Summary
Using high throughput chemical and genetic screening, STF-118804, an inhibitor of nicotinamide phosphoribosyltransferase, was identified as a cell type-specific inhibitor of mixed-lineage leukemia with MLL chromosomal rearrangements (Matheny et al., 2013). The approach was powerful as is the potential for NAD as a specific cancer target.
Philadelphia chromosome-positive chronic myelogenous leukemia remains the paragon of genotype-targeted cancer pharmacology. The identification of ABL kinase activation by BCR-ABL fusion and the availability of STI-571 as an ABL-targeted inhibitor has allowed hundreds of thousands of US patients to be treated with a specific agent (Druker, 2008). Childhood acute lymphoblastic leukemias (ALL) that are refractory to treatment frequently contain chromosome translocations of the MLL gene (de Boer et al., 2013; Muntean and Hess, 2012). The MLL gene encodes a histone H3 lysine 4 methyltransferase that is essential for HOX gene expression during development. Although MLL translocations result in loss of the endogenous methyltransferase domain, oncogenic MLL fusion proteins are associated with a set of transcriptional elongation promoting proteins and with the histone H3 Lys79 methyltransferase DOT1L, which further enhance target gene expression(de Boer et al., 2013; Muntean and Hess, 2012). In addition, the N-terminal menin-binding motif of MLL and MLL fusions is essential for interactions with menin, the lens epithelium-derived growth factor and chromatin, and is required for leukemogenesis (Muntean and Hess, 2012). Thus, multiple MLL-associated factors are potential drug targets for MLL-rearranged ALL (de Boer et al., 2013; Muntean and Hess, 2012).
Motivated by the severity of the malignancy and the wealth of potential drug targets, Cleary and co-workers conducted a cellular high throughput screen for lead compounds that would inhibit proliferation of MLL-rearranged ALL and possess little activity against ALL cell lines without MLL gene rearrangement. STF-118804 was identified in such screens. To identify the molecular target of this compound, they infected an MLL-rearranged cell line with pools of a ~9300 gene-targeting shRNA library, which contains ~25 knockdown reagents per gene, and identified the shRNAs that were depleted by selection with STF-118804. The data indicated that silencing of the nicotinamide phosphoribosyltransferase gene (NAMPT) produced hypersensitivity to the chemical probe (Matheny et al., 2013).
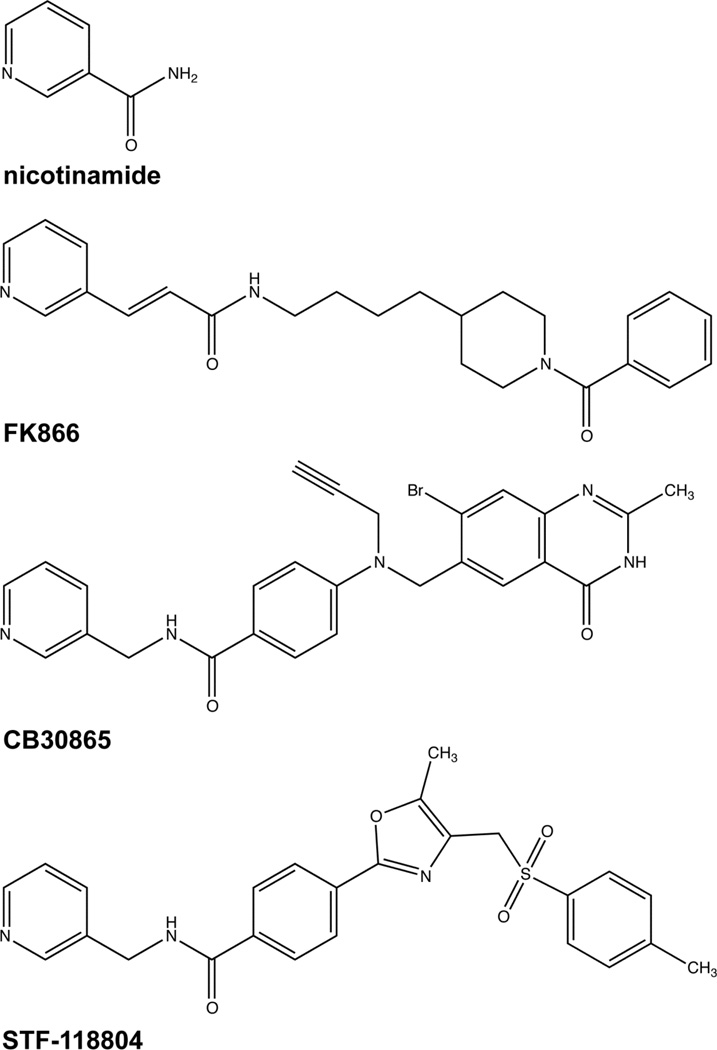
NAMPT is the enzyme required for salvage biosynthesis from nicotinamide to nicotinamide adenine dinucleotide (NAD). Nicotinamide, like the other NAD precursor vitamins, nicotinate and nicotinamide riboside, is a water soluble 3-pyridyl-containing metabolite. STF-11804 bears a strong resemblance to Nampt inhibitors FK866 and CB30865. All three are 3-pyridyl compounds with a hydrophobic extension (Figure 1). NAD and its reduced and phosphorylated forms, NADH, NADP and NADPH, are essential for function of multiple oxidoreductases in central carbon metabolism. This is termed the redox function of NAD. In addition, NAD is a consumed substrate of Sir2-related protein lysine deacetylases (sirtuins) and ADPribose-transfer enzymes including poly (ADPribose) polymerase (PARP). Nonredox functions of NAD are mediated by enzymes, such as sirtuins and PARP, that degrade the NAD dinucleotide to nicotinamide and ADPribosyl products, necessitating nicotinamide salvage in order to maintain NAD homeostasis (Belenky et al., 2007).
Figure 1.
Chemical structures of 3-pyridyl compounds, nicotinamide, STF-118804, FK866 and CB30865
NADH is so central to life that many common cell viability assays measure it as a proxy for proliferation. Cellular reduction of tetrazolium salts such as 3-(4, 5-dimethyl-2-thiazolyl)-2, 5-diphenyl-2H-tetrazolium bromide (MTT) and phenoxazine dyes such as resazurin (sold as Cell Titer Blue) is commonly employed in microtiter assays of proliferation and viability. Because these assays principally report on NADH-dependent reduction rather than cell viability, they are ideally suited to identify NAD-targeting compounds. Thus, the identification of a Nampt inhibitor as antiproliferative could have been a trivial result because inhibition of NAD salvage necessarily limits the amount of NADH available to reduce resazurin, independent of whether cells are viable. However, the investigators clearly showed that MLL-rearranged cell lines are hypersensitive to STF-11804-induced apoptosis (Matheny et al., 2013). Why then are rearranged MLL cells so sensitive to targeting NAD?
Just as NAD is involved in redox biology and non-redox biology, mechanisms for genotype-specific cytotoxicity fall into two classes. MLL fusion complexes play established roles in epigenetic programming. If MLL-rearranged cells induce the activity of NAD-consuming enzymes such as sirtuins and PARP, the resulting flux from NAD to nicotinamide would be higher and such cells might be hypersensitive to inhibition of nicotinamide salvage. Similarly, if maintenance of the malignant phenotype depends on activity of an NAD-consuming enzyme, then NAD depletion might rob a sirtuin or a PARP of its limiting substrate.
The redox functions of NAD are also sources of potential genotype-specific mechanisms. Cancer cells tend to rely less on oxidative metabolism than surrounding cells, largely because they are less driven by ATP production than production of nucleotides and lipids (Vander Heiden et al., 2009). A ramp up of glycolysis, a ramp down of citric acid cycle enzyme gene expression, and sensitivity to the glycolytic inhibitor 2-deoxy-glucose have been reported for ALL cells (Boag et al., 2006). This would suggest that the NAD requirements for glucose fermentation, i.e., glyceraldehyde-3-phosphate dehydrogenase and lactate dehydrogenase, are targeted by Nampt inhibitors.
There is further potential for genotype-specificity in Warburgian models because it has been shown that p53 mutations in ALL are largely restricted to cases with MLL rearrangement (Lanza et al., 1995). Ribose-5-phosphate synthesis through the pentose phosphate pathway (PPP), which is required for nucleotide biosynthesis, depends on cytosolic NADP and produces cytosolic NADPH for fatty acid synthesis and detoxification of reactive oxygen species. Strikingly, p53 inhibits glucose-6-phosphate dehydrogenase (G6PDH), such that p53-normal cells are inhibited in the use of glucose to generate NADPH, nucleotides and fatty acids, while p53-mutant cells are derepressed for the PPP(Jiang et al., 2011). Consistent with targets in glucose fermentation and the PPP being inhibited by NAD targeting, FK866 is reported to lead to a sharp decline in nucleocytosolic NAD prior to affecting mitochondrial NAD pools (Pittelli et al., 2010). However, some NAD must be available for redox cycling in mitochondria to produce citrate for the subsequent appearance of cytosolic citrate and conversion to cytosolic acetyl-coA for fatty acid synthesis (Ghanta et al., 2013).
Now that Nampt has been identified as an ALL target, further tumor profiling is warranted to identify the range of malignancies that are sensitive. An exciting possibility is that the therapeutic window for Nampt inhibitors can be modulated nutritionally: low NAD precursor vitamins might be warranted for the most aggressive chemotherapy, while provision of nicotinate or nicotinamide riboside might be warranted either to wash out from chemotherapy or to protect nonmalignant tissues from collateral damage (Bogan and Brenner, 2008).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Belenky P, Bogan KL, Brenner C. NAD+ metabolism in health and disease. Trends Biochem. Sci. 2007;32:12–19. doi: 10.1016/j.tibs.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Boag JM, Beesley AH, Firth MJ, Freitas JR, Ford J, Hoffmann K, Cummings AJ, de Klerk NH, Kees UR. Altered glucose metabolism in childhood pre-B acute lymphoblastic leukaemia. Leukemia. 2006;20:1731–1737. doi: 10.1038/sj.leu.2404365. [DOI] [PubMed] [Google Scholar]
- Bogan KL, Brenner C. Nicotinic acid, nicotinamide, and nicotinamide riboside: a molecular evaluation of NAD+ precursor vitamins in human nutrition. Annu. Rev. Nutr. 2008;28:115–130. doi: 10.1146/annurev.nutr.28.061807.155443. [DOI] [PubMed] [Google Scholar]
- de Boer J, Walf-Vorderwülbecke V, Williams O. In focus: MLLrearranged leukemia. Leukemia. 2013;27:1224–1228. doi: 10.1038/leu.2013.78. [DOI] [PubMed] [Google Scholar]
- Druker BJ. Translation of the Philadelphia chromosome into therapy for CML. Blood. 2008;112:4808–4817. doi: 10.1182/blood-2008-07-077958. [DOI] [PubMed] [Google Scholar]
- Ghanta S, Grossmann RE, Brenner C. Mitochondrial protein acetylation as a cell-intrinsic, evolutionary driver of fat storage: Chemical and metabolic logic of acetyl-lysine modifications. Crit. Rev. Biochem. Mol. Biol. 2013 doi: 10.3109/10409238.2013.838204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Du W, Wang X, Mancuso A, Gao X, Wu M, Yang X. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat. Cell Biol. 2011;13:310–316. doi: 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanza C, Gaidano G, Cimino G, Coco, Lo F, Basso G, Sainati L, Pastore C, Nomdedeu J, Volpe G, Parvis G. p53 gene inactivation in acute lymphoblastic leukemia of B cell lineage associates with chromosomal breakpoints at 11q23 and 8q24. Leukemia. 1995;9:955–959. [PubMed] [Google Scholar]
- Matheny CJ, Wei MC, Bassik MC, Donnelly AJ, Kampmann M, Iwasaki M, Piloto O, Solow-Cordero DE, Bouley DM, Rau R, et al. Next-Generation NAMPT inhibitors identified by sequential high-throughput phenotypic chemical and functional genomic screens. Chemistry & Biology. 2013;20 doi: 10.1016/j.chembiol.2013.09.014. inpress. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntean AG, Hess JL. The Pathogenesis of Mixed-Lineage Leukemia. Annu. Rev. Pathol. Mech. Dis. 2012;7:283–301. doi: 10.1146/annurev-pathol-011811-132434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittelli M, Formentini L, Faraco G, Lapucci A, Rapizzi E, Cialdai F, Romano G, Moneti G, Moroni F, Chiarugi A. Inhibition of nicotinamide phosphoribosyltransferase: cellular bioenergetics reveals a mitochondrial insensitive NAD pool. J Biol Chem. 2010;285:34106–34114. doi: 10.1074/jbc.M110.136739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]