrils extending over 10–20 μm in length, with a quantifiable D-banding spacing variation of 0.2%. Occasional defects in the fibrils distribution have also been observed, likely indicating fibrillar fusion events.
Introduction
Collagen in its form of fibrous protein plays a very important role in defining the structural and mechanical properties in most vertebrate tissues, ranging from bone to tendon, cornea, and skin. Mechanical properties of these tissues such as tensile or shear strengths, compressive or torsional loads are controlled by well-defined adaptations of the fibrous anisotropic collagen fibrils present within the scaffold to their native environment (1–6). A particular case of adaptation is tissue growth, where the collagen fibrils alter their intra- and interfibrillar cross-links, degree of mineralization, and fibril diameter depending on the fibril environment (7–10). Research in this field would benefit from the ability to obtain high resolution images of these tissues in their native state for different stages of the growth process. However, imaging complex biological samples in the native state is a challenging task because of the need to penetrate inside tissue without disturbing it. Electron microscopy is the most powerful method for high resolution (11) but requires sample desiccation, fixation, and sectioning, which may disrupt the subtle order that can be central to the function of the tissue. Fluorescence microscopy allows access to a wide range of fluorescent tags that can bind to specific chemical states within the tissue. However, visible light has a short penetration length, and the sample needs to be sufficiently transparent and permeable to diffusion of the fluorophore species.
X-rays can penetrate millimeter-sized blocks of tissue in situ without the need for sectioning. A good selection of imaging modalities is available (12), but all are limited by the relatively weak contrast between the tissues biochemical constituents. Staining strategies, or the use of soft x-ray microscopy (in the water window x-ray energies between the absorption edges of carbon and oxygen, 280 and 530 eV, respectively), can be used to increase the contrast. However, the staining procedures are likely to bring the sample to an unnatural state. Moreover, soft x-rays can only be used for thin samples due to the limited penetration depth; and at these energies, samples suffer a higher rate of radiation damage (13).
For highly ordered biological samples, x-ray diffraction can lead to excellent contrast in images because only the ordered parts of the sample contribute to the signal, which are readily distinguished from the disordered parts between the samples. In this sense, it can be compared to the structural-component sensitivity of fluorescent labeling, but with sensitivity to periodic structures rather than chemical composition. Using diffraction contrast, that is, dark-field contrast, the scanning transmission x-ray microscope has provided high-contrast images, but this technique is limited in resolution to the smallest probe size that can be created by x-ray optics. A step beyond the scanning transmission x-ray microscope is to measure and interpret the diffraction pattern obtained from each position of the probe on the sample as a local image with higher resolution. However, this requires solving a phase problem to invert the diffraction pattern, as x-ray detectors can only record the intensity of the diffracted wave, and the phase information is lost, preventing the possibility to obtain an image from the sample. This can be overcome by using a coherent x-ray beam and properly selected overlap ratio between the probe positions used to obtain the diffraction patterns. Using this procedure, the phase problem is overdetermined and can be solved by using phase retrieval algorithms. This method, called x-ray ptychography (14–16), is used here in Bragg projection mode (17,18) to image the position of individual collagen fibrils within an intact extract of rat tail tendon. In this mode, the sample, in Bragg condition, is scanned laterally to the incident beam. In the case of tissues, the Bragg condition is achieved in the small angle regime, thus the sample is placed perpendicularly to the incident beam, and the experiment is carried out in transmission geometry. The resulting images reconstructed by Bragg projection ptychography reveal structural information related to the tissue organization such as fibril diameter, fibril-to-fibril variations of the collagen D-spacing, and the possible evidence of fibril fusions.
We have previously reported our successful recording of synchrotron coherent x-ray diffraction (CXD) patterns from intact rat tail tendons at the coherent diffraction beamline 34-ID-C of the Advanced Photon Source (19). These patterns showed good fringe visibility because of the high x-ray beam coherence, but still retained a curved arc-shaped envelope of the speckles, which was due to disruption of the alignment of the fibrils within the tendon due to sample handling. Subsequently, we developed procedures to support the tendons under slight tension, clamped inside a wet cell, and maintained in a buffered solution. Improved diffraction patterns were recorded at the I-22 beamline of Diamond Light Source (20). These first order diffraction patterns showed an essentially linear array of speckles, again with good visibility, indicating that the collagen fibrils were close to fully aligned within the area of the coherent x-ray beam, 20 μm in diameter. The I-22 x-ray beam was insufficiently stable to obtain meaningful ptychography data so that, before this work, we had been unsuccessful in inverting the diffraction patterns to images of the collagen arrangement within the tendons. This work reports that these limitations have now been overcome.
CXD exploits the extremely small source size of third-generation synchrotron radiation sources. A sufficiently small aperture, placed just before the sample, selects the spatially coherent portion of the incident beam. A silicon (111) double-crystal monochromator ensures temporal coherence. Third-generation sources use x-ray undulators (21), which ensure that a sufficient photon flux, around 108 photons per second, passes these coherence-defining components to allow satisfactory measurement of diffraction from weakly scattering biological samples such as the collagen fibrils within a rat tail tendon. Diffraction from the sample is then a mutual interference of all illuminated regions, which contains information about the relative position of every part. When measured correctly, CXD has an assembly of very fine speckles modulating the smooth continuous diffraction pattern of a disordered sample. Ptychography algorithms, such the one used in this study, can be fed with CXD patterns to retrieve an image of the sample. When the used CXD patterns correspond to a single Bragg peak from the sample, corresponding to a given hierarchical structure, the image would contain the information of the relative positions of all the structures of this type. In the particular case of tendon, when using ptychography algorithms on the collagen D-banding Bragg peak, the reconstructed image shows the relative positions of all the contributing fibrils, providing significant information about the tissue internal structure.
Materials and Methods
Native tendon samples preparation
The samples, kept frozen until dissection, consisted of bundles of fibrils carefully extracted from the tail tendons of young healthy rats. The fibril bundles (10 mm long and 200–300 μm diameter) were gently pulled off with fine tweezers and stored in phosphate buffered solution (7.4 pH) at 4°C before the experiment. For the synchrotron measurements, one single bundle was mounted in a brass cell as described in previous experiments (20), gently clamping the bundle to keep the fibrils parallel without extra stretching. The cell was then filled with phosphate buffered solution to keep the fibrils wet and in physiological conditions during the data acquisition.
Gold lithographed model sample
A model sample simulating the collagen fibrils D-banding periodicity and typical sizes was used during the experiment to determine the probe function (see Fig. 3). The model sample was a nanofabricated phase plate of gold motifs (ladder structures of 7 blocks each one, each block 200 × 100 nm with a period of 200 nm so the total motif size is 100 × 1300 nm). Blocks were 600 nm deep, giving a phase shift of 1 rad at 8.5 keV. The periodicity of 200 nm was chosen to mimic the third order collagen Bragg peak (as lithographed structures of 67 nm size were too unstable). The sample was fabricated following previously reported methods (22).
Figure 3.
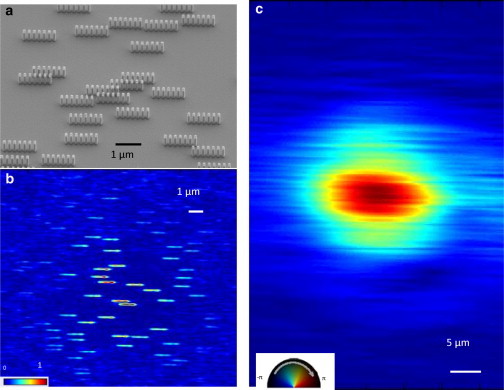
Reconstruction images of the gold lithographed model object and the illumination probe. (a) Scanning electron microscopy image of the sample, where the ladder motifs simulating the collagen d-banding periodicity are clearly shown. (b) Bragg projection ptychography reconstruction of a 17 × 17 μm area of the sample. The image shows the amplitude of the reconstruction (that is, the sample density) normalized to 1. (c) Probe amplitude and phase recovered at the same time as the sample image with the ePIE algorithm. Complex color scale: brightness indicates amplitude, color indicates phase.
Coherent x-ray ptychography data acquisition
Ptychographic data sets were collected at the coherent small angle x-ray scattering beamline (cSAXS beamline) at the Swiss Light Source (SLS), Paul Scherrer Institut (Switzerland). The beam energy was set to 8.5 keV (wavelength λ = 0.146 nm) and a Rh-coated plane mirror was used to reject higher harmonics. A 20 μm diameter pinhole used as transverse coherence aperture was located 30 cm in front, or upstream, of the sample. We installed a second 100 μm diameter pinhole 13 cm downstream of the first one, to remove the scattering coming from the 20 μm aperture. The sample was mounted on a two-dimensional (2D) piezoelectric translation stage (Physik Instrumente, PI, Karlsruhe, Germany). The detector, an optically coupled charge-coupled device camera with a columnar CsI scintillator and 1300 × 1340 pixels of 20 μm size (PI-SCX 1300 from Princeton Instruments, Trenton, NJ) was located 7.21 m downstream from the sample. Given the sample-to-detector distance and the area of the diffraction, the pixel size of the reconstructions was 51.4 nm.
The tendon sample was scanned in a 26 × 3 mesh grid with steps of 2 μm, covering an area of 72 × 26 μm. At each position, a diffraction pattern was recorded with 60 s exposure time, before the onset of radiation damage, as we established in previous work (20). Background of the diffraction patterns was subtracted by using data from an empty cell experiment, where the same experiment was performed with a brass cell filled with phosphate buffered solution and without the tendon sample. Given the thickness of the brass cell, the x-rays scattered by the buffer solution were not coherent with the scattering from the sample, hence the background was added incoherently and we could remove it without affecting the collagen sample data. Similar scans with much shorter exposure times (2 s) were performed on the gold lithographed sample.
Image reconstruction using ptychography algorithm ePIE
A modified version of the ePIE algorithm (23) was used for reconstructing the images from the corrected diffraction patterns. This iterative reconstruction method requires as input the measured diffraction data, and initial guesses for the probe and object functions. Although the algorithm can succeed with no knowledge of the object, a reasonably good guess of the probe function is needed; the more accurate this guess is, the quicker the algorithm will converge. In the case of weakly scattering biological objects for which the probe is harder to reconstruct, a reference object can be used to obtain the probe (24), or to validate the obtaining probe after the reconstruction (25). Here, diffraction data from the gold lithographed sample was used to obtain the complex probe function, with a model of the propagated wave from the 20 μm pinhole as the initial probe guess. Starting from a constant-valued object updated every iteration, the probe update was started after iteration 20, and the algorithm was considered to be well converged after 500 iterations. The reconstructed probe function was then used in the reconstruction of the tendon sample, without updating, and the object function was reconstructed starting from a constant value. Little benefit was found in iterating beyond 500 iterations. For the reconstruction of the collagen data, a 2D array containing the (001) collagen Bragg peak and 20 extra neighbor pixels was extracted from each diffraction pattern. This array was then embedded in a 1024 × 1024 array and centered by calculating the center of mass of the peak intensities. This bigger array was subsequently used in the ePIE algorithm for the Bragg projection ptychography reconstruction of the collagen data.
Results and Discussion
Fibril diameter estimated from the first order diffraction peaks
Diffraction patterns measured from a fresh and unexposed region of a whole rat tail tendon, suspended in buffered solution, are shown in Fig. 1. The high visibility of speckles is a reflection of the high degree of coherence of the x-ray beam and the stability of the setup. The diffraction is at the location of the first order layer line of the collagen periodicity, d = 67 nm, with a corresponding momentum transfer of nm−1. Along the Q-direction (up the page in Fig. 1), the peak is very sharp with the same width of a single speckle, which corresponds to the inverse size of the beam, μm−1. Transverse to the Q-direction there are many peaks, spread along an almost perfect straight line. As discussed in our previous work (20), these two features indicate that the fibrils within the tendon extend fully over the 20 μm size of the beam defined by the pinhole and that they are almost perfectly parallel.
Figure 1.
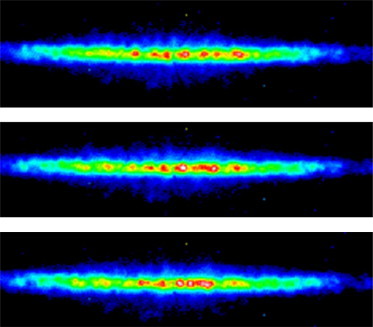
CXD patterns recorded from overlapping regions of a rat tail tendon immersed in buffered solution. They show an approximately one-dimensional speckle pattern in the direction transverse to the momentum transfer. Between frames, the sample is shifted relative to the beam by a fraction of its diameter, precisely 2 μm, resulting in similar, but not identical, diffraction patterns at each location. Intensity in arbitrary units.
The appearance of the diffraction in Fig. 1 contains information that allows us to construct a detailed schematic model of the sample ultrastructure. The pattern is essentially one-dimensional (1D), indicating that it arises from the lateral interference of a finite number of fibrils each with a different relative position, as illustrated by the cartoon in Fig. 2. Each fibril, assumed to be a perfectly periodic grating (with the D-banding as the basic repeat), traverses the beam but has a different offset of the internal grating structure along its length. According to the current high-resolution structural model of the collagen fibril (26), each polypeptide strand spans 4.46 repeats of the grating period, D. This forms a regular array of dense folded protein segments separated by less dense linker regions, believed to be more flexible. These triple-helical folds align the filaments side by side within each fibril, so that the entire fiber is modulated coherently with more and less dense protein-filled bands, called D-bands. These bands are also used to determine the location of mineralization nucleation events as collagen becomes used as a scaffold for bone (27,28). They can also be visualized by a number of techniques, for example transmission electron microscopy (27) and atomic force microscopy (29,30). To explain the formation of a 1D speckle pattern in Fig. 2, consider that each fibril in the assembly contributes a strong first order diffraction peak, with an intensity given by the structure factor of the modulated protein bands, which is sharp along the radial Q-direction and broadened by the finite size of the fibrils in the transverse direction. The phase of that structure factor is then given by the position of the fibril relative to its neighbors, as suggested by the colors in Fig. 2, and when a collection of fibrils diffract coherently within the illuminate area of the beam, the structure factors interfere to give a 1D speckle pattern, as observed.
Figure 2.
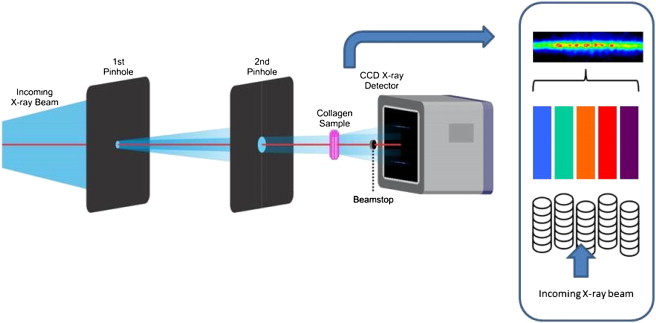
Schematic view of the experimental geometry, showing the two pinhole apertures, and the location of the sample, with its major symmetry axis perpendicular to the beam (axis (001) of the fibrils). The insight shows a schematic interpretation of the parallel alignment of the collagen fibrils within the tendon. Within each fibril all the collagen molecules are aligned into D-bands of modulated density, with 67 nm spacing, which diffract x-rays. The D-band position, which offsets between adjacent fibrils within the structure can be represented as phases, indicated by blocks of color. The relative phases of the structure factor of the diffraction signal from each fibril combine together to give a one-dimensionally modulated interference pattern, as seen in the data.
The dimensions of the region that diffracts coherently within the examined tendon therefore can be estimated from the raw diffraction patterns in Fig. 1. The transverse distribution of peaks, each with the width of one speckle, is enclosed by an envelope of width 6.5 μm−1. This envelope, before it becomes modulated by the speckles, is due to the Fourier transform of the smallest structure diffracting coherently in the tendon. This structure, that has a size (diameter) in real space of μm, is therefore one single fibril bundle.
Reconstruction of the probe function
Ptychographic data were obtained by scanning the sample in a 26 × 3 rectangular array of positions across the probe, moving in 2 μm steps. To minimize the radiation damage, it was determined empirically that this was the greatest number of positions that allowed the diffraction pattern to be satisfactorily reproduced at the end of the scan. Fig. 1 shows three frames of the scan with a clear evolution of the diffraction pattern between frames. To reconstruct an image of the scanned region of the sample, it was necessary to have a good starting estimate of the probe, meaning the amplitude and phase distribution of the wave field at the sample position. This estimate is updated in the ePIE procedure by constraining the Fourier transform (i.e., the far-field diffraction pattern) of the product of the probe and the unknown object (both complex functions) to be equal in magnitude to the square root of the measured diffraction intensity at every pixel. Because of the overlap redundancy, there is sufficient information to constrain the probe and object uniquely. However, it is known from experience that the convergence is only good when the probe estimate is close to the correct function (31). It is also known that regular grids are a poor choice of scan pattern because they can lead to nonunique factorizations, whereby the probe and a repeating copy of the object become intermixed (32).
To obtain a good estimate of the probe in our experiments, we also measured a test object placed in the same plane and using the same experimental configuration (24,25). The test object was designed to simulate a domain structure resembling collagen. Electron beam lithography was used to print small blocks of gold onto a silicon nitride membrane in linear arrays with 200 nm spacing, three times the D-band spacing of real collagen. These arrays were placed randomly on the membrane in a dilute pattern. Ptychography data were measured by scanning this test sample through the beam in the same way as the real sample and the first order peak of the linear arrays was extracted for analysis. The reconstruction was carried out using the same ePIE algorithm. The resulting image, shown in Fig. 3, provides a blurred image of each model array seen as an elliptical strip of amplitude surrounded by empty space; the individual blocks making up the array are not expected to be resolved because only the first order diffraction peak was used. But each strip has a well-defined phase that is related to its position relative to its neighbors, for the same reason as the collagen fibrils shown in Fig. 2. These phases were found to correlate accurately with the designed positions in the structure (31). The amplitude and phase of the probe, obtained at the same time, shown in Fig. 3 c, has the pattern expected from wave propagation through a circular pinhole in classical optics; the distortion from ideal can be attributed to imperfections in the laser-drilled structure of the actual pinhole used. This probe function was used as a fixed illumination function in the subsequent reconstruction of the collagen data.
Reconstructed image of the collagen distribution
Ptychographic reconstruction of the actual collagen data was then possible, but required a very large number of iterations (typically 500 to 1000, see Materials and Methods section) on a personal computer enhanced with graphical processor unit. The slow convergence is thought to be due to imperfections in the data, such as detector artifacts, background contributing to reduced visibility of the speckles, and probe-sample position errors. Because we were close to the radiation damage limit, which we established at the beginning of the experiment by monitoring the changes in the diffraction pattern of a continuously irradiated sample, it is likely there may have been some internal motion of the sample’s microstructure during the exposure that would appear as reduced visibility. Sample motion between exposures would contribute to probe position errors, also known to impair convergence of the ePIE algorithm (33,34). The resulting images were heavily striped by a strong phase ramp extending across the entire field of view of the sample, with an equal-and-opposite phase ramp across the probe. This behavior represents an inherent ambiguity of the ptychography method. The ramp ambiguity is easy to see in the ptychography formalism: at every probe position, the amplitude of the measured diffraction pattern is expected to agree with the Fourier transform of the product of the probe with the selected position on the sample. A phase ramp on this product is prohibited by the correct centering of the diffraction data, which would become shifted if a ramp were present. However, opposite phase ramps on the probe and the sample, which cancel out in their product, are allowed, as found in our image. We therefore ran a second set of reconstructions of the collagen data set where the probe was not allowed to be updated. This resulted in a better image without strong global phase ramps. The resulting image of the scanned region of the tail tendon is shown in Fig. 4.
Figure 4.
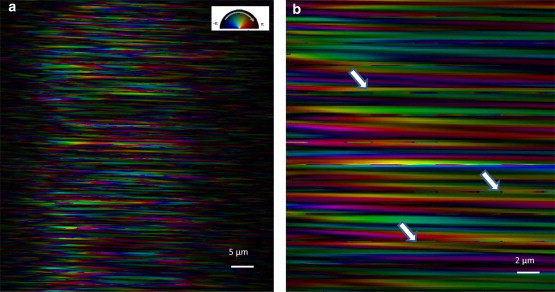
(a) Final image of a 25 × 10 μm region of the tendon. Only the collagen contributes to the image, obtained in dark field using the first order Bragg peak. The image is displayed in the complex color scale as indicated in the inset: brightness indicates amplitude, color indicates phase. Phase variations indicate relative shifts of the collagen D-band structure with respect to each other. A shift of one D-band spacing, or 67 nm, maps onto a full 2π rotation of the phase. (b) A zoom in the image. White arrows indicate signatures of fibril fusion events.
In Fig. 4, the strong fibrous texture can be identified immediately with the expected structure of the tendon, with the expected widths of ∼1 μm for each of the imaged blocks. It can clearly be seen that the phase is constant along some of the fibrils and starts to vary in other parts. This phase variation can be directly interpreted as local strain in the sense of local variations of the D-band spacings. Understanding strain is a major goal of Bragg CXD methods applied to materials science with a formal relationship between the observed phase and the local displacement, , at each position inside a crystal (35,36):
| (1) |
is the momentum transfer of the Bragg reflection used to measure the CXD pattern, which is the nm−1 here. This formula, which applies to all forms of Bragg CXD imaging, allows us to interpret the phase variations seen in Fig. 4 quantitatively in terms of local variations of the D-band spacing. A histogram of these local variations is shown in Fig. 5, where the phase gradients over 10 μm of the fibrils reconstructed in Fig. 4 have been converted using Eq. 1 to D-band spacing variations. The fibrils with the steepest gradients in Fig. 4 change phase by (in either direction) over ∼10 μm, although much smaller phase gradients are more typical (see Fig. 5). A phase gradient of radians over 10 μm means a D-spacing variation of 0.134 nm per lattice period (using Eq. 1), implying an observed maximum variation in D-spacing of 0.2%.
Figure 5.
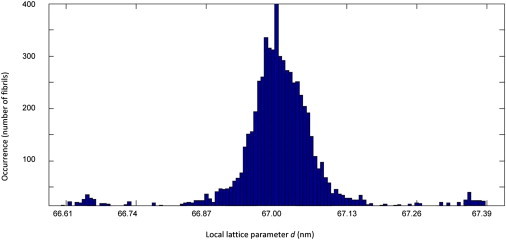
Histogram of local lattice parameters (local value of the D-band spacing), calculated by using the phase gradient of the fibrils in the reconstructed image (Fig. 4) over 10 μm and following Eq. 1. is the momentum transfer of the Bragg reflection used to measure the CXD pattern, which is the nm−1 here. The maximum occurrence of 3000 events around 67 nm was cropped for display purposes. To see this figure in color, go online.
Collagen fibrils packing and fibril fusions
So far we have been discussing the image as if it represents a 2D view of the collagen fibrils within the tendon, but this is not a complete account for the sample. The extract of the tendon that we examined had a circular cross section of ∼200 μm, containing a large number of smaller fibrils throughout its thickness. The x-rays penetrate the entire structure and contribute an equal signal to the CXD pattern from any depth. The interference between the top and bottom of the sample is limited by the longitudinal coherence length of the beam, which is only ∼0.5 μm for a Si(111) monochromator. However, the optical path length difference between the rays diffracting from the top and bottom of the tendon at the first order Bragg peak is only 1.1 nm, so our experiment is fully coherent in the longitudinal direction. We can safely assume that the packing of the fibrils in the depth direction is the same as in the direction transverse to the beam; therefore, the structure seen in the image is a projection along the beam of all the fibrils.
If we assume the contribution to the total diffraction signal from each fibril has the same structure factor amplitude with a different phase, determined by its shift along the fibril bundle axis, we can consider the resulting image to be a phased projection of all the fibrils. We can consider, for the sake of simplicity, the image at a given pixel to be the sum of N structure factors of uniform magnitude A and random phase, one for each of the N fibrils encountered through the depth. This is a classical random walk problem in the complex plane, for which the sum has a magnitude and a random phase. This random phase model can explain the image in Fig. 4 rather well: for N = 100, the image amplitude contrast would be around or ∼10% and its variation with position would be about the same as the width of one fibril, as seen in the direction transverse to the fibrils. Along the fibril direction, the maxima and minima of the interference would depend on the degree of alignment of the fibrils, but the apparent length would also be about the same as the persistence of an individual fibril. This argument suggests that the phased projection image resembles in a loose way the actual arrangement of the fibrils within the tendon, but that care must be taken with quantitative interpretation.
The arrangement of fibrils visible in Fig. 4 shows mostly continuous strands spanning the image, but not all of them completely traverse the field of view. There is a distribution of lengths apparent in the image, mostly in the range 10–20 μm in length. The gradual fading out at the ends is probably due to dephasing of overlapping strands in the random-phase summation, therefore represents a structural persistence length of the material, indicating the typical length over which the fibrils remain straight and preserve their interference through the depth.
Fig. 4 might also show some evidence that the fibril fusion process, believed to be associated with the tissue assembly pathway, acts on this length scale of a few tens of microns. Fibril fusion is postulated as the mechanism for tissue growth, where fibrils grow by the formation of subfibrils that laterally entwine with other subfibrils (37–39). This mechanism, observed by Cisneros et al. (38), is based on three conditions: a), close contact; b), parallel alignment; and c), registry of banding patterns. In their in vitro approach, Cisneros et al. presented how adjacent collagen fibrils could either fuse laterally or by their ends. In more than half of all the cases of fibril fusion observed, the fused area showed an almost perfect register in the banding periodicity. Furthermore, if a mismatch between the fusing fibrils would occur, it would present itself as a shift in the D-banding periodicity between fibrils, that is, in a local strain of the periodic lattice of fibrils, visible in the reconstructed diffraction data as a phase shift. The phase shifts are easily observed in Fig. 4 (white arrows), denoting end-to-end fusion of two consecutive fibrils.
Conclusion
In summary, we have applied what we believe to be the new phase-contrast imaging method of Bragg projection x-ray ptychography to obtain images of the distribution of fibrils within an intact tendon tissue, with minimal sample preparation. By using only the Bragg peak to obtain the image, we have removed all contributions of the nonperiodic components of the cells involved. We have interpreted the images in terms of the structural perfection of the collagen-containing portion of the material, finding a fibril persistence length of 10–20 μm and a fiber-to-fiber variation of D-spacing of 0.2%. The reconstructed images might also show evidence of the extension of the mechanism of fibril fusions up to the scale of some tens of nm. Our results show that Bragg CXD imaging methods, particularly ptychography, can be very useful to quantitatively characterize the packing order of highly hierarchical biological samples.
Acknowledgments
This work was partly funded by the Engineering and Physical Sciences Research Council (EPSRC) Basic Technology grant No. EP/E034055: Ultimate Imaging.
Footnotes
Felisa Berenguer's and Cameron M. Kewish’s present address is Synchrotron Soleil, Gif-sur-Yvette, France.
Joan Vila-Comamala’s and Fucai Zhang’s present address is Diamond Light Source, Research Complex at Harwell, Harwell Oxford Campus, Didcot, United Kingdom.
This is an Open Access article distributed under the terms of the Creative Commons-Attribution Noncommercial License (http://creativecommons.org/licenses/by-nc/2.0/), which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
References
- 1.De Caro L., Altamura D., Giannini C. Rat-tail tendon fiber SAXS high-order diffraction peaks recovered by a superbright laboratory source and a novel restoration algorithm. J. Appl. Cryst. 2013;46:672–678. [Google Scholar]
- 2.Silver F.H., Freeman J.W., Landis W.J. Molecular basis for elastic energy storage in mineralized tendon. Biomacromolecules. 2001;2:750–756. doi: 10.1021/bm0100615. [DOI] [PubMed] [Google Scholar]
- 3.Landis W.J., Librizzi J.J., Silver F.H. A study of the relationship between mineral content and mechanical properties of turkey gastrocnemius tendon. J. Bone Miner. Res. 1995;10:859–867. doi: 10.1002/jbmr.5650100606. [DOI] [PubMed] [Google Scholar]
- 4.Fratzl P., Gupta H.S., Roschger P. Structure and mechanical quality of the collagen-mineral nano-composite in bone. J. Mater. Chem. 2004;14:2115–2123. [Google Scholar]
- 5.Buehler M.J. Nature designs tough collagen: explaining the nanostructure of collagen fibrils. Proc. Natl. Acad. Sci. USA. 2006;103:12285–12290. doi: 10.1073/pnas.0603216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parry D.A.D., Barnes G.R.G., Craig A.S. A comparison of the size distribution of collagen fibrils in connective tissues as a function of age and a possible relation between fibril size distribution and mechanical properties. Proc. R. Soc. Lond. B Biol. Sci. 1978;203:305–321. doi: 10.1098/rspb.1978.0107. [DOI] [PubMed] [Google Scholar]
- 7.Hulmes D.J.S. Building collagen molecules, fibrils, and suprafibrillar structures. J. Struct. Biol. 2002;137:2–10. doi: 10.1006/jsbi.2002.4450. [DOI] [PubMed] [Google Scholar]
- 8.Parkinson J., Kadler K.E., Brass A. Self-assembly of rodlike particles in two dimensions: a simple model for collagen fibrillogenesis. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics. 1994;50:2963–2966. doi: 10.1103/physreve.50.2963. [DOI] [PubMed] [Google Scholar]
- 9.Knott L., Bailey A.J. Collagen cross-links in mineralizing tissues: a review of their chemistry, function, and clinical relevance. Bone. 1998;22:181–187. doi: 10.1016/s8756-3282(97)00279-2. [DOI] [PubMed] [Google Scholar]
- 10.Bailey A.J. Molecular mechanisms of ageing in connective tissues. Mech. Ageing Dev. 2001;122:735–755. doi: 10.1016/s0047-6374(01)00225-1. [DOI] [PubMed] [Google Scholar]
- 11.Starborg T., Kalson N.S., Kadler K.E. Using transmission electron microscopy and 3View to determine collagen fibril size and three-dimensional organization. Nat. Protoc. 2013;8:1433–1448. doi: 10.1038/nprot.2013.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakdinawat A., Attwood D. Nanoscale x-ray imaging. Nat. Photonics. 2010;4:840–848. [Google Scholar]
- 13.Howells M.R., Beetz T., Starodub D. An assessment of the resolution limitation due to radiation-damage in x-ray diffraction microscopy. J. Electron Spectrosc. Relat. Phenom. 2009;170:4–12. doi: 10.1016/j.elspec.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodenburg J.M., Hurst A.C., Johnson I. Hard-x-ray lensless imaging of extended objects. Phys. Rev. Lett. 2007;98:034801–034804. doi: 10.1103/PhysRevLett.98.034801. [DOI] [PubMed] [Google Scholar]
- 15.Thibault P., Dierolf M., Pfeiffer F. High-resolution scanning x-ray diffraction microscopy. Science. 2008;321:379–382. doi: 10.1126/science.1158573. [DOI] [PubMed] [Google Scholar]
- 16.Godard P., Carbone G., Chamard V. Three-dimensional high-resolution quantitative microscopy of extended crystals. Nat. Commun. 2011;2:568–573. doi: 10.1038/ncomms1569. [DOI] [PubMed] [Google Scholar]
- 17.Hruszkewycz S.O., Highland M.J., Fuoss P.H. Imaging local polarization in ferroelectric thin films by coherent x-ray Bragg projection ptychography. Phys. Rev. Lett. 2013;110:177601–177605. doi: 10.1103/PhysRevLett.110.177601. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi Y., Suzuki A., Ishikawa T. Bragg x-ray ptychography of a silicon crystal: visualization of the dislocation strain field and the production of a vortex beam. Phys. Rev. B. 2013;87:121201–121204. [Google Scholar]
- 19.Berenguer de la Cuesta F., Wenger M.P.E., Robinson I.K. Coherent X-ray diffraction from collagenous soft tissues. Proc. Natl. Acad. Sci. USA. 2009;106:15297–15301. doi: 10.1073/pnas.0905151106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berenguer de la Cuesta F., Bean R.J., Robinson I.K. Collagen imaged by coherent x-ray diffraction: towards a complementary tool to conventional scanning SAXS. J. Phys. Conf. Ser. 2010;247:012004. [Google Scholar]
- 21.Doniach S., Hodgson K., Winick H. Early work with synchrotron radiation at stanford. J. Synchrotron Radiat. 1997;4:380–395. doi: 10.1107/S0909049597012235. [DOI] [PubMed] [Google Scholar]
- 22.Gorelick S., Guzenko V.A., David C. Direct e-beam writing of dense and high aspect ratio nanostructures in thick layers of PMMA for electroplating. Nanotechnology. 2010;21:295303–295310. doi: 10.1088/0957-4484/21/29/295303. [DOI] [PubMed] [Google Scholar]
- 23.Maiden A.M., Rodenburg J.M. An improved ptychographical phase retrieval algorithm for diffractive imaging. Ultramicroscopy. 2009;109:1256–1262. doi: 10.1016/j.ultramic.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 24.Dierolf M., Thibault P., Pfeiffer F. Ptychographic coherent diffractive imaging of weakly scattering specimens. New J. Phys. 2010;12:035017. [Google Scholar]
- 25.Giewekemeyer K., Thibault P., Salditt T. Quantitative biological imaging by ptychographic x-ray diffraction microscopy. Proc. Natl. Acad. Sci. USA. 2010;107:529–534. doi: 10.1073/pnas.0905846107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orgel J.P., Irving T.C., Wess T.J. Microfibrillar structure of type I collagen in situ. Proc. Natl. Acad. Sci. USA. 2006;103:9001–9005. doi: 10.1073/pnas.0502718103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jantou-Morris V.M.A.H., Horton M.A., McComb D.W. The nano-morphological relationships between apatite crystals and collagen fibrils in ivory dentine. Biomaterials. 2010;31:5275–5286. doi: 10.1016/j.biomaterials.2010.03.025. [DOI] [PubMed] [Google Scholar]
- 28.Nudelman F., Pieterse K., Sommerdijk N.A.J.M. The role of collagen in bone apatite formation in the presence of hydroxyapatite nucleation inhibitors. Nat. Mater. 2010;9:1004–1009. doi: 10.1038/nmat2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bozec L., Odlyha M. Thermal denaturation studies of collagen by microthermal analysis and atomic force microscopy. Biophys. J. 2011;101:228–236. doi: 10.1016/j.bpj.2011.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bozec L., van der Heijden G., Horton M. Collagen fibrils: nanoscale ropes. Biophys. J. 2007;92:70–75. doi: 10.1529/biophysj.106.085704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bean, R. J. 2012. Imaging of domain structures with coherent x-ray diffraction. PhD thesis. University College London.
- 32.Thibault P., Dierolf M., Pfeiffer F. Probe retrieval in ptychographic coherent diffractive imaging. Ultramicroscopy. 2009;109:338–343. doi: 10.1016/j.ultramic.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 33.Maiden A.M., Humphry M.J., Rodenburg J.M. An annealing algorithm to correct positioning errors in ptychography. Ultramicroscopy. 2012;120:64–72. doi: 10.1016/j.ultramic.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 34.Zhang F., Peterson I., Rodenburg J.M. Translation position determination in ptychographic coherent diffraction imaging. Opt. Express. 2013;21:13592–13606. doi: 10.1364/OE.21.013592. [DOI] [PubMed] [Google Scholar]
- 35.Pfeifer M.A., Williams G.J., Robinson I.K. Three-dimensional mapping of a deformation field inside a nanocrystal. Nature. 2006;442:63–66. doi: 10.1038/nature04867. [DOI] [PubMed] [Google Scholar]
- 36.Robinson I.K., Harder R. Coherent X-ray diffraction imaging of strain at the nanoscale. Nat. Mater. 2009;8:291–298. doi: 10.1038/nmat2400. [DOI] [PubMed] [Google Scholar]
- 37.Cheema U., Chuo C.-B., Brown R.A. Engineering functional collagen scaffolds: cyclical loading increases material strength and fibril aggregation. Adv. Funct. Mater. 2007;17:2426–2431. [Google Scholar]
- 38.Cisneros D.A., Hung C., Muller D.J. Observing growth steps of collagen self-assembly by time-lapse high-resolution atomic force microscopy. J. Struct. Biol. 2006;154:232–245. doi: 10.1016/j.jsb.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 39.Christiansen D.L., Huang E.K., Silver F.H. Assembly of type I collagen: fusion of fibril subunits and the influence of fibril diameter on mechanical properties. Matrix Biol. 2000;19:409–420. doi: 10.1016/s0945-053x(00)00089-5. [DOI] [PubMed] [Google Scholar]


