Abstract
Follicular lymphoma (FL) is an incurable malignancy1, with transformation to an aggressive subtype being a critical event during disease progression. Here we performed whole genome or exome sequencing on 10 FL-transformed FL pairs, followed by deep sequencing of 28 genes in an extension cohort and report the key events and evolutionary processes governing initiation and transformation. Tumor evolution occurred through either a ‘rich’ or ‘sparse’ ancestral common progenitor clone (CPC). We identified recurrent mutations in linker histones, JAK-STAT signaling, NF-κB signaling and B-cell development genes. Longitudinal analyses revealed chromatin regulators (CREBBP, EZH2 and MLL2) as early driver genes, whilst mutations in EBF1 and regulators of NF-κB signaling (MYD88 and TNFAIP3) were gained at transformation. Collectively, this study provides novel insights into the genetic basis of follicular lymphoma, the clonal dynamics of transformation and suggests that personalizing therapies to target key genetic alterations within the CPC represents an attractive therapeutic strategy.
Follicular lymphoma (FL), the most common indolent non-Hodgkin’s lymphoma, remains a significant clinical burden as the majority of patients undergoes multiple relapses and eventually develops resistance to standard therapies. Furthermore, a subset of patients transform (tFL) to the more aggressive diffuse large B cell lymphoma (DLBCL), associated with poor clinical outcomes2-4. Recent genetic profiling and case studies of donor-derived FL following stem cell transplantation suggest the putative existence of a ‘long-lived’ tumor-initiating progenitor cell compartment, from which successive disease events occurred5-8. An in-depth characterization and chronicling of the underlying genetic events from FL to tFL will guide us in developing effective targeted therapies. To address this, we conducted whole-genome (WGS) or exome sequencing (WES) of sequential FL-tFL and matched germline (GL) samples from 10 cases (Fig. 1a and Online Methods), with deep targeted sequencing of 28 genes in an extension cohort (Supplementary Fig. 1 and Supplementary Table 1).
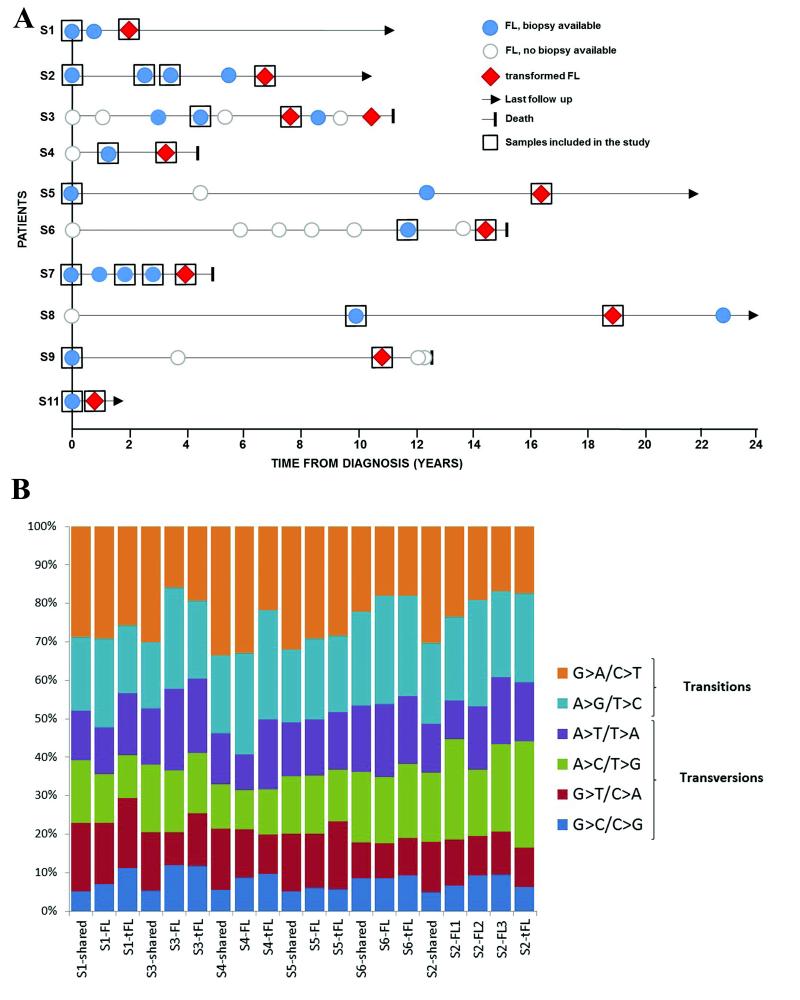
Figure 1. Clinical timelines and somatic mutation profiles for the discovery cases.
(a) Biopsy information with disease event timelines for the 10 WGS and WES cases. (b) Distribution of base substitution patterns of all somatic single nucleotide variants (SNVs) identified in the 6 paired FL-tFL genomes. The somatic SNVs for each case were sorted into three categories: shared (variants present in both FL and tFL samples), FL-specific and tFL-specific. Aberrant somatic hypermutation (aSHM) did not contribute to the majority of variants detected, with the exception of previously described gene targets (Supplementary Table 13).
For the 10 index cases (6 WGS, 4 WES), the sequencing yielded a mean genomic and exomic coverage of 37x and 110x, respectively, with 96% of the targeted bases covered at >10-fold coverage (Supplementary Table 2). In the 6 WGS cases, we detected approximately 10000 somatic variants per tumor (Supplementary Table 3). G>A/C>T transitions were the most common nucleotide substitutions, consistent with other cancer types (Supplementary Fig. 2a). We observed a higher frequency of transversions in the transformation-specific variants (Fig. 1b and Supplementary Fig. 2b; P = 0.006), reflected in the lower transition/transversion (Ti/Tv) ratios (P = 0.008; Supplementary Fig. 2c). Focusing on protein-altering changes, we identified between 21 and 143 non-synonymous somatic variants per sample with an increased frequency of mutations at transformation in the majority of cases (Supplementary Fig. 3). In total, 1560 protein-altering variants affecting 908 genes were detected across the 10 cases, comprising missense changes (84.8%), short indels (8.9%) and nonsense mutations (6.3%) (Supplementary Table 4). We verified 235 SNVs and 48 indels from 293 variants, corresponding to 118 genes, thereby achieving a concordance of 98% and 88%, respectively (Supplementary Table 5). Interrogation of the 6 WGS cases identified copy number alterations (CNAs), novel structural variants, fusion transcripts and recurrent mutations within the non-coding regions (5′UTR, 3′UTR and promoter regions) (Supplementary Fig. 4 and Supplementary Table 6).
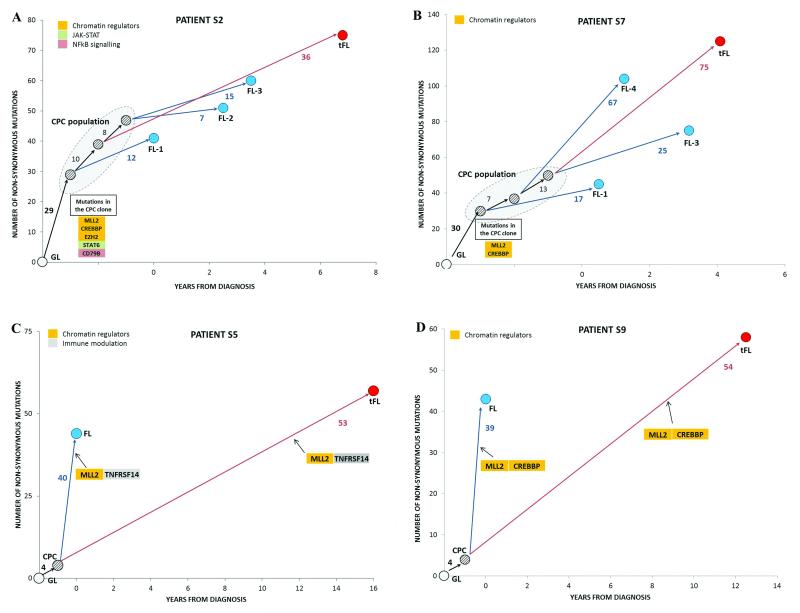
To determine the evolutionary relationship between the 10 paired FL-tFLs, phylogenetic trees were constructed for each patient using the somatic variants detected (non-synonymous only and all variants). Each tree revealed branching rather than linear evolution, with the ‘trunk’ representing genetic events shared by the FL tumor(s) and its paired tFL, thus supporting the presence of an ancestral CPC clone. Two distinct patterns of evolution from the CPC emerged. Firstly, all cases, with the exception of S5 and S9, had a high clonal semblance between the paired FL and tFL tumor which we refer to as evolution through a ‘rich’ ancestral CPC (Figs. 2a,b, Supplementary Figs. 5a-f and 6). The mutations harbored within each CPC clone demonstrated an enrichment for genes involved in chromatin regulation, with concurrent mutations in MLL2, a histone methyltransferase responsible for histone H3-lysine 4 (H3K4) trimethylation and other histone-modifying genes (CREBBP, EP300, EZH2 and MEF2B)9-12. Apart from frequent mutations in histone-modifiers, other mutational targets within the 8 ‘rich’ CPC clones included genes involved in immune modulation (B2M, CD58, TNFRSF14), JAK-STAT (SOCS1 and STAT6) and BCR/NF-κB signaling (BCL10, CARD11, CD79B), many of which have been previously reported in DLBCL10-16. In contrast, a unique pattern of evolution was seen in cases S5 and S9 where only 4 non-synonymous mutations were shared between the FL and tFL sample (referred to as evolution through a ‘sparse’ CPC) (Figs. 2c,d). Intriguingly in both cases, different MLL2 (S5 and S9), TNFRSF14 (S5) and CREBBP (S9) mutations occurred in the diagnostic FL and tFL biopsies, appearing to have arisen from independent clones and reflecting a convergent pattern of evolution. These observations strongly support tumoral dependency on CREBBP, MLL2 and TNFRSF14 alterations during lymphomagenesis and progression.
Figure 2. Clonal evolution patterns of FL to tFL for four cases.
In each case, a phylogenetic tree was constructed using non-synonymous mutations and illustrated alongside the corresponding event timeline. All trees have shared mutations (‘trunk’), which represents a common ancestral origin, the CPC, and unique ‘branches’ depicting the mutations present in that biopsy alone. The length of individual branches denotes the number of mutations separating the two disease events. S2 (a) and S7 (b), both with 3 FL (blue) and a single tFL (red) biopsy show divergent clonal evolution with a ‘rich’ CPC pool from which subsequent events arise. S5 (c) and S9 (d) illustrating a ‘sparse’ CPC (4 shared mutations between FL and tFL in both cases) and convergent evolution, in which similar genetic alterations occurs independently in separate branches of the tree. All paired tumors were confirmed to be clonally-related with BCL2-IGH rearrangement analyses. The pattern of clonal evolution was identical with all somatic variants included (Supplementary Fig. 6). Functional annotation of the mutated genes was based on previously reported functions10-16.
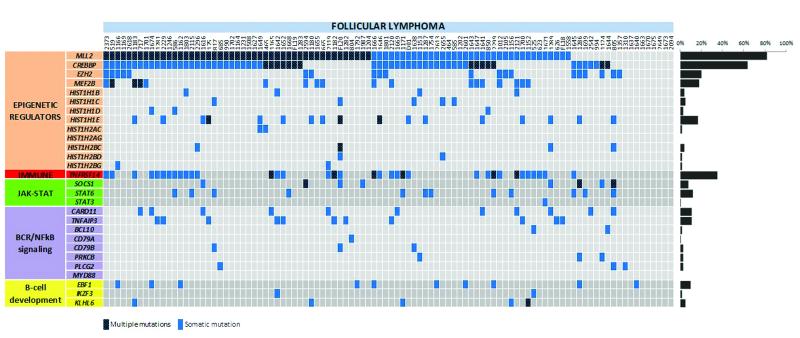
To evaluate the mutation prevalence of genes identified in our discovery cohort, we performed deep targeted resequencing of 28 candidate genes (mainly prioritizing genes involved in functional processes not previously implicated in FL (Supplementary Tables 7, 8)), in an extension cohort of 100 independent FL biopsies and 32 paired FL-tFL cases (including the 10 index cases), yielding a mean depth of 840x. A prominent feature of the genetic landscape was the presence of mutations targeting genes involved in chromatin regulation through histones and histone modifications (Fig. 3 and Supplementary Fig. 7), confirming previous studies9-12. Notably, over 70% of cases had concurrent mutations in at least 2 of the histone-modifying enzymes screened (CREBBP, EZH2, MEF2B and MLL2). MLL2 was mutated in 82% of our cohort, with approximately half of the cases harboring multiple mutations. Mutations in the histone acetyltransferase, CREBBP, and the histone methyltransferase, EZH2, were higher than earlier studies at 64% and 20%, respectively9-11.
Figure 3. Distribution of mutations in 28 genes in FL.
The heat map indicates the case-specific, concurrent and mutually exclusive mutation patterns in 100 FL cases. Each column represents an individual affected case and each row denotes a specific gene assigned into one of five functional categories as labeled on the left. The bar graph on the right shows the frequency of mutations found per gene across all samples. A heat map corresponding to the paired FL-tFL series is included in Supplementary Fig. 7b.
Notably, linker histones were recurrently mutated in FL. These encode proteins that facilitate the folding of higher-order chromatin structures and regulate access of histone-modifying enzymes and chromatin remodeling complexes to their target genes17-19. Overall, we observed mutations affecting at least one H1 gene in 28% of our series, with HIST1H1C and HIST1H1E being the most frequently mutated (Fig. 3 and Supplementary Table 9). Most of the H1 mutations were missense and clustered within the highly conserved globular domain (Supplementary Fig. 8), targeting residues directly involved in DNA binding20-23. To assess the functional relevance of these mutations, we generated H1 TKO (triple knockout)/hH1cWT and H1 TKO/hH1cS102F cell lines by over-expression of wild-type and S102F HIST1H1C mutant in H1c/H1d/H1e triple null mouse embryonic stem cells (H1 TKO ESCs)18 (Supplementary Fig. 9). The S102F HIST1H1C alteration demonstrated dramatically impaired association with chromatin relative to wild-type (Figs. 4a,b), suggesting that these mutations most likely lead to a loss-of-function phenotype by reducing the binding affinity and residence time of these H1 subtypes in chromatin compromising chromatin compaction and specific gene regulation.
Figure 4. Functional analyses of linker histone, HIST1H1C, and EBF1 mutations.
(a) Reversed phase HPLC (RP-HPLC) profiles of histones extracted from chromatin isolated from hH1cWT or hH1cS102F expressing H1 TKO mouse ESC cells. The hH1cS102F mutant demonstrated a higher hydrophobicity compared to WT. (b) The ratio of individual H1 variants (and total H1) to the nucleosome of the indicated ESC cells. The ratio is calculated56,57 from the HPLC analysis shown in (a) and demonstrates that the total H1 levels in H1 TKO/hH1cS102F ESCs were reduced compared to H1 TKO/hH1cWT, as a result of a poorer association of FLAG-hH1cS102F histones with chromatin (only 35% of FLAG-hH1c). (c) Schematics of the domains and location of mutations in EBF1 detected by targeted resequencing (NCBI protein reference sequence: NP_076870.1). DBD, DNA-binding domain; IPT, immunoglobulin domain; HLH, helix-loop-helix. Coding mutations were colored green (missense), yellow (nonsense) or red (frameshift). Mutations at the same amino acid residues in different cases are depicted. (d) Quantitative reverse transcription PCR (Q-RT-PCR)-based gene expression from Ebf1-deficient primary murine pre-pro-B-cells transduced with wild-type or mutant Ebf1. All 3 Ebf1 mutants demonstrate similar expression levels. Both DBD-mutants (G171D and S238T) exhibit abrogation of target gene activation. Igll1 and Cd79b transcripts were normalized to the control (pMYs) vector whilst Ebf1 transcripts of mutant Ebf1 constructs were normalized to wt Ebf1.
We observed frequent mutations in components of the JAK-STAT signaling pathway: STAT6 (12%) and SOCS1 (8%) (Supplementary Fig. 10). Similar SOCS1 and STAT6 mutations have been described in other germinal center (GC) lymphomas24-25 and shown to contribute to constitutive STAT6 activation and promotion of tumor cell survival in lymphoma cell lines26-28.
Constitutive activation of the anti-apoptotic NF-κB signaling pathway, caused by mutations affecting positive and negative regulators, is an established feature of the activated B-cell-like subtype of DLBCL (ABC DLBCL). In contrast, these mutations occur significantly less frequently in GC B-cell-like DLBCL (GCB DLBCL)13-14,29-30. Interestingly, we detected mutually exclusive mutations in this pathway in one-third of FLs with a predilection for CARD11 (11%) and TNFAIP3 (11%) (Fig. 3). The type and location of the detected aberrations were reminiscent of ABC DLBCL, with the exception of CD79B (Supplementary Fig. 10) where the FL variants were frameshift mutations affecting the immunoreceptor tyrosine-based activation motif (ITAM) domain in contrast to the predominant missense mutations at the Y196 residue reported in ABC DLBCL14. Given the number of emerging therapeutic agents targeting this signaling cascade, such treatments may be beneficial to a preselected subset of FL patients harboring mutations in this pathway.
Mutations in genes important in B-cell development were identified in 17% of our cohort (Fig.3 and Supplementary Fig. 10). We centered on early B-cell factor 1 (EBF1), a transcription factor frequently deleted in high-risk acute lymphoblastic leukemia31. EBF1 operates as a homodimer binding the consensus sequence 5′TCCCNNGGGA32,33, with more than half of the mutations in our series affecting the DNA-binding domain (DBD) at conserved residues (Fig. 4c). Binding of EBF1 to target genes can elicit different responses including transcriptional activation, repression or H3K4me2 modification34. In particular, transcriptional activation of Igll1, encoding the λ5 surrogate light chain of the pre-B-cell receptor has been used as an assay to assess the binding and function of EBF133. Therefore, we tested the effects of three mutations, G171D and S238T (localized within the DBD) and R381S (within the helix-loop-helix domain, HLH) for their ability to activate the Ebf1 target genes Igll1 and Cd79b in Ebf1-deficient primary murine pre-pro-B-cells (Fig. 4d). G171D and S238T mutant lines demonstrated markedly impaired up-regulation of both Igll1 and Cd79b, most likely reflecting a reduced binding to regulatory elements in their respective promoters. The HLH mutant, R381S, showed a more modest reduction of target gene expression. Taken together, the data suggest that the EBF1 mutations detected in FL lead to loss of function and a reduction in EBF1 target gene expression.
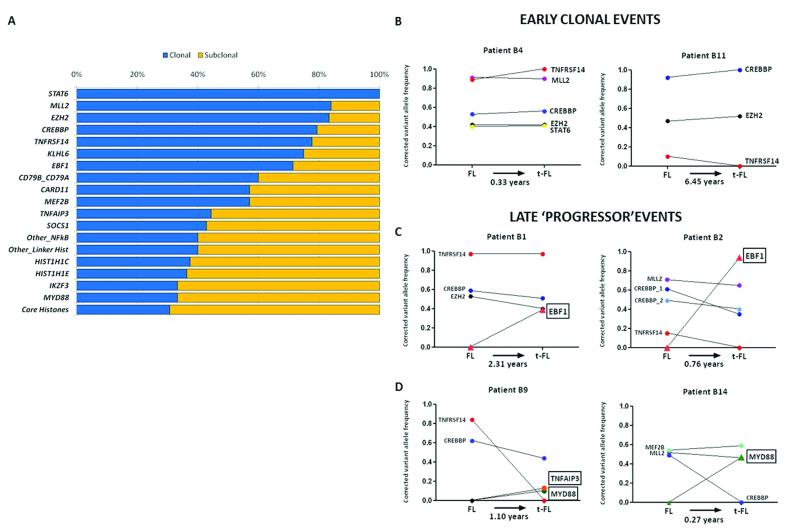
To discriminate early from late genetic events, we integrated variant allele frequencies (VAFs) generated from our high-depth sequencing data with SNP array copy number profiles on 29 cases (26 FL-tFL pairs and 3 diagnostic FLs), using the ABSOLUTE algorithm35. This scheme allowed us to correct for variations in tumor content, ploidy and loci-specific CNAs therefore establishing accurate clonal or subclonal status of mutations in our 28 selected genes. Mutations present in nearly all tumor cells (clonal) would suggest early events and therefore represent initiating ‘driver’ genes. Our results showed that mutations in the histone-modifying genes (MLL2, CREBBP and EZH2) as well as mutations in STAT6 and TNFRSF14 were predominantly clonal events (Fig. 5a). These results expand upon the findings of a recent study implicating CREBBP as an early event in FL36. Interestingly, many of these ‘driver’ genes (CREBBP, MLL2, TNFRSF14) harbored compound heterozygous mutations and were frequently accompanied by either deletions or acquired uniparental disomy (aUPD), serving as a pathogenic mechanism to render tumor cells homozygous for a pre-existing abnormality (Supplementary Fig. 11). Moreover, our longitudinal (paired FL-tFL) analyses demonstrated that mutations in these genes were stable and remained clonally dominant irrespective of therapy, in the majority of cases, as the disease progressed (Fig. 5b and Supplementary Fig. 12) reaffirming that these represent key events within the founding CPC that repopulates the tumor at transformation. Targeted resequencing in additional FL-tFL pairs confirmed the findings in our discovery cohort demonstrating predominantly evolution via ‘rich’ CPCs (Supplementary Figs. 7, 12).
Figure 5. Early and late mutations in FL and tFL.
(a) Clonality profiles of 28 recurrent gene mutations. In cases with genes harboring multiple mutations, the mutation with the highest VAF was considered. (b) Longitudinal analyses of paired FL-tFLs show early initiating mutations with corrected variant allele frequencies that were unchanged with disease progression in two representative cases. (c) Acquisition of clonal EBF1 mutations at transformation illustrated with two examples. (d) Acquisition of NF-κB genes at transformation, particularly MYD88 which demonstrated no mutations in the extension cohort of 100 FLs, suggesting these are predominantly late genetic events during the disease course.
We next evaluated the FL-tFL pairs for novel transformation-associated events. Through copy number profiling, we found an increase in the regions affected by CNAs and importantly, an enrichment of particular CNAs at transformation, such as amplifications of EZH2, MDM2, MYC and REL (Supplementary Fig. 13 and Supplementary Table 10). EBF1 mutations were acquired and restricted to just the transformation biopsies in 5 of 32 cases, with VAFs indicating clonal events (Fig. 5c). Remarkably, we also identified acquisition of alterations affecting regulators of NF-κB signaling at transformation, specifically mutations in MYD88 and TNFAIP3 (Fig. 5d and Supplementary Fig. 12). Four of the five MYD88 mutations identified were restricted to the transformation biopsy, of which none harbored the L265P mutation common to other B-cell lymphomas30,37 (Supplementary Fig. 10). In all cases, these ‘progressor’ variants did not pre-exist in the antecedent FL biopsies suggesting they were gained during the most recent clonal expansion leading to the transformation event. The acquisition of these late genetic events underscores the need for temporal mutational profiling as we move towards an era of precision medicine. It was particularly striking that the time interval from the antecedent FL biopsy to transformation in these cases was brief (range: 0.27-2.41 years), raising the possibility that acquisition of particular genetic events is selectively advantageous by promoting a higher fitness of the clone that drives transformation.
In summary, we report the most comprehensive sequencing effort in FL to date. The mutational landscape of FL is predominantly epigenetically ‘addicted’ but highlights co-occurring aberrations of genes involved in B-cell development, JAK-STAT and NF-κB signaling. By longitudinal profiling, we provide unequivocal evidence for a reservoir ancestral population, enriched in early driver genes, that propagates successive disease events. We did not identify a single compelling genetic event responsible for transformation but instead demonstrate that acquisition of distinct genetic alterations may prompt the onset of aggressive disease. The frequency of relapses suggests that the CPC is resistant to our standard therapies and that adopting a stratified treatment approach targeting specific early genetic lesions identified within the putative CPC may ultimately offer the best chance of eradicating these cells and cure of FL.
ONLINE METHODS
Patient samples
Ten patients with FL were selected based on the availability of sequential tumor lymph node biopsies (a sample with FL and a subsequent sample with histological transformation to DLBCL (tFL)) and matched germline (GL) consisting of remission bone marrows or peripheral blood specimens. The clonality between each of the paired FL and tFL samples was confirmed by BCL2-IGH breakpoint analysis, as previously described38. Clinical characteristics of these patients are presented in Supplementary Table 1. Overall, 34 samples (10 paired FL/tFL/GL and 2 additional relapsed FL samples for case S2 and S7) were evaluated. An extension cohort of 22 additional paired FL/tFL and 100 independent FL cases were included in our validation experiments (Supplementary Table 8). All biopsies were previously histologically reviewed to confirm diagnosis and tumor content before DNA extraction from fresh frozen tumor samples. Tumor content was further estimated using the ABSOLUTE36 (version 1.0.4) and ASCAT39 (version 2.1) algorithms. Written consent was obtained for collection and use of specimens for research purposes with ethical approval obtained from the Institutional Review Board (10/H0704/65 and 06/Q0605/69).
Whole genome sequencing (WGS)
5μg of genomic DNA from matched tumor and GL samples were fragmented to average 300 bp insert-size libraries and sequenced using the HiSeq 2000 platform with paired-end reads of 100 bp, according to manufacturer’s protocols (Illumina, San Diego, CA). Detailed coverage data for all cases are shown in Supplementary Table 2.
WGS read mapping and somatic mutation detection
Raw sequencing data were aligned to the reference human genome (hg19) using the Illumina ELANDv2 aligner. The alignments were sorted and converted into BAM format. PCR duplicates were removed, clusters of poorly aligned and anomalous reads were de-novo assembled into contigs using the CASAVA (v1.8) package (see URLs). We implemented the Strelka algorithm40 to identify somatic single nucleotide variants (SNVs) and indels for matched tumor-GL samples, selected for its sensitivity of mutation detection with varying tumor purities. After candidate indel detection and realignment, somatic variants were called and post-call filtering was performed. Only the somatic calls originating from homozygous reference alleles in the normal samples were considered. The joint somatic quality score Q ≥ 15 for SNVs and ≥ 30 for indels was applied for all post-filtering calls. The complete list of variants is detailed in Supplementary Table 4.
Whole exome sequencing and analysis
Capture libraries were constructed from 3μg of tumor and GL DNA using the Agilent SureSelectXT Human All Exon V4 kit. Enriched exome libraries were multiplexed and sequenced on the Illumina HiSeq 2500 to generate 100bp paired-end reads (Supplementary Table 2). Sequencing reads were aligned to the reference genome, using Burrows-Wheeler Aligner (BWA)41. SAM to BAM conversion and marking of PCR duplicates were performed using the Picard tools (version 1.86), followed by local realignment around indels and base quality score recalibration using the Genome Analysis Toolkit (GATK)42 (v2.3.9). Somatic SNVs and indels were identified using the Strelka pipeline.
Annotation and recurrently mutated genes
All post-filtered somatic SNVs and short indels were annotated using the SNPnexus tool43. The Ensembl database (v63) was used for gene/transcript identification and detection of amino acid changes. Any variants overlapping with common and GL polymorphisms present in the dbSNP 132 and the HapMap databases were masked. The allele frequencies for individual variants in both GL and tumor samples were measured by read counts. Somatic variants identified in both whole genome and exome sequencing is included in Supplementary Table 4.
Copy number and structural variant analyses from whole genome sequencing data
Copy number variants (CNV) were identified using VarScan244 (v2.3.5) with each tumor sample matched against GL. Samples were segmented using the circular binary segmentation (CBS) algorithm45 implemented in the R/Bioconductor package DNAcopy (v2.12), which allowed for undoing splits if the ratio between tumor and normal copy number was within 3 standard deviations. Structural variants were called using Pindel46 (v0.2.4t), with the exception of translocations, using the default parameters. Only calls that had zero supporting reads in the normal sample but ≥ 8 supporting reads in the tumor sample were retained. Translocations (fusion transcripts) were identified using DELLY47 and Breakdancer48 algorithms on paired-end reads only retaining SVs called by both methods, with ≥ 5 supporting reads in the tumor.
Identification of recurrent mutations in promoter regions
Based on the Ensembl gene/transcript annotation, the somatic variants in the region of −2,000 to +250 bp (within 5′ UTR) relative to transcription start site (TSS) for each transcript of protein coding genes were identified across all WGS cases. Recurrently mutated promoter regions among multiple patients were identified and mutation hotspots in these regions were also noted (Supplementary Table 6).
Mutation and indel validation
SNVs and indels were confirmed by bidirectional Sanger sequencing, fluorescence-based fragment size analysis or targeted deep sequencing (Fluidigm-MiSeq), as detailed below (Supplementary Table 5). Concordance of selected variants called by WGS, WES and validation targeted deep sequencing with estimated VAFs are shown in Supplementary Fig. 14.
Phylogenetic analysis
Evolutionary trees were constructed for each of the 10 WGS/WES patients based on the distance matrix between GL, FL and tFL samples derived from the numbers of somatic variants (all variants, and nonsynonymous SNVs and indels) from each biopsy, using the Neighbor-Joining algorithm49 implemented in the PHYLIP package (see URLs). Once the consensus phylogenetic tree was determined, it was redrawn starting from GL leading to the putative CPC, then to FL and tFL samples, with the branch length proportional to the number of somatic changes i.e. genetic distance between the samples; and plotted against the clinical timelines.
High density SNP arrays
Copy number analysis and regions of copy neutral loss of heterozygosity (cnLOH, also known as acquired uniparental disomy, aUPD) was performed on a cohort of 26 paired FL-tFL and 3 diagnostic FL DNA samples (including 7 of our discovery cases) using the Affymetrix SNP 6.0 microarrays. Further details are given in the Supplementary Note.
Targeted resequencing
Twenty-eight candidate genes (Supplementary Table 8) were sequenced in the extension cohort cases using the Access Array™ platform (Fluidigm). All samples were screened in duplicate, with a number of matched GL and normal tonsil DNA included as controls in each run. The access array amplification was performed in a multiplex format using genomic DNA (50ng) according to the manufacturer’s recommendation. The multiplexed library pools were deep sequenced using the Illumina MiSeq. After demultiplexing and FASTQ file generation of the raw data, reads were aligned to the reference genome using the BOWTIE2 algorithm50. SAMtools51 was used to generate sorted BAM files for each sample, and the VarScan 2 tool was used to examine the pileup file to call variants, including SNPs and short indels, implementing a minimum threshold variant allele frequency (VAF) of 5% and a Fisher’s Exact Test p < 0.05. Only bases meeting the minimum base quality of 20 from reads meeting the minimum mapping quality of 20 were considered. A minimum read depth of 10 at a position was required to make calls and variants with >90% reads supporting one strand were excluded. Only concordant variants present in both duplicates or confirmed by independent Sanger sequencing were retained. Identified variants were annotated using SNPnexus to exclude those reported in dbSNP132 and to identify variants that had non-synonymous consequences or affected the splice sites.
Estimating clonality of mutations
Tumor purity, ploidy and absolute DNA copy numbers were determined from Affymetrix SNP6.0 data (n = 26 FL-tFL pairs and 3 diagnostic FLs) by implementing the ABSOLUTE algorithm (Supplementary Table 11). The ASCAT algorithm was also used to derive tumor purity and ploidy to ensure a high concordance across both algorithms (Supplementary Table 11). These estimates were used to compute and classify individual somatic mutations obtained from our deep sequencing analyses as clonal or subclonal, based on the posterior probability that the cancer cell fraction (CCF) exceeded 0.95, as previously reported52 (Supplementary Table 12).
Identification of recurrent targets of aberrant somatic hypermutation (aSHM)
SHM hotspots typically occur within the 2-kb window downstream of the transcription start site (TSS) and we have focused our analyses on these regions53. The total number of SNVs and the average mutation density across the 14 WGS tumor samples were investigated. For each gene, the ratio of transition to transversion mutations, ratio of mutations at C:G to A:T sites and the percentage of SNVs within the hotspot motif, WRCY, their associated p-values, and the SHM indicator were calculated in accordance to previous studies54. The probability of observing the number of SNVs or more for each target gene was calculated based on the Poisson distribution given the expected number of SNVs per SHM target region if those were uniformly randomly distributed in all target genes. A list of SHM gene targets is included in Supplementary Table 13.
EBF1 mutagenesis, retroviral expression and target gene analysis
Three FL-derived mutants (G171D and S238T localized within the DBD and R381S within the HLH domains) were generated to assess their ability to activate Ebf1 target gene expression. Ebf1-deficient pre-pro-B-cell lines (cultured on OP9 feeders in the presence of IL-7, Flt3 ligand and SCF) were transduced with wild-type or mutant Ebf1-GFP expressing bicistronic retroviruses as described previously34. GFP-positive cells were sorted 24 h after transduction and expression was confirmed by immunoblotting (data not shown). Total RNA was extracted using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. Quantitative RT-PCR analysis was performed in triplicates on a 7500 Fast Real-Time PCR system (Applied Biosystems) using Power SYBR Green master mix to measure the mRNA expression of the Ebf1 target genes Igll1 and Cd79b33,34.
Generation of hH1c and hH1c/S102F overexpressing clones in H1c/H1d/H1e triple knockout embryonic stem cells (H1 TKO ESCs)
The FLAG-hH1c and FLAG-hH1c/S102F overexpression plasmids were constructed by replacing the mouse H1d coding sequence with FLAG-hH1c or FLAG-hH1c/S102F coding sequences in a 5-kb fragment encompassing the mouse H1d upstream and downstream regulatory sequences which were inserted into a vector containing a Blasticidin-resistant gene. Plasmid DNA (20μg) was transfected into 2×107 H1 TKO ESCs, as previously described55. A total of 48 clones (24 clones for each vector transfection) resistant to Blasticidin were picked and screened by immunoblotting using an anti-FLAG antibody (Sigma-Aldrich). Cell clones with the highest levels of FLAG-hH1c and FLAG-hH1c/S102F were selected for further analysis.
Preparation and analysis of histones
Chromatin was prepared from ESC and histones were extracted with 0.2 N sulfuric acid, according to previously described protocols56-58. Approximately 50μg of total histones was injected into a C18 reverse phase column (Vydac) on an AKTA UPC10 system (GE Healthcare). The effluent was monitored at 214 nm, and the peak areas were analyzed with AKTA UNICORN (v5.11) software. The A214 values of the H1 and H2B peaks were adjusted according to the respective peptide bonds, and the H1/nucleosome ratio was calculated, as previously described57-58.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to the patients for donating tumor specimens as part of this study. We would like to thank T. Chaplin-Perkins and B. Young for performing the SNP6.0 array experiments as part of the Affymetrix Array Core Facility and G. Clark at London Research Institute for automated DNA sequencing. This study was predominantly funded by Cancer Research UK through the Genomic Initiative and Programme grant to J.F., also supported by Leukaemia and Lymphoma Research (grant to J.F.) and OTKA K-76204 grant (grant to A.M.). Y.F. is a recipient of the Georgia Cancer Coalition Distinguished Scholar Award and C.P. and Y.F. are in part, supported by the NIH grant (GM085261 to Y.F.). C.B. is a recipient of the European Hematology Association (EHA) Partner fellowship (2009/1) and was supported by the European Union and the State of Hungary, co-financed by the European Social Fund in the framework of TÁMOP 4.2.4. A/1-11-1-2012-0001 ‘National Excellence Program’. J.O. is a recipient of the Kay Kendall Leukaemia Fund (KKLF) Junior Clinical Research Fellowship (KKL 557).
Footnotes
COMPETING FINANCIAL INTERESTS: We declare no competing financial interests.
URLs:
CASAVA, http://support.illumina.com/sequencing/sequencing_software/casava.ilmn;
PHYLIP package, http://evolution.genetics.washington.edu/phylip.html;
Affymetrix, http://www.affymetrix.com/support/technical/byproduct.affx?product=genomewidesnp_6
Accession codes: Sequencing data have been deposited at the European Genome-phenome Archive under accession number EGAS00001000399. Affymetrix SNP 6.0 data have been deposited at the Gene Expression Omnibus (GEO) database under the accession number GSE42525.
References
- 1.Swenson WT, et al. Improved survival of follicular lymphoma patients in the United States. J Clin Oncol. 2005;23:5019–26. doi: 10.1200/JCO.2005.04.503. [DOI] [PubMed] [Google Scholar]
- 2.Al-Tourah AJ, et al. Population-based analysis of incidence and outcome of transformed non-Hodgkin’s lymphoma. J Clin Oncol. 2008;26:5165–9. doi: 10.1200/JCO.2008.16.0283. [DOI] [PubMed] [Google Scholar]
- 3.Montoto S, Fitzgibbon J. Transformation of indolent B-cell lymphomas. J Clin Oncol. 2011;29:1827–34. doi: 10.1200/JCO.2010.32.7577. [DOI] [PubMed] [Google Scholar]
- 4.Montoto S, et al. Risk and clinical implications of transformation of follicular lymphoma to diffuse large B-cell lymphoma. J Clin Oncol. 2007;25:2426–33. doi: 10.1200/JCO.2006.09.3260. [DOI] [PubMed] [Google Scholar]
- 5.Carlotti E, et al. Transformation of follicular lymphoma to diffuse large B-cell lymphoma may occur by divergent evolution from a common progenitor cell or by direct evolution from the follicular lymphoma clone. Blood. 2009;113:3553–7. doi: 10.1182/blood-2008-08-174839. [DOI] [PubMed] [Google Scholar]
- 6.Eide MB, et al. Genomic alterations reveal potential for higher grade transformation in follicular lymphoma and confirm parallel evolution of tumor cell clones. Blood. 2010;116:1489–97. doi: 10.1182/blood-2010-03-272278. [DOI] [PubMed] [Google Scholar]
- 7.Ruminy P, et al. S(mu) mutation patterns suggest different progression pathways in follicular lymphoma: early direct or late from FL progenitor cells. Blood. 2008;112:1951–9. doi: 10.1182/blood-2007-11-124560. [DOI] [PubMed] [Google Scholar]
- 8.Weigert O, et al. Molecular ontogeny of donor-derived follicular lymphomas occurring after hematopoietic cell transplantation. Cancer Discov. 2012;2:47–55. doi: 10.1158/2159-8290.CD-11-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bödör C, et al. EZH2 Y641 mutations in follicular lymphoma. Leukemia. 2011;25:726–9. doi: 10.1038/leu.2010.311. [DOI] [PubMed] [Google Scholar]
- 10.Morin RD, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–5. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morin RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pasqualucci L, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–95. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lenz G, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319:1676–9. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 14.Davis RE, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463:88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pasqualucci L, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43:830–7. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, et al. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A. 2013;110:1398–403. doi: 10.1073/pnas.1205299110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bednar J, et al. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci U S A. 1998;95:14173–8. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Croston GE, Kerrigan LA, Lira LM, Marshak DR, Kadonaga JT. Sequence-specific antirepression of histone H1-mediated inhibition of basal RNA polymerase II transcription. Science. 1991;251:643–9. doi: 10.1126/science.1899487. [DOI] [PubMed] [Google Scholar]
- 19.Fan Y, et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 20.Brown DT, Izard T, Misteli T. Mapping the interaction surface of linker histone H1(0) with the nucleosome of native chromatin in vivo. Nat Struct Mol Biol. 2006;13:250–5. doi: 10.1038/nsmb1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goytisolo FA, et al. Identification of two DNA-binding sites on the globular domain of histone H5. EMBO J. 1996;15:3421–9. [PMC free article] [PubMed] [Google Scholar]
- 22.Ramakrishnan V, Finch JT, Graziano V, Lee PL, Sweet RM. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993;362:219–23. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- 23.Vyas P, Brown DT. N- and C-terminal domains determine differential nucleosomal binding geometry and affinity of linker histone isotypes H1(0) and H1c. J Biol Chem. 2012;287:11778–87. doi: 10.1074/jbc.M111.312819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mottok A, et al. Inactivating SOCS1 mutations are caused by aberrant somatic hypermutation and restricted to a subset of B-cell lymphoma entities. Blood. 2009;114:4503–6. doi: 10.1182/blood-2009-06-225839. [DOI] [PubMed] [Google Scholar]
- 25.Ritz O, et al. Recurrent mutations of the STAT6 DNA binding domain in primary mediastinal B-cell lymphoma. Blood. 2009;114:1236–42. doi: 10.1182/blood-2009-03-209759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baus D, et al. STAT6 and STAT1 are essential antagonistic regulators of cell survival in classical Hodgkin lymphoma cell line. Leukemia. 2009;23:1885–93. doi: 10.1038/leu.2009.103. [DOI] [PubMed] [Google Scholar]
- 27.Mottok A, Renné C, Willenbrock K, Hansmann ML, Bräuninger A. Somatic hypermutation of SOCS1 in lymphocyte-predominant Hodgkin lymphoma is accompanied by high JAK2 expression and activation of STAT6. Blood. 2007;110:3387–90. doi: 10.1182/blood-2007-03-082511. [DOI] [PubMed] [Google Scholar]
- 28.Ritz O, et al. STAT6 activity is regulated by SOCS-1 and modulates BCL-XL expression in primary mediastinal B-cell lymphoma. Leukemia. 2008;22:2106–10. doi: 10.1038/leu.2008.85. [DOI] [PubMed] [Google Scholar]
- 29.Compagno M, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459:717–21. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ngo VN, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470:115–9. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harvey RC, et al. Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood. 2010;116:4874–84. doi: 10.1182/blood-2009-08-239681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hagman J, Belanger C, Travis A, Turck CW, Grosschedl R. Cloning and functional characterization of early B-cell factor, a regulator of lymphocyte-specific gene expression. Genes Dev. 1993;7:760–73. doi: 10.1101/gad.7.5.760. [DOI] [PubMed] [Google Scholar]
- 33.Treiber N, Treiber T, Zocher G, Grosschedl R. Structure of an Ebf1:DNA complex reveals unusual DNA recognition and structural homology with Rel proteins. Genes Dev. 2010;24:2270–5. doi: 10.1101/gad.1976610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Treiber T, et al. Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription- independent poising of chromatin. Immunity. 2010;32:714–25. doi: 10.1016/j.immuni.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 35.Carter SL, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–21. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Green MR, et al. Hierarchy in somatic mutations arising during genomic evolution and progression of follicular lymphoma. Blood. 2013;121:1604–11. doi: 10.1182/blood-2012-09-457283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Treon SP, et al. MYD88 L265P somatic mutation in Waldenström’s macroglobulinemia. N Engl J Med. 2012;367:826–33. doi: 10.1056/NEJMoa1200710. [DOI] [PubMed] [Google Scholar]
- 38.Ladetto M, et al. A validated real-time quantitative PCR approach shows a correlation between tumor burden and successful ex vivo purging in follicular lymphoma patients. Exp Hematol. 2001;29:183–93. doi: 10.1016/s0301-472x(00)00651-2. [DOI] [PubMed] [Google Scholar]
- 39.Van Loo P, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci U S A. 2010;107:16910–5. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saunders CT, et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–7. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 41.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler etc. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dayem Ullah AZ, Lemoine NR, Chelala C. SNPnexus: a web server for functional annotation of novel and publicly known genetic variants (2012 update) Nucleic Acids Res. 2012;40:W65–70. doi: 10.1093/nar/gks364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koboldt DC, et al. VarScan2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–76. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–72. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 46.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rausch T, et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen K, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6:677–81. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 50.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Landau DA, Wu CJ. Chronic lymphocytic leukemia: molecular heterogeneity revealed by high-throughput genomics. Genome Med. 2013;5:47. doi: 10.1186/gm451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pasqualucci L, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–6. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 54.Khodabakhshi AH, et al. Recurrent targets of aberrant somatic hypermutation in lymphoma. Oncotarget. 2012;3:1308–19. doi: 10.18632/oncotarget.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y, et al. Histone h1 depletion impairs embryonic stem cell differentiation. PLoS Genet. 2012;8:e1002691. doi: 10.1371/journal.pgen.1002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cao K, et al. High-resolution mapping of h1 linker histone variants in embryonic stem cells. PLoS Genet. 2013;9:e1003417. doi: 10.1371/journal.pgen.1003417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fan Y, Skoultchi AI. Genetic analysis of H1 linker histone subtypes and their functions in mice. Methods Enzymol. 2004;377:85–107. doi: 10.1016/S0076-6879(03)77005-0. [DOI] [PubMed] [Google Scholar]
- 58.Medrzycki M, Zhang Y, Cao K, Fan Y. Expression analysis of mammalian linker-histone subtypes. J Vis Exp. 2012;(61):3577. doi: 10.3791/3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.