Abstract
RNA interference (RNAi) is a powerful new tool for the selective ablation of gene expression, facilitating loss-of-function studies. However, appropriate controls are considered essential to confirm the specificity of RNAi experiments. The most stringent control is rescue of the target gene in a form that is refractory to RNAi. To facilitate rescue of the target gene, we have created improved dual expression lentiviral vectors with the ability to simultaneously drive expression of a shRNA for RNA interference and a rescue transgene in a single vector system. In proof-of-principle experiments, we ablated > 90% of target gene expression by targeting either the open reading frame, or the 3′ UTR region. Target gene expression was successfully rescued with a cDNA containing silent third-codon point mutations in the targeted region or with native cDNA when the 3′ UTR was targeted. Finally, expression of the rescue transgene can be manipulated by positional cloning and appropriate promoter selection. The dual expression lentiviral vectors described here represent a versatile strategy for confirming the integrity of RNAi experiments and may facilitate functional analyses even in the absence of an established gain-of-function model system.
Keywords: RNA interference, rescue, lentiviral vector
INTRODUCTION
The recent discovery that double-stranded RNA molecules can regulate the expression of genes has had a profound impact on experimental biology and related fields [1]. Loss-of-function studies represent a key experimental approach for understanding the biology and function of specific proteins and/or genes. RNA interference (RNAi) has proven to be an efficient and cost-effective methodology for loss-of-function studies and is now widely considered to be the method of choice for the selective ablation of gene expression [2]. Typically synthetic short interfering RNA (siRNA) [3], or short hairpin RNA (shRNA) [4, 5] are used to specifically target the destruction of the corresponding sequence-specific mRNA. However, RNAi is not completely understood in mammalian cells and double-stranded RNA molecules appear to mediate their effects through surprisingly diverse mechanisms. Recent reports suggest that double-stranded RNA molecules can induce interferon response genes [6, 7], or even silence non-target genes [8, 9]. Consequently, appropriate controls are considered to be absolutely essential to confirm the specificity of this experimental technique [10].
The most definitive and unambiguous control for RNAi experiments is rescue of the targeted gene in a form that is refractory to RNAi [10]. This is commonly accomplished by using a cDNA that has one or more silent mutations in the region targeted by the double-stranded RNA [11, 12]. Alternatively, the 3′-untranslated region of the gene of interest can be targeted, as the rescue cDNA typically does not require the native untranslated region for expression. Although rescue of the targeted gene represents an unequivocal specificity control, technical challenges limit the applicability of this approach. Commonly, RNAi and rescue of the targeted gene are performed in two steps, requiring two transfection and/or transduction events. Slight differences in transfection and/or transduction efficiency can lead to inconsistent RNAi and rescue of the targeted gene resulting in inconclusive findings. To facilitate consistent RNAi and rescue of the target gene, we have created improved dual expression lentiviral vectors with the ability to simultaneously drive expression of a shRNA for RNA interference and a rescue transgene in a single vector system. We demonstrate here that this vector system can be used to efficiently ablate gene expression, with concurrent rescue of the targeted gene. Further, expression of the rescue transgene can be manipulated by positional cloning and appropriate promoter selection. Significant concerns persist about the specificity of RNAi, with the perception that many RNAi experiments lack the controls required for unambiguous interpretation of results. We believe that this versatile system of improved dual expression lentiviral vectors will encourage the routine use of target gene rescue in RNAi experiments, facilitating the interpretation of these experiments.
MATERIALS AND METHODS
Dual expression lentiviral vectors
pSicoR and related lentiviral vectors allow for conditional or stable expression of shRNA for RNA interference in cell lines and transgenic mice [13]. The vectors were generously provided by Tyler Jacks, Ph.D. at Massachusetts Institute of Technology (Cambridge, MA), but are now also available at Addgene (Cambridge, MA). To facilitate cloning of a rescue transgene into the lentiviral vectors, a multiple cloning site was generated by cloning a synthetic oligonucleotide with additional restriction sites (BamHI-XmaI-SwaI-NheI-EcoRI-NotI-BamHI) into the BamHI site following the shRNA insertion site.
RNA interference
Candidate shRNA sequences specific for EpCAM were identified using online RNAi design programs and cloned into the pSicoR vector as previously described [13]. Control and specific gene sequences are available upon request.
EpCAM cDNA cloning and mutagenesis
The EpCAM cDNA was initially amplified by PCR and subsequently cloned into the pcDNA3.1 vector. Silent point mutations were introduced into the EpCAM cDNA in the shRNA target region to render it resistant to shRNA. The silent point mutations were generated using the QuikChange site-directed mutagenesis kit with Pfu-ultra as the polymerase (Stratagene; La Jolla, CA). The primers used for mutagenesis are available upon request. The EpCAM truncation deletion mutants EpCAM-26 (AA 1-26), EpCAM-265 (AA 1-265), and EpCAM-288 (AA 1-288) were generated with standard molecular biology techniques.
Expression of rescue transgenes
The native EpCAM cDNA, EpCAM cDNA resistant to shRNA, or EpCAM cDNA truncation deletion mutants were cloned into the pSicoR parental vectors at the AgeI-NheI site, or the newly generated multiple cloning site (XmaI-NheI). All shRNA and cDNA cloning events were confirmed by restriction digest and sequence analysis.
Lentivirus generation and transduction
Lentivirus was generated as described [13]. Briefly, HEK-293T cells were transfected with 1.6 μg of pSicoR experimental or control vector, 1.2 μg of Δ8.9, and 0.2 μg VSVG in 30 μL Opti-MEM media. After overnight culture, 2 mL of serum containing media was added and the incubation was allowed to continue for an additional 48hrs. Viral supernatants were filtered through a 0.45 μm Millipore filter, and used directly to infect cells with Protamine sulfate (Sigma, Saint Louis, MO) at a concentration of 6 μg/mL.
Invasion assays
Invasion through reconstituted synthetic basement membrane (Matrigel) was performed as recommended by the manufacturer (BD Biosciences, San Jose, CA). Cells that traversed the filter were counted by light and/or fluorescent microscopy by a technician blinded to the experimental conditions.
Real Time PCR
Total RNA was extracted using TRIZOL (Invitrogen, Carlsbad, CA). For cDNA synthesis, we utilized the MMLV reverse transcriptase kit (Invitrogen) and generated cDNA from 1000 ng of total RNA. Amplification of EpCAM truncation deletion mutants EpCAM-26, EpCAM-266, and EpCAM-314 were carried out with specific primers. Relative expression values were normalized to GAPDH and 18S rRNA using ΔΔCT to calculate relative expression values.
Immunoblotting
Cells were washed twice in PBS and lysed with M-PER lysis buffer (Pierce, Rockford, IL) supplemented with protease inhibitor cocktail (Boehringer, Indianapolis, IN) at 4°C for 30 min. Lysates were centrifuged at 10,000 RPM for 15 min at 4°C and the supernatants were collected. Approximately 20 μg of protein was loaded with loading buffer, and electrophoresed on a 12% NuPAGE gel (Invitrogen). The protein was subsequently transferred onto PVDF membrane and probed with EpCAM and β-actin primary antibodies (Santa Cruz Biotechnology, Santa Cruz, CA). The signal was detected using the appropriate horseradish peroxidase-conjugated secondary antibody and the ECL immunoblotting detection system (GE Healthcare, Piscataway, NJ).
Flow Cytometry
Cells were preincubated with PE-conjugated EpCAM antibody (BD Biosciences, Franklin Lakes, NJ) for 20 minutes at room temperature in FACS buffer (0.1% BSA in PBS), washed three times with PBS and resuspended into FACS buffer. Analyses were performed using a FACS Calibur flow cytometer (BD Biosciences). Finally, data were analyzed using CELLQuest software (BD Biosciences).
RESULTS
Our overall strategy was to construct an improved dual expression vector with the ability to simultaneously drive expression of a shRNA for RNA interference and a rescue transgene in a single vector system. The vector is based on the pSicoR vector system [13]. To facilitate cloning of the rescue transgene we introduced a multiple cloning site (XmaI-SwaI-NheI-EcoRI-NotI) into the vector at the BamHI site. Representative dual expression lentiviral vectors used in this study are illustrated schematically in Figure 1. In an initial step, HEK-293 cells were transduced with a 65-kD EpCAM-YFP fusion protein. Following antibiotic selection, we obtained a relatively pure population of fluorescent HEK-293 cells as measured by flow cytometry (data not shown) and fluorescent microscopy (Figure 2A). These cells were then transduced with a dual expression vector encoding shRNA targeting the EpCAM ORF and an EpCAM cDNA resistant to shRNA. Fluorescent microscopy and immunoblot analysis clearly demonstrate that the dual expression vector is able to specifically ablate expression of the 65-kD YFP-EpCAM fusion protein with concomitant rescue expression of the 40-kD native EpCAM protein (Figure 2).
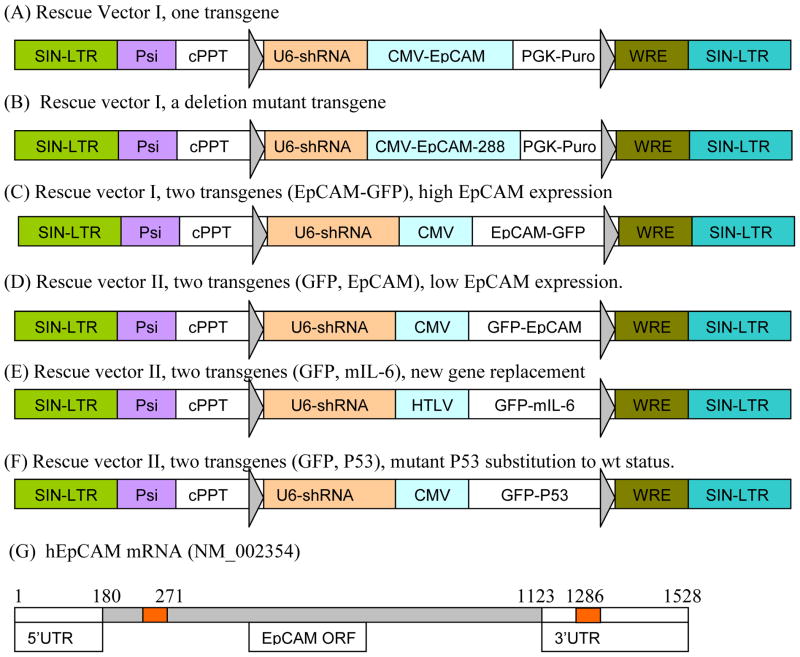
Figure 1. Strategies for generation of dual expressing lentiviral vectors for concurrent RNA interference and rescue using rescue vector I (one transgene) and rescue vector II (two transgenes).
(A) Dual expression lentiviral vector expressing shRNA targeting EpCAM ORF (271) and expressing EpCAM cDNA resistant to shRNA. (B) shRNA targeting EpCAM at 3′UTR rescued with truncated EpCAM cDNA. (C, D) shRNA targeting EpCAM ORF (271) with GFP-EpCAM expression; and GFP-EpCAM expression (E) Gene replacement with shRNA targeting mEpCAM and expressing GFP-mIL-6 cDNA. (F) Mutant P53 substitution; by shRNA to knockdown expressing shRNA resistant GFP-wt-P53. (G) Schematic representations of hEpCAM mRNA structure and shRNA target regions, shRNA were designed to target sequences in the ORF (271) or 3′UTR (1286) of EpCAM. Lentiviral vectors used in A–B are pSicoR-puro and C–F is pLL3.7.
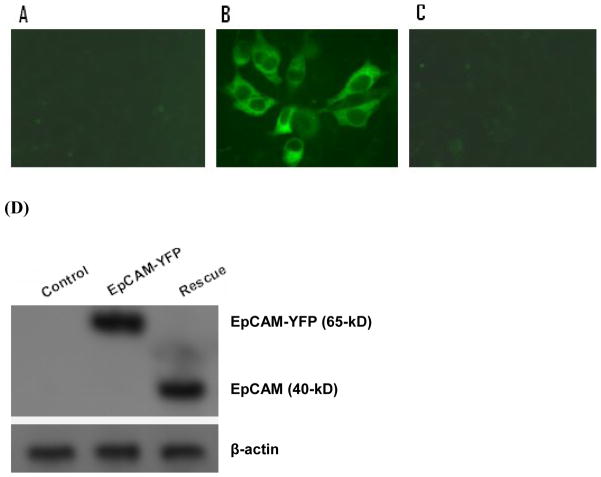
Figure 2. Validation of dual expression lentiviral vector RNA interference and rescue in HEK-293 cells.
EpCAM null, HEK-293 (A), HEK-293 cells were stably transduced with an EpCAM-YFP fusion protein (65-kD, B). Specific ablation of EpCAM-YFP fusion protein with the rescue vector 1 (C). Fluorescence image was assessed under 40 x magnifications with a Olympus-BX51 fluorescence microscope. Immunoblot confirming specific ablation of EpCAM–YFP (65-kD) and expression of native EpCAM cDNA (40-kD) resistant to shRNA (D). Lane 1: HEK-293 cells; Lane 2: HEK-293 cells stably transduced with EpCAM-YFP; Lane 3: HEK-293 cells stably transduced with EpCAM-YFP, and rescued with dual expression vector.
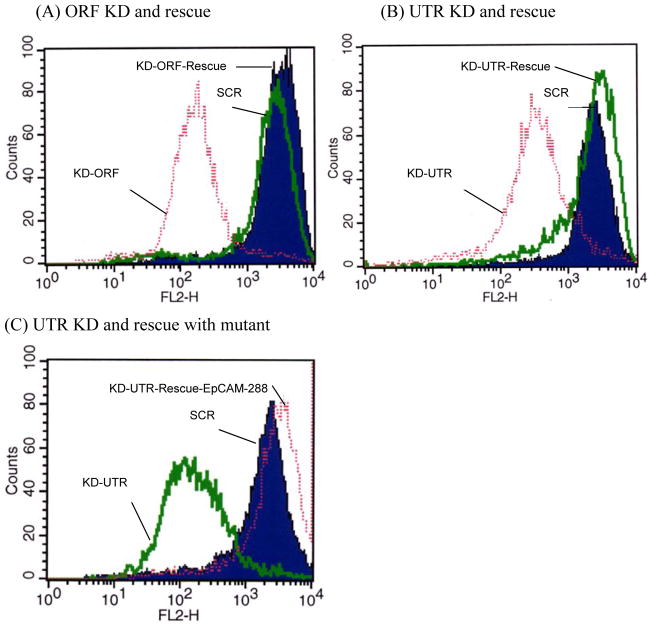
Next, we determined the ability of dual expression vectors to specifically ablate and rescue EpCAM expression in breast and gastric cancer cell lines known to overexpress endogenous EpCAM. Three different experimental vectors and corresponding appropriate controls were designed and assessed. The first vector system targets EpCAM in the ORF at base 271. The rescue cDNA contains a silent mutation and is refractory to RNA interference. Expression of EpCAM is reduced > 90% by the vector targeting EpCAM without a rescue transgene, and rescued to native levels with the dual expression vector (Figure 3A). In the second vector system, the shRNA targets EpCAM in the 3′ UTR at base 1286, and is rescued by wildtype EpCAM cDNA. Again, flow cytometry demonstrates the efficacy of this strategy with > 90% reduction in EpCAM expression levels with the vector expressing the shRNA alone, and rescue to native levels with the dual expression construct (Figure 3B). The third vector system targets EpCAM in the 3′ UTR at base 1286, but is rescued by a truncation deletion mutant, EpCAM-288 (Figure 3C). This truncation mutant lacks the 26 amino acid cytoplasmic tail domain of EpCAM and may prove valuable in defining the functional biology of EpCAM in breast cancer, specifically the role of the cytoplasmic tail in signaling. In all three vector systems, the expression of the rescue transgene was directly comparable to wildtype EpCAM expression. This is not always the case, and we have established techniques that provide the ability to control the expression of the rescue transgene as detailed below.
Figure 3. RNAi rescue for overexpressing cells.
Quantitative analysis of EpCAM expression following transduction with dual expression lentiviral vectors for RNA interference and rescue. Flow cytometry analysis of AGS gastric cancer cells following transduction with dual expression lentiviral vectors. In panels A and B EpCAM is targeted in the ORF or in the 3′ UTR and rescued with EpCAM cDNA resistant to the shRNA. (A) EpCAM KD at open reading frame (ORF) and express RNAi resistant EpCAM (B) EpCAM KD at UTR and express wild type in frame EpCAM coding cDNA (C) EpCAM is targeted in the 3′ UTR and rescued with a truncation mutant of 288 amino acids.
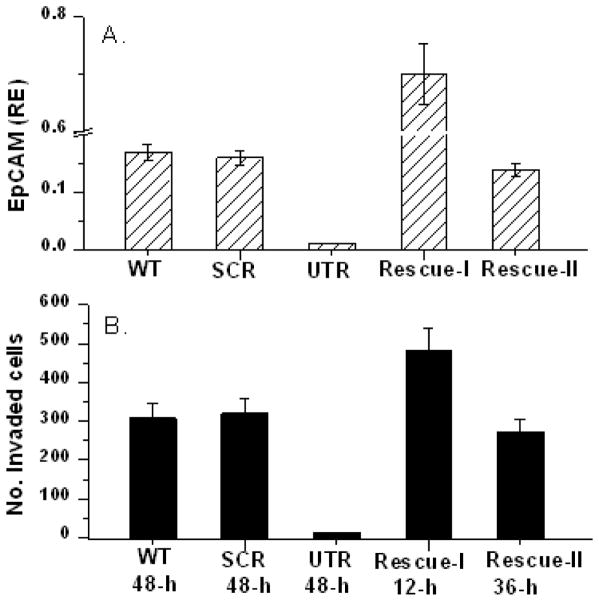
To determine if the dual expression lentiviral vector system can be used to rescue the functional phenotype associated with specific ablation of EpCAM expression, we performed Boyden chamber invasion assays. As shown in Figure 4, specific ablation of EpCAM gene expression with shRNA targeting the 3′ UTR results in a dramatic decrease in the ability of MDA-231 cells to invade through the basement membrane. Rescue of EpCAM expression with two different dual expression lentiviral vectors restored the ability of MDA-231 cells to invade through the basement membrane. Of particular note, EpCAM expression is significantly higher in U6-EpCAM(1286)/CMV-wtEpCAM cells than in wildtype cells (130-fold higher at the mRNA level and 20-fold higher at the protein level, data not shown). Overexpression of EpCAM in these cells is associated in a significant increase in the invasiveness of MDA-231 cells (cell invasion is observed in less than 24 hours, compared to 96 hours for wildtype or control cells). EpCAM expression with the second vector, U6-EpCAM (1286)/CMV-GFP-wtEpCAM, is comparable to wildtype MDA-231 cells (data not shown), and invasion also appears to be comparable. These studies confirm that rescue of EpCAM expression with dual expression lentiviral vectors restores the functional phenotype of the parental cells, but also underscores the importance of the expression level of the rescue transgene. We have used “positional effect” cloning to regulate the expression of the rescue transgene as detailed below.
Figure 4. RNAi rescue for low protein expressing cells. Rescuing gene expression at physiological amount restores cells phenotype.
MDA-MB-231 cells were transduced with the dual expression lentiviral vectors as indicated: WT, non-transduced SCR, UTR-KD, Rescue-I (UTR-KD, CMV-EpCAM), Rescue-II (UTR-KD, CMV-GFP-EpCAM). (A). q-RT-PCR for EpCAM expression, (B) Number of cells invaded with indicated lentiviral vector for KD or rescue. The time point indicated at which maximum number of invaded cells are recorded to assess the invasion time point. Rescue vector I overexpress EPCAM showing early invasion time point than in wildtype or Rescue-II cells.
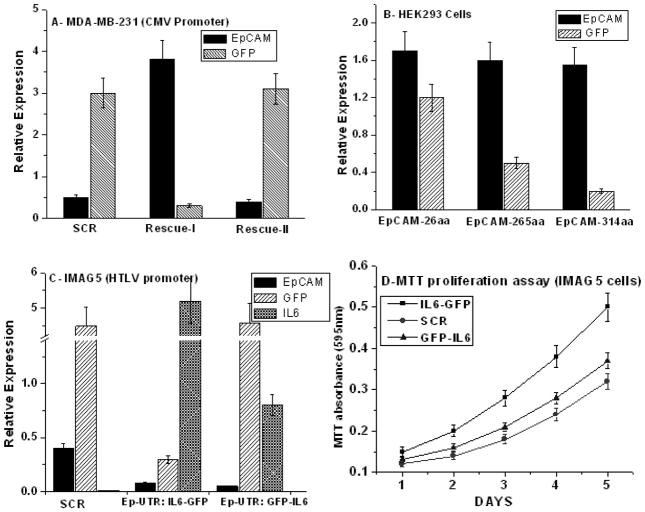
There is a significant difference in EpCAM expression between the two vectors U6-EpCAM(1286)/CMV-wtEpCAM and U6-EpCAM(1286)/CMV-GFP-wtEpCAM. In the second construct both GFP and EpCAM are under the control of the CMV promoter, with EpCAM positioned after the GFP gene. We believe that the expression of EpCAM is decreased relative to the first construct because of “positional effect” [14]. To formally test this hypothesis we performed a series of experiments where the order of rescue transgenes, the size of rescue transgenes, and the specific promoters were varied and the impact on GFP, EpCAM and other rescue transgenes was investigated. In the first series of experiments the order of the rescue transgenes on EpCAM and GFP expression was determined. Figure 5A demonstrates clearly that cloning of the rescue transgene immediately behind the CMV promoter is associated with higher relative expression. For instance EpCAM expression is approximately 10-fold higher if it is cloned immediately behind the CMV promoter. In a second series of experiments this “positional effect” was explored in more detail by cloning the GFP gene behind the full-length or truncated versions of the EpCAM gene. Figure 5B clearly shows that as the GFP construct is displaced farther from the CMV promoter by the first rescue transgene, expression levels are decreased. Specifically, expression levels of GFP following EpCAM-265 and EpCAM-314 are decreased 55% and 78%, respectively, compared to the relatively short EpCAM-26 mutant which encodes only the cytoplasmic tail of EpCAM. EpCAM expression levels remain relatively consistent. To verify this observation in a separate model, we tested GFP and mIL-6 under the control of the HTLV promoter. Consistent with previous data, Figure 5C demonstrates that the order of the rescue transgenes behind the HTLV promoter determines relative expression levels. Further, there is evidence to suggest that the HTLV promoter is stronger than the CMV promoter as both rescue transgene expression levels were 30–50% higher than with CMV promoter (data not shown). Taken together, the expression data suggest that these dual expression lentiviral vectors can be manipulated by positional cloning and/or promoter selection to achieve desired levels of rescue transgene expression.
Figure 5. Expression levels of the second “rescue” transgene are dependent on length from promoter (“positional effect”) in the dual expression lentiviral vector.
(A) MDA-MB-231 cells were transduced with the dual expression lentiviral vectors encoding EpCAM and GFP under the CMV promoter (vectors C and D figure 1): SCR, Rescue-I (CMV-EpCAM-GFP) and Rescue-II (CMV-GFP-EpCAM). (B) EpCAM negative cells HEK-293 cells were transduced with the indicated dual expression lentiviral vectors encoding truncated forms of EpCAM with GFP under the CMV promoter: EpCAM-26, EpCAM-265, EpCAM-314 (C) Mouse cell line IMGE5 cells were transduced with the indicated dual expression lentiviral vectors: SCR, HTLVCMV-IL6-GFP, and HTLV-GFP-IL6. Overexpression of IL-6 by traditional rescue decrease proliferation.
DISCUSSION
The most unequivocal specificity control in any RNAi experiment is rescue of the targeted gene at physiological levels in a form refractory to RNAi. Several strategies have been proposed to rescue gene expression in RNAi experiments. The most commonly proposed strategy is reintroduction of the gene in a form refractory to RNAi using either a cDNA construct [11, 12, 15], or a bacterial artificial chromosome (BAC) [16]. To date cDNA and BAC rescue have been performed using separate rescue constructs and we believe that this represents a major limitation of this strategy. Rescue using a separate construct requires that two independent events must occur: (1) efficient transfection/transduction or selection of the siRNA, or shRNA construct and (2) efficient transfection/transduction or selection of the rescue construct. As these are two independent events, results can be easily confounded by differences in transfection/transduction or selection efficiency. The dual expression lentiviral vector system described here is able to circumvent the limitations of separate RNAi and rescue constructs, with the ability to simultaneously drive expression of a shRNA for RNAi and a rescue transgene in a single vector system.
In RNAi experiments, target gene rescue should be within the physiologic range. Overexpression or underexpression of the target gene may not be associated with rescue of the observed phenotype. Another advantage of the dual expression lentiviral vector system described here is the ability to regulate target gene expression through the use of positional effect and/or promoter selection. It has been well documented in other model systems that genes driven off the same promoter show differences in expression based upon their position relative to a given promoter [14]. We demonstrate here that this phenomenon can be exploited to facilitate manipulation of rescue transgene expression by positional cloning. Rescue transgene expression clearly decreases as the distance from the promoter increases (Figure 5). Promoter selection provides a second tool to regulate rescue transgene expression. Our results demonstrate that the HTLV promoter is associated with greater rescue transgene expression than the CMV promoter. Therefore, both positional effect and promoter selection are versatile tools for manipulation of rescue transgene expression.
Although this vector system was originally designed to facilitate the interpretation of loss-of-function studies, it also has potential applications in gain-of-function studies. In some model systems, selective ablation of gene expression results in a dramatic phenotype, but no gain-of-function models are available. For example, in loss-of-function studies, we have demonstrated that specific ablation of EpCAM expression results in decreased breast cancer invasion in vitro [17]. However, overexpression of EpCAM in breast epithelial cells does not confer an invasive phenotype. Invasion is a complex biologic process that is not readily recapitulated by the addition of a single gene. The dual expression lentiviral vector system described here has provided, for the first time, a gain-of-function model to study EpCAM in breast cancer biology. Selective ablation of EpCAM expression and concurrent rescue restores the invasive potential of MDA-231 breast cancer cells in vitro. This gain-of-function model provides the opportunity to study the structural elements of EpCAM that confer this phenotype through structure/function analyses with truncation and/or site-directed mutants.
Loss-of-function studies have been instrumental in unraveling the biology and function of genes. Observing the phenotype of cells following selective ablation of gene expression often provides unique and significant insights into gene function. In this report we describe a versatile dual expression lentiviral vector system that is capable of efficient RNAi and regulation of targeted gene rescue. This strategy is applicable to most cell types and has the potential to greatly facilitate the routine use of target gene rescue in RNAi experiments.
Acknowledgments
We thank Tyler Jacks, Ph.D. and Andrea Ventura, M.D., Ph.D. for kindly providing the pSico and pSicoR parental vectors.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 2.McManus MT, Sharp PA. Gene silencing in mammals by small interfering RNAs. Nat Rev Genet. 2002;3:737–747. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- 3.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 4.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 5.Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL, Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet. 2003;34:263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- 7.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 8.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 9.Scacheri PC, Rozenblatt-Rosen O, Caplen NJ, Wolfsberg TG, Umayam L, Lee JC, Hughes CM, Shanmugam KS, Bhattacharjee A, Meyerson M, Collins FS. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:1892–1897. doi: 10.1073/pnas.0308698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Editors Whither RNAi? Nat Cell Biol. 2003;5:489–490. doi: 10.1038/ncb0603-490. [DOI] [PubMed] [Google Scholar]
- 11.Jiang Y, Price DH. Rescue of the TTF2 knockdown phenotype with an siRNA-resistant replacement vector. Cell Cycle. 2004;3:1151–1153. [PubMed] [Google Scholar]
- 12.Lassus P, Rodriguez J, Lazebnik Y. Confirming specificity of RNAi in mammalian cells. Sci STKE 2002. 2002:PL13. doi: 10.1126/stke.2002.147.pl13. [DOI] [PubMed] [Google Scholar]
- 13.Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, Van Parijs L, Jaenisch R, Jacks T. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci U S A. 2004;101:10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mizuguchi H, Xu Z, Ishii-Watabe A, Uchida E, Hayakawa T. IRES-dependent second gene expression is significantly lower than cap-dependent first gene expression in a bicistronic vector. Mol Ther. 2000;1:376–382. doi: 10.1006/mthe.2000.0050. [DOI] [PubMed] [Google Scholar]
- 15.Kojima S, Vignjevic D, Borisy GG. Improved silencing vector co-expressing GFP and small hairpin RNA. Biotechniques. 2004;36:74–79. doi: 10.2144/04361ST02. [DOI] [PubMed] [Google Scholar]
- 16.Kittler R, Pelletier L, Ma C, Poser I, Fischer S, Hyman AA, Buchholz F. RNA interference rescue by bacterial artificial chromosome transgenesis in mammalian tissue culture cells. Proc Natl Acad Sci U S A. 2005;102:2396–2401. doi: 10.1073/pnas.0409861102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osta WA, Chen Y, Mikhitarian K, Mitas M, Salem M, Hannun YA, Cole DJ, Gillanders WE. EpCAM is overexpressed in breast cancer and is a potential target for breast cancer gene therapy. Cancer Res. 2004;64:5818–5824. doi: 10.1158/0008-5472.CAN-04-0754. [DOI] [PubMed] [Google Scholar]