Two-step transcriptomic profiling of bone-resorbing OCs versus nonresorbing MGCs generated a list of 115 genes potentially involved in bone resorption. Of these, RhoE was investigated. Its role in podosome dynamics is central for OC migration, SZ formation, and, ultimately, bone resorption.
Abstract
The function of osteoclasts (OCs), multinucleated giant cells (MGCs) of the monocytic lineage, is bone resorption. To resorb bone, OCs form podosomes. These are actin-rich adhesive structures that pattern into rings that drive OC migration and into “sealing-zones” (SZs) that confine the resorption lacuna. Although changes in actin dynamics during podosome patterning have been documented, the mechanisms that regulate these changes are largely unknown. From human monocytic precursors, we differentiated MGCs that express OC degradation enzymes but are unable to resorb the mineral matrix. We demonstrated that, despite exhibiting bona fide podosomes, these cells presented dysfunctional SZs. We then performed two-step differential transcriptomic profiling of bone-resorbing OCs versus nonresorbing MGCs to generate a list of genes implicated in bone resorption. From this list of candidate genes, we investigated the role of Rho/Rnd3. Using primary RhoE-deficient OCs, we demonstrated that RhoE is indispensable for OC migration and bone resorption by maintaining fast actin turnover in podosomes. We further showed that RhoE activates podosome component cofilin by inhibiting its Rock-mediated phosphorylation. We conclude that the RhoE-Rock-cofilin pathway, by promoting podosome dynamics and patterning, is central for OC migration, SZ formation, and, ultimately, bone resorption.
INTRODUCTION
Osteoclasts (OCs) are multinucleated giant cells (MGCs) of the monocytic lineage that are responsible for bone resorption. They differentiate and fuse from mononucleated precursors such as monocytes (Mos) or immature dendritic cells (DCs) in the presence of receptor activator of nuclear factor κB ligand (RANKL) and macrophage colony-stimulating factor (M-CSF; Rivollier et al., 2004; Speziani et al., 2007; Boyce, 2013). Once differentiated, mature OCs resorb the bone matrix by secreting protons and proteases, such as tartrate-resistant acidic phosphatase (TRAP), cathepsin-K, and matrix metalloprotease 9 (MMP9), onto the underlying bone matrix. A circular adhesive superstructure called the “sealing zone” (SZ) allows these molecules to be confined within the resorption pit (lacuna) for efficient bone degradation (Duong et al., 1998).
Besides OCs, other types of MGCs can also be generated from immature DCs under various conditions. DCs stimulated by interleukin-17 (IL-17) and potentiated by interferon-γ (IFN-γ) lead to DC-17γ-MGCs (Coury et al., 2008). In addition to resulting from cell–cell fusion of the same precursors (i.e., DCs), DC-17γ-MGCs have OC-like characteristics, namely the expression of bone-degrading proteases such as TRAP, MMP9, and cathepsin-K (Coury et al., 2008), as well as form adhesive structures called “podosomes” (Olsson Akefeldt et al., 2013).
Podosomes are integrin-based, actin-rich, dot-like structures present in OCs, DCs, and macrophages (Zambonin-Zallone et al., 1988; Linder and Kopp, 2005). They are the structural units of the OC SZ (Luxenburg et al., 2007; Ma et al., 2010). A widely accepted model of intrapodosomal architecture in monocytic cells depicts two distinct domains within podosomes: a peripheral domain made of a loose F-actin meshwork with adhesion and adaptor molecules such as αVβ3 integrin, paxillin, vinculin, and myosin II, and a central domain comprising a tightly connected F-actin network with adaptor molecules such as cortactin and Arp2/3 (Linder and Aepfelbacher, 2003; Chabadel et al., 2007). When formed in OCs, podosomes are first assembled in clusters that self-organize into “rings” and finally either into SZs on mineralized substrates or “SZ-like” structures (also known as “podosome belts”) on nonmineralized substrates. Besides driving the saltatory migration typical of OCs, podosome rings have been described as short-lived patterns, with lifespans of several minutes, mediating the transition from clusters to SZ-like patterns (Hu et al., 2011). The latter are more stable structures, with lifespans of several hours (Destaing et al., 2003; Saltel et al., 2004; Bruzzaniti et al., 2005; Jurdic et al., 2006; Ma et al., 2008). Of interest, the lifespan of individual podosomes decreases by at least twofold with the transition from clusters to rings/SZ-like structures, indicating differential regulation during podosome “remodeling” (Luxenburg et al., 2007). The regulation of podosome assembly and disassembly, as well as their inner architecture and stability, is of central importance to their patterning. Therefore fast and dynamic polymerization and depolymerization of actin, the scaffold of podosomes, has to be tightly regulated in OCs. For example, knockout of gelsolin, a high-affinity actin-severing and capping protein, results in the inability of OCs to form podosomes (Chellaiah et al., 2000a). In the same context, slowing the rate of actin turnover in OCs with cytochalasin D, an actin-capping molecule, leads to overstabilized podosomes with longer lifespans and seemingly larger podosome cores (Luxenburg et al., 2012). Another important mechanism that regulates podosome size, stability, and patterning is actomyosin-II contractility around podosome cores (Meddens et al., 2013).
Several studies provided data as to how podosomes are regulated at the molecular level in OCs. Src is an indispensable regulator of phosphorylation of podosomal proteins such as cortactin and gelsolin (Luxenburg et al., 2006; Destaing et al., 2008). In addition, control of OC podosome patterning by small GTPases, namely RhoA and Rac1/2, is well documented. Rac activation after integrin-dependent signaling is essential to formation of a functional SZ. Indeed, mice lacking both Rac1 and Rac2 display massive osteopetrosis due to the inability of OCs to form SZs and, subsequently, resorb bone (Croke et al., 2011). Supporting this result, deletion of Rac GTPase exchange factors Vav3 and Dock5 in mice also presents osteopetrotic phenotypes explained by an OC functional defect (Faccio et al., 2005; Vives et al., 2011). Furthermore, RhoA is critical for maintaining OC polarization and SZ formation on a mineralized matrix (Chellaiah et al., 2000b; Destaing et al., 2005). The complete molecular scheme of the actin cytoskeleton regulation needed to adequately support bone resorption by OCs is still largely incomplete. Studies of Rho-Rock signaling in OC and OC-like cells show that this pathway can temporally modulate podosome arrangement during osteoclastogenesis and in a substrate-dependent manner, but the signaling downstream of Rock in OCs is unclear (Chellaiah et al., 2000b; Ory et al., 2000, 2008).
In this study, we sought to find new genes implicated in the regulation of the actin cytoskeleton of bone-resorbing OCs. Our approach aimed at finding genes that are highly expressed in mature OCs by comparing the transcriptomic profiles of human primary Mo-derived OCs (Mo-OCs) and DC-derived OCs (DC-OCs) on one hand with those of Mos, DCs, and DC-17γ-MGCs with OC-like traits on the other. As a result, we obtained a short list of actin-regulating genes that were up-regulated in mature OCs and therefore considered as candidates for functional studies. We focused our investigation on the role of one candidate, RhoE (also known as RnD3), in bone resorption using a genetic knockout model. Given its already described function in other actin structures such as focal adhesions and stress fibers, we investigated the implication of RhoE in OC podosome organization, patterning, and actin dynamics, as well as in OC migration. Finally, given that RhoE interaction with Rock-I has been widely shown to be a central pathway of RhoE function (Riento et al., 2003, 2005), we investigated how RhoE signals to the OC cytoskeleton through Rock-I.
RESULTS
Nonresorbing human MGCs exhibit OC-like characteristics but are unable to resorb bone
Knowing that DC-17γ-MGCs are multinucleated giant cells expressing OC degradation markers such as TRAP, cathepsin-K, and MMP9, we first asked whether they were able to resorb the mineral matrix in comparison with bone-resorbing DC-OCs. Then we investigated their actin-containing adhesion structures. Initially differentiated on tissue culture dishes, mature DC-OCs and mature DC-17γ-MGCs were detached on days 5 and 13 of culture, respectively, and seeded on apatite–collagen complex (ACC), a bone-mimicking substrate (Shibutani et al., 2000; Saltel et al., 2004). Cells were then stimulated with M-CSF and RANKL to promote resorption. Confocal microscopy of these cells fixed and stained for F-actin showed that DC-OCs, as expected, exhibited circular actin-rich SZs and were able to resorb the mineralized matrix. DC-17γ-MGCs, however, were unable to resorb the matrix and displayed circular actin structures that were significantly less frequent and smaller than with OC SZs (Figure 1, A and B). We therefore named the latter actin structures “dysfunctional SZs” because of their inability to ensure proper degradation of the mineral matrix.
FIGURE 1:
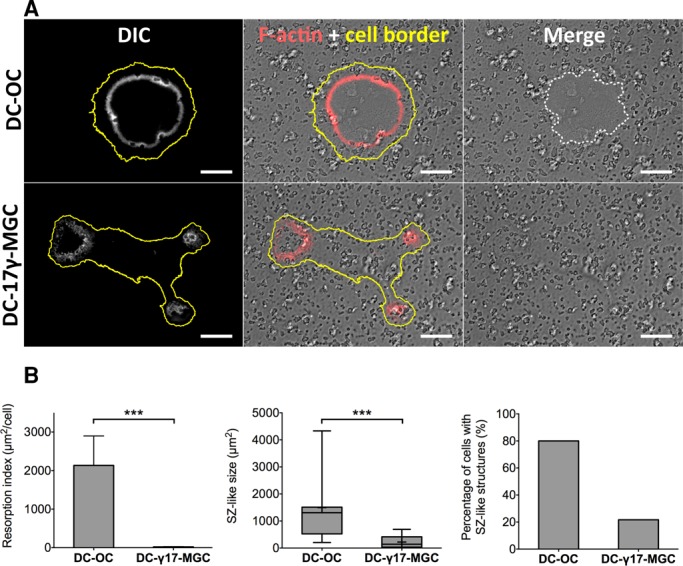
DC-17γ -MGC, like OCs, are able to make podosomes but not functional SZs for resorption. (A) Cells seeded on a resorbable ACC matrix. As expected, DC-OCs form SZs (F-actin is in white in the left and red in the middle) and are able to resorb bone (resorbed area outlined with a white dotted line, right). DC-17γ -MGCs make dysfunctional SZ structures and are unable to resorb the matrix. Yellow line outlines the cell periphery. Scale bar, 20 μm. (B) Quantification of F-actin–containing structures and resorption areas per cell. Left, percentage of fixed DC-OCs exhibiting SZs compared with DC-17γ-MGCs exhibiting dysfunctional SZ structures. Middle, box-and-whiskers plot showing surface area of SZ and dysfunctional SZs. Represented on the plot are minimum and maximum values (whiskers), mean (+), median (middle line in gray box), and 25 and 75% quartiles (limits of gray box). Right, histogram of the resorption index (area of resorbed matrix per cell) of the two cell types. Error bars are SEM.
To gain further insight into cytoskeletal organization in DC-17γ-MGCs, we compared their actin organization with that of human Mo-OCs and DC-OCs on glass. We labeled these cells for actin and cortactin—a component of the podosome core—as well as for paxillin and vinculin—components of the podosome cloud. We acquired images with a confocal microscope and visualized protein colocalization of one podosome marker at a time with actin by displaying channel merges and pixel-to-pixel multiplication of normalized and nonsaturated micrographs. We found that, in podosome cores, cortactin colocalized perfectly with actin of Mo-OCs and DC-OCs, as well as in DC-γ17-MGCs. Vinculin and paxillin precisely colocalized with actin in podosome clouds in all three cell types (Figure 2). These results showed that DC-17γMGCs, albeit displaying bona fide podosomes, are unable to efficiently organize SZs, which could explain their inability to resorb bone.
FIGURE 2:
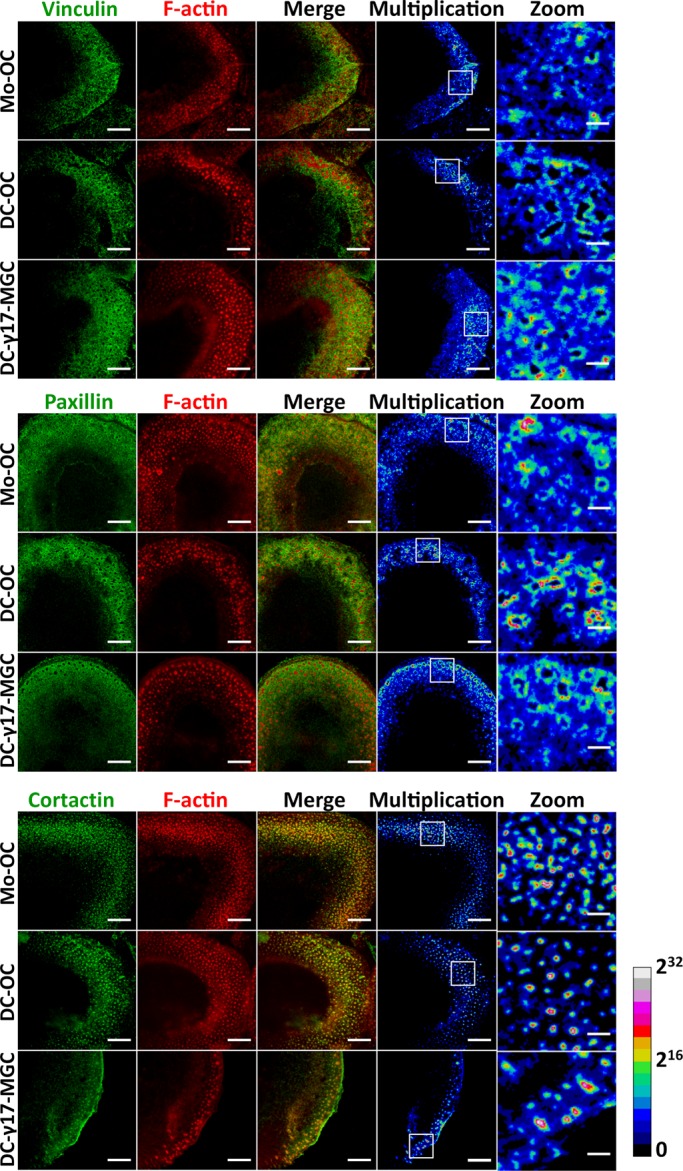
Cytoskeleton organization in DC-17γ-MGC compared with Mos and DC-OCs. Cells seeded on glass showing colocalization of paxillin or vinculin (green) with F-actin (red) at the podosome cloud (top and middle, respectively) and cortactin (green) with F-actin (red) at the podosome core. Multiplication images here calculated pixel to pixel from F-actin combined with one podosomal protein at a time and are represented in 32-bit float images with a ratio lookup table. The calibration bar indicates the correspondence of false color to 32-bit pixel fluorescence intensities. Scale bar, 10 μm (first four columns), 2 μm (magnified square insets).
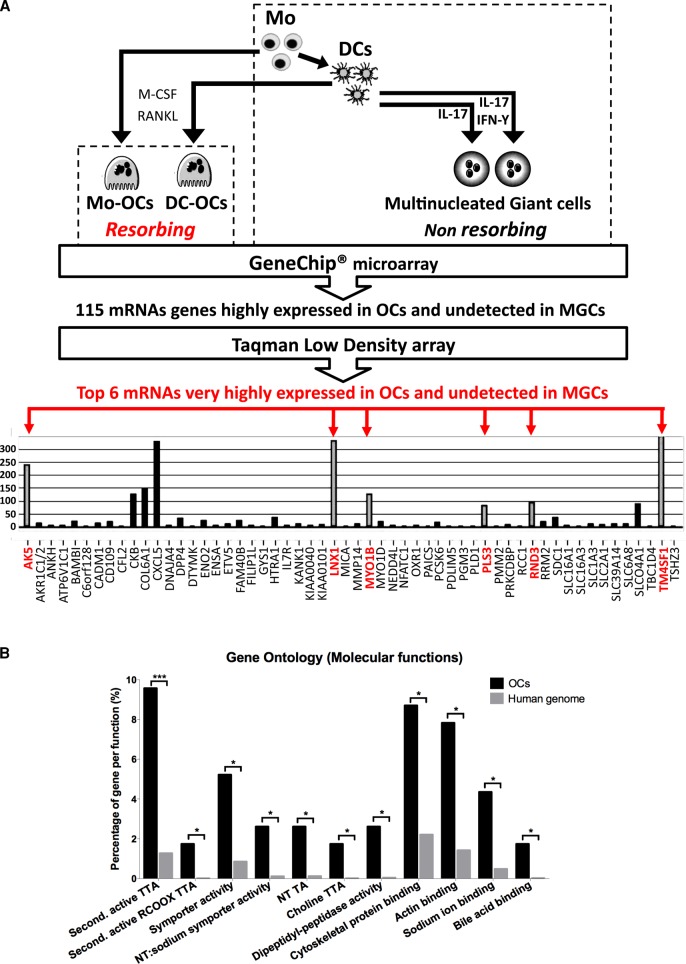
Comparative transcriptomics reveals osteoclast-specific genes
To identify new genes involved in OC-mediated bone resorption while excluding genes involved in myeloid cell-cell fusion, we performed transcriptomic profiling of human bone-resorbing Mo-OCs and DC-OCs, their respective precursor cells (i.e., Mos and DCs), and nonresorbing DC-derived MGCs. DC-derived MGCs included MGCs derived from DCs by stimulation with IL-17 and DC-17γ-MGCs (Figure 3A). Blood monocytes from healthy human donors were differentiated into either DCs with M-CSF and IL-4 or OCs with M-CSF and RANKL. DCs were then further differentiated in OCs (DC-OCs) with M-CSF and RANKL or DC-17γ-MGCs with IL-17 and INF-γ. Total RNAs were extracted from these six culture conditions from two to four independent healthy donors and separately analyzed by GeneChip Microarray technology using 54,675 distinct probes. Given the high number of probe sets used, a gene was considered as expressed in a cell type if its mean expression was superior to the overall gene expression in the same cell type (see Materials and Methods). This allowed us to establish a differential list of 115 genes that were expressed only in OCs (Figure 3 and Supplemental Figures S1 and S2). Among these 115 genes, we found several that were known regulators of OC functions: NFACT1, the OC-specific transcription factor (Takayanagi, 2007; reviewed in Takayanagi et al., 2002); the creatine kinase CKB and the proton pump ATP6v1c1, both important for OC podosome ring formation (Chang et al., 2008; Feng et al., 2009); and CXCL5, a secreted protein that is part of the OC chemokine signature (Moreaux et al., 2011). We considered these findings as a validation of our approach. Resorbing and nonresorbing cultured cell types were all maintained in cell fusion–inducing culture conditions. Consequently, any genes known for their induction of cell–cell fusion (such as DC-STAMP, ATP6v0d2, or OSCAR) were eliminated from this differential analysis. We therefore considered these 115 genes as new potential candidates for the investigation of OC functions. The “molecular functions” annotation of this list compared with the human genome, as determined by Gene Ontology vocabulary, underlined enrichment in “Cytoskeletal protein binding” and its subcategory “Actin binding” (Figure 3B).
FIGURE 3:
Workflow and results of the comparative transcriptomic study used to probe for OC-specific genes. (A) In vitro treatments used for differentiation and fusion of human Mos into resorbing OCs, of immature DCs into resorbing OCs, and of DCs into nonresorbing MGCs by addition of IL-17 and/or IFN-γ. GeneChip Microarray technique was applied to total RNAs obtained from each of cell type, followed by mathematical comparison to sort out 115 mRNAs highly and exclusively expressed in Mo-OCs and DC-OCs. Validation of the up-regulation of the 115 genes in OCs by Taqman low-density array performed on cDNAs from Mos, DCs, Mo-OCs, DC-OCs, and DC-17γ-MGCs. Due to the high cutoff value for detection, only 56 of 115 genes were found to be expressed in OCs. Among these, six genes (gray columns, names in red) were undetected in Mos, DCs, or in MGCs. (B) Enriched Gene Ontology molecular functions in the 115 OC-specific genes compared with the human genome identified by the online tool FatiGO. NT, neurotransmitter; second, secondary; TA, transporter activity; TTA, transmembrane transporter activity.
We aimed at further validating candidates that would be preferentially involved in cytoskeletal regulation of bone resorption as opposed to genes instead implicated in other OC functions. Owing to their cytoskeletal properties, DC-17γ-MGCs were retained as the DC-derived MGC reference model to be compared with OCs. We determined RNA expression values of the previously identified 115 genes in Mo-OCs, DC-OCs, DC-17γ-MGCs, and their precursors (i.e., Mos and DC) by Taqman low-density array (TLDA), a high-throughput quantitative real-time PCR. We then averaged the mean expression of a gene (Xc) from several human blood donors and used the limma test to determine whether Xc was significantly higher in Mo- and DC-OCs jointly than in nonresorbing Mos, DCs, and DC-17γ-MGCs. This method allowed us to spot six genes that were highly expressed in OCs (gene names in red in Figure 3A) but not even detected in the other cells types. These six genes are TM4SF1, PLS3 (also known as T-fimbrin), ARHE (also known as RHOE or RND3), LNX1, AK5, and MYO1B. Three of these six genes (PLS3, MYO1B, and RHOE) were already described as regulators of actin structures in several cell types, whereas PLS3 protein is a component of podosomes in Mo-OCs (Babb et al., 1997). Given the importance of Rho-GTPases in the regulation of podosomes in OCs, we focused on further investigating RhoE role in OCs.
RhoE is essential for bone resorption but not osteoclast differentiation
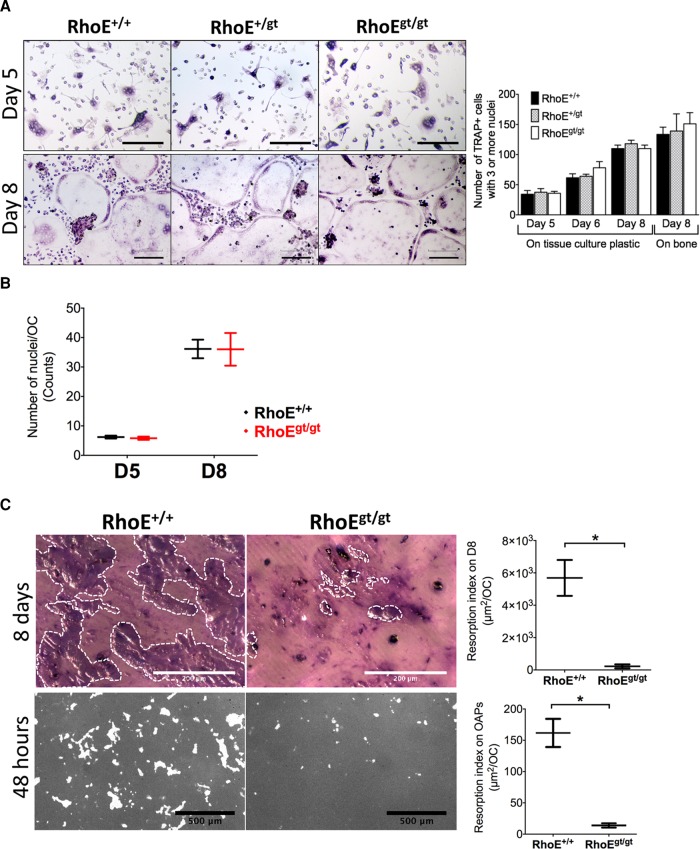
RhoE was shown to be a regulator of focal adhesions and stress fibers in different cell types (Guasch et al., 1998; Klein and Aplin, 2009). Before investigating its role in OC functions, we first assayed the implication of RhoE in ex vivo osteoclastogenesis using primary murine precursors isolated from a RhoE gene trap (gt) mouse model (Mocholi et al., 2011). Because these animals rarely survive until day 15 after birth, we used either fetal liver (FL) or P14 bone marrow as a source of OC precursors. We differentiated primary OCs from FL precursors from RhoE+/+, RhoE+/gt, and RhoEgt/gt embryonic day–15.5 (E15.5) littermates in the presence of M-CSF and RANKL on culture-treated dishes and cortical bone slices. Differentiation of FL-derived OCs (FL-OCs) on days 5, 6, and 8 postseeding was assayed by TRAP staining. On both substrates, no differences in the number of TRAP-positive cells with three or more nuclei were observed between the different genotypes (Figure 4A). In the same assay, we also counted the number of nuclei per OC on days 5 and 8 of culture on plastic wells as a measure of OC precursor fusion (Figure 4B). Again, we observed no difference in the absence of RhoE expression. We therefore concluded that RhoE does not play a role in osteoclastogenesis.
FIGURE 4:
RhoE is dispensable for primary murine OC differentiation and fusion but is required for bone resorption. (A) Micrographs showing TRAP staining of FL-OCs differentiated with M-CSF and RANKL on tissue culture–treated plates after 5, 6, and 8 d of culture. Scale bars, 200 μm. Histogram shows a time-scale quantification of the number of OCs (TRAP-positive cells with three or more nuclei) per well on tissue culture plastic and bone slices. (B) Number of nuclei per OC as indicator of OC fusion on tissue culture–treated plastic. (C) Top, micrographs and plot showing resorption index of OCs seeded for 8 d on bone. Resorbed bone is stained with toluidine blue (violet surface outline with white dotted lines). Scale bar, 200 μm. Graphs show mean ± SEM. Bottom, micrographs and plot showing resorption index of OCs seeded for 48 h on a bone-mimicking substrate (Osteo Assay plates). Unresorbed matrix is stained with silver nitrate (gray). Resorbed surface is not stained (white). Scale bar, 500 μm.
To evaluate the role of RhoE in OC-mediated bone resorption, we performed resorption assays in two different conditions. First, precursors from RhoE+/+ and RhoEgt/gt littermates were differentiated directly on cortical bone slices. On day 8 of differentiation, the cultures were stopped, and the resorption index was determined from surface area measurement of toluidine blue–positive resorption pits and TRAP staining of OCs on the same slices. RhoEgt/gt OCs resorbed bone ∼25 times less than wild-type OCs (Figure 4C, top). In the second condition, we detached day-4 RhoE+/+ and RhoEgt/gt FL-derived OCs (FL-OCs) that were initially differentiated on culture-treated plates and seeded them for 48 h on an Osteo Assay substrate (OAS) plate, a bone-mimicking substrate. We counted the number of TRAP-positive OCs per well and, in replicate wells, quantified the total resorbed area per well with silver nitrate staining. Hence we calculated the mean resorption index per OC. Again, RhoEgt/gt OCs resorbed the OAS matrix ∼12 times less than RhoE+/+ OCs (Figure 4C, bottom). These results accounted for an essential role of RhoE in bone resorption in vitro.
Small sealing zones and defective podosome organization in RhoE-deficient osteoclasts
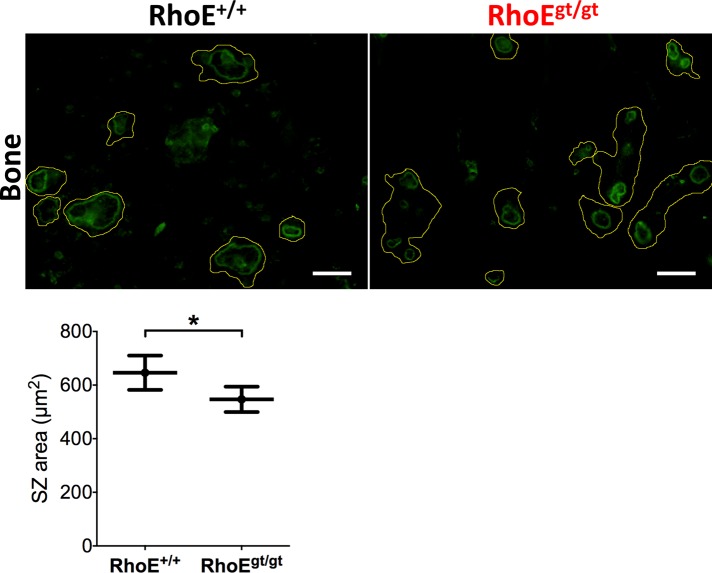
RhoE is involved in the control of actin cytoskeleton in other cell types (Guasch et al., 1998; Klein and Aplin, 2009). We therefore investigated its role in SZ formation, given the critical importance of this actin structure in the bone resorption process. Day-4 RhoE+/+ and RhoEgt/gt FL-OCs were lifted from culture-treated plates and seeded for 24 h on bone slices. These OCs were then fixed, stained for actin, and imaged using confocal microscopy. Then, using the software ImageJ, we manually determined peripheral outlines of SZs from acquired micrographs, allowing the quantification of SZ surface areas. There was a slight yet significant 15% decrease in mean SZ size in RhoEgt/gt OCs compared with RhoE+/+ OCs (Figure 5). This result indicates that RhoE expression is dispensable for SZ formation but is needed to maintain normal SZ size.
FIGURE 5:
RhoEgt/gt OCs exhibit smaller SZs. Micrographs of F-actin–stained FL-OCs seeded on cortical bone slices showing smaller SZs sizes. Yellow lines show cell periphery inferred from overcontrasted images. SZs were determined by their circular shape and strong fluorescence. SZ outlines were manually drawn, and inner surface areas were quantified. Calculated mean ± SEM is plotted. Scale bar, 50 μm.
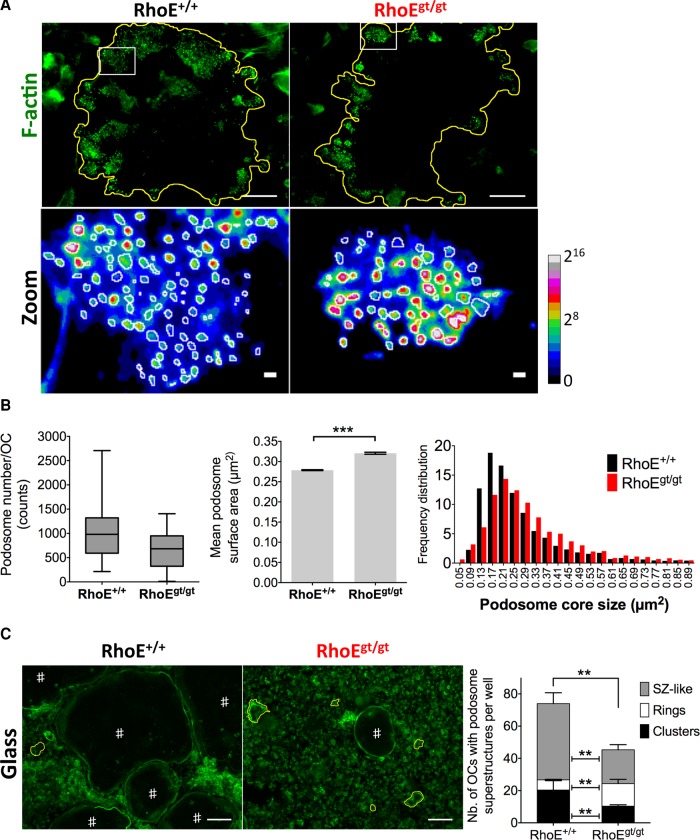
Because of the observed effect of RhoE knockdown on SZ size, we investigated its importance for podosome formation. Bone marrow (BM)–derived OCs (BM-OCs) were fixed on day 4, fluorescently stained for F-actin, and imaged with confocal microscopy. Based on observed local fluorescence intensity, F-actin staining of OCs allowed not only identification of individual podosomes within a cluster, but also discrimination between the two subdomains of podosomes: the core, with high fluorescence intensity, and the cloud with low intensity, according to Chabadel et al. (2007) and van den Dries et al. (2013b; Figure 6A). We therefore applied a method of fast and curated fluorescence intensity–based quantification of podosome core surface area. Briefly, once OC cell periphery was manually determined, we applied the “Find Peaks” plug-in in ImageJ with a set of fixed parameters (see Materials and Methods) to count podosome numbers per cluster-presenting OC and the size of individual podosome cores. We found that the mean number of podosomes per OC was decreased in RhoEgt/gt OCs (645 podosomes/cell) compared with RhoE+/+ OCs (1018 podosomes/cell). Podosome core size was increased, however, in the absence of RhoE expression, as shown by mean values and frequency distribution of size (Figure 6B). The effect of RhoE depletion on podosome size and number was not accompanied by a change in cell spreading as determined by quantification of RhoE+/+ and RhoEgt/gt OC surface (Supplemental Figure S3A). These data suggest that RhoE regulates podosome formation and size.
FIGURE 6:
Podosome size, number, and collective patterning are regulated by RhoE in OCs. (A) Top, micrographs of BM-OC podosomes stained for F-actin, showing reduced number of podosomes in RhoEgt/gt OCs. The yellow line in the zoomed images (bottom) shows cell periphery (top). Bottom, zoom of white insets in top lane using ratio lookup table colors. White circles depict peripheries of podosome cores as identified by local intensity thresholds using ImageJ (see Materials and Methods) followed by manual curating. The calibration bar indicates the correspondence of false color to 16-bit pixel fluorescence intensities (B) Left, whisker-box plot depicts minimum and maximum number of podosomes per cell (whiskers), 25 and 75% quartiles (lower and upper limits of boxes), and median (midline of boxes). Middle, histogram showing mean podosome core size with SEM bars. Left, frequency distribution histogram of podosome core sizes, showing a shift toward bigger sizes in RhoEgt/gt OCs. Horizontal axis shows the centers of intervals. (C) Micrographs of F-actin–stained FL-OC cultures. #iOCs with SZ-like patterns. SZ-like patterns are several hundred micrometers in diameter and delineate the cell periphery. The yellow lines outline OCs that have podosome rings. Rings, similar to SZ-like patterns, are also circular superstructures but are only tens of micrometers in diameter. Histogram shows counts of clusters, rings. and SZ-like patterns per well. Note the decrease in RhoEgt/gt OCs in stable and metastable podosome superstructures, that is, clusters and SZ-like patterns, respectively, as well as the increase of transient superstructures, that is, rings. Scale bar, 100 μm.
The regulation of podosomes at the unit level by RhoE prompted us to investigate its possible role in collective podosome patterning. To do so, we quantified the number of OCs with clusters, rings, and SZ-like structures per well. According to previous studies, we applied the terms “rings” and “SZ-like” patterns to circular podosome superstructures present in OCs seeded on nonmineralized substrates (Saltel et al., 2004; Destaing et al., 2005; Jurdic et al., 2006; Luxenburg et al., 2007; Biosse Duplan et al., 2013). However, the distinction between these two superstructures lies in the fact that rings are small patterns (several tens of micrometers in diameter. as indicated in Figure 7A, discussed later), and they are formed in a part of the cell. “Sealing zone–like” patterns (also known as “belts”) are giant circular patterns (up to several hundreds of micrometers in diameter, as indicated in Figure 6C), closely aligned with the entire cell periphery and present in much bigger OCs. We would also specify that, whereas podosome clusters and rings are common to OCs seeded on bone and nonmineralized substrates (e.g., culture-treated glass or plastic), SZs are specific to bone-adherent OCs, and, finally, SZ-like patterns are formed by OCs on nonmineralized substrates. The total number per well of OCs exhibiting any of these podosomal structures significantly decreased in RhoEgt/gt OCs (45/well) compared with RhoE+/+ OCs (75/well), whereas the total number of OCs per well was not altered, as previously shown by the TRAP assay. In more detail, the mean number of OCs with clusters decreased from 20/well in wild-type cultures to 10/well in RhoEgt/gt, and SZ-like structures from 47/well to 21/well, in the same order. The number of OC with rings increased, however, to 14/well in RhoEgt/gt OCs compared with six/well for wild-type OCs (Figure 6C). In summary, RhoE depletion led to a decrease in clusters and SZ-like patterns, both known as stable superstructures with lifespan of several hours, but favored the increase of transient, short-lived rings. The alterations in podosome size and number in RhoEgt/gt OCs could account for the defects observed in cluster formation and subsequent transition of clusters into rings and then SZ-like superstructures. Indeed, it was suggested that collective podosome patterning depends on intrinsic podosome organization (Destaing et al., 2008).
FIGURE 7:
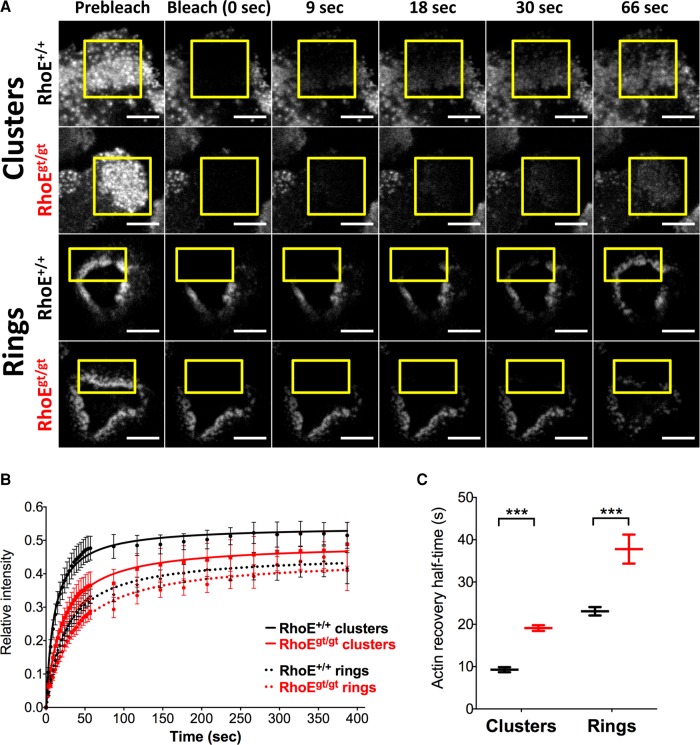
FRAP of LifeAct-mCherry reveals slower turnover in podosome clusters and rings in RhoEgt/gt OCs. (A) Micrographs of time frames pre- and postphotobleaching of LifeAct-mCherry–expressing FL-OCs on day 6. Yellow insets show bleached areas. Scale bar, 10 μm. (B) Mean fluorescence recovery of photobleached areas normalized to mean fluorescence of the same area before photobleaching and to the nonbleached remainder of the cell during recovery. Data were gathered from n > 12 samples per genotype and per structure from two independent cultures. Each culture was differentiated using a pool of precursors from at least three animals of the same genotype. Dots are means of mean relative fluorescence. Lines are fitted curves. Bars are SD. (C) F-actin recovery half-times calculated from recovery curves. Recovery half-times are increased in clusters from 9.8 ± 1.4 s in RhoE+/+ to 19.7 ± 3.5 s in RhoEgt/gt OCs and in rings from 24 ± 3 s in RhoE+/+ to 34.4 ± 3.8 s in RhoEgt/gt.
RhoE controls actin turnover in podosome clusters and rings
The aberrant podosome sizes, numbers, patterned superstructures, and OC-mediated bone resorption observed in RhoEgt/gt OCs raises a question: how does RhoE regulates podosomes? To address this point, we first examined the expression levels of structural podosome components by assaying surface expression of the integrin subunits αV and β3 (Supplemental Figure S3B), as well as the total protein levels of cortactin, vinculin, and paxillin (Supplemental Figure S4), in RhoE+/+ and RhoEgt/gt day-5 BM-OCs. We found that RhoE deletion did not affect the expression levels of these essential podosome components. We therefore suspected that, instead of regulating the expression of the structural components of podosomes, RhoE could act on podosomes by modulating their actin dynamics. We performed fluorescence recovery after photobleaching (FRAP) experiments on actin in podosome clusters and rings of day-6 FL-OCs 48 h after their infection with a LifeAct-mCherry–coding lentivirus to label actin (Figure 7A). We fitted relative fluorescence intensity data with a nonexponential equation of recovery (Negi and Olson, 2006) to minimize constraints inherent to mathematical functions (e.g., an exponential function). In wild-type OCs, mean actin turnover half-time in clusters and rings was 9.8 ± 1.4 and 19.7 ± 3.5s, respectively. This showed that rings normally exhibit slower actin turnover. These values were significantly increased in RhoEgt/gt OC for both structures: 24 ± 3 s in clusters and 34.4 ± 3.8 s in rings (Figure 7B). We concluded that RhoE is involved in maintaining fast actin turnover in podosomes.
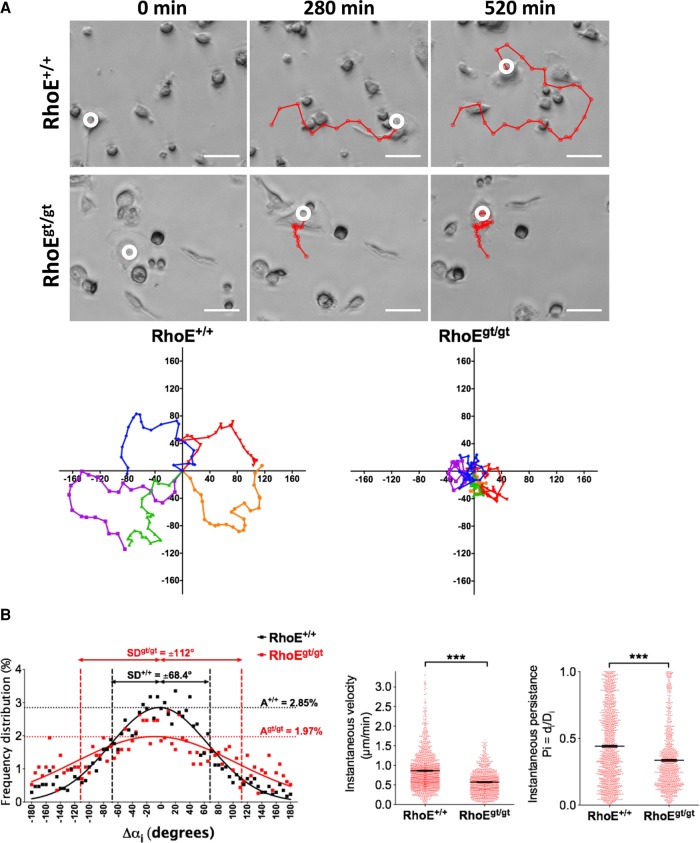
Depletion of RhoE results in impaired OC migration
Having demonstrated a role of RhoE in podosome actin dynamics and podosome stability and patterning, we hypothesized that RhoE would be implicated in OC migration. To test this hypothesis, we assayed OC random migration. Differentiating Sp-OCs expressing or not RhoE were detached on day 4 of culture and replated on culture-treated dishes at identical low confluence (5 × 104 cells/well) to minimize OC fusion events. Time-lapse movies were then acquired with phase-contrast microscopy, and randomly migrating OCs were manually tracked. As visualized in single-OC migration tracks (Figure 8A), wild-type OCs displayed typical saltatory migration. This mode of migration is consistent with displacement in a straightforward direction followed by a recognizable angular turn, as we reported in a previous study (Hu et al., 2011). Saltatory migration allowed OCs to randomly cover a large surface area. RhoEgt/gt OCs displayed aberrant migration tracks: they turned in circles because of frequent consecutive angular turns to one side (Figure 8A). The frequency distribution of the instantaneous angle change (Δαi) of several wild-type OC migration tracks nicely fitted a Gaussian distribution (R2 = 0.91) in which the amplitude, indicating the frequency of straightforward movement (i.e., when Δαi is close to 0°), was 2.85 ± 0.07% and the absolute SD (i.e., the interval containing the most frequent 34% single-sided angle changes close to the mean) was 68.4 ± 2.01°. Of interest, values from RhoEgt/gt OCs deviated from the Gaussian distribution (R2 = 0.58), with an amplitude of 1.97 ± 0.08% and an absolute SD of 115 ± 6.51°. The mean of Δαi Gaussian distributions did not vary between the two genotypes (−2.35 ± 1.99° for RhoE+/+ OCs and −8.76 ± 5.31° for RhoEgt/gt OCs) because, in our quantification, we pooled several individual tracks with negative or positive Δαi values. However, amplitudes and SDs were significantly different, denoting decrease of the frequency of straightforward displacements and increase of the frequency of angular turns during OC migration in the absence of RhoE expression. This aberrant distribution of angle changes therefore resulted in a significantly lower instantaneous persistence of RhoEgt/gt OCs (Pi = 0.335) compared with that of RhoE+/+ OCs (Pi = 0.419; Figure 8B). In addition, quantification of the instantaneous velocity of displacement (Vi) showed a significant decrease from 0.86 μm/s ± 0.01 in RhoE+/+ OCs to 0.57 μm/s ± 0.38 in RhoEgt/gt OCs (Figure 8B). These data demonstrate that RhoE is a key regulator of OC migration directionality and velocity.
FIGURE 8:
RhoEgt/gt OCs show impaired migration. (A) Top, micrographs of time-lapse imaging showing Sp-OC migration tracks (red). Scale bar, 40 μm. Bottom, representative rose plots of 5 RhoE+/+ and 5 RhoEgt/gt OC migration tracks for a total duration of 8 h/track. (B) Plots of migration parameters of migrating OCs (RhoE+/+, n = 84 tracks, 1870 time points; RhoEgt/gt, n = 39 tracks, 1214 time points). Left, instantaneous velocity of displacement (i.e., speed between two consecutive positions). Middle, instantaneous persistence of OC migration (Pi = di/Di). Right, frequency distribution of angle change between two consecutive movements. Raw values are represented with dots and fitted with a Gaussian function. Error bars are SEM.
RhoE inhibits Rock-mediated cofilin phosphorylation
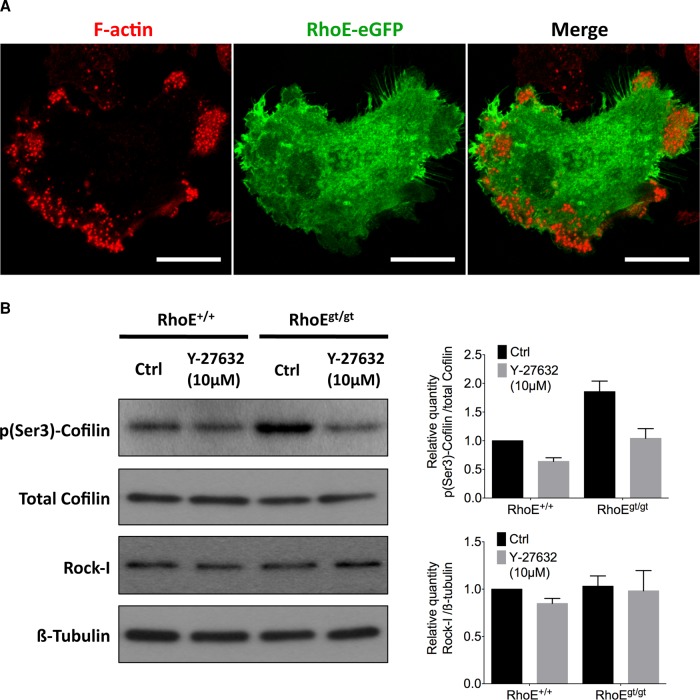
Having shown a role of RhoE in podosome dynamics and organization, we sought to find its subcellular localization and the molecular mechanism through which it signals to podosomes. Day-5 BM-OCs overexpressing RhoE–enhanced green fluorescent protein (eGFP) were fixed, stained with phalloidin, and imaged with a confocal microscope. Of interest, RhoE did not colocalize with podosomes per se but was instead ubiquitous in the cytoplasm, with certain enrichments at membrane borders (Figure 9A). We therefore considered that RhoE probably acts indirectly on actin in podosomes.
FIGURE 9:
RhoE is cytoplasmic and inhibits Rock-I from phosphorylating cofilin. (A) Micrograph of a day-5 BM-OC expressing RhoE-eGFP (green) 24 h posttransfection and stained for F-actin (red), showing ubiquitous, cytoplasmic localization of RhoE. (B) Immunoblotting of total cofilin, phosphorylated cofilin at Ser-3 (p(Ser-3)-cofilin), Rock-I, and β-tubulin from total cell lysate of day-5 BM-OCs that were serum induced for 2 h with or without 10 μM Y-27632 (a Rock inhibitor) after 2 h of serum starvation. Quantification of protein expression of p(Ser-3)-cofilin normalized to total cofilin reveals 1.86-fold increase in RhoEgt/gt OCs compared with RhoE+/+ OCs without affecting overall Rock-I levels. Diagrams show mean ± SEM from three independent experiments.
It has been shown that RhoE acts on actin in focal adhesions and stress fibers via its inhibition of Rock-I kinase activity. Hence we looked, in mature OCs, for Rock-I phosphorylation targets that could mediate the effects of RhoE on podosome size and actin dynamics, such as the myosin-II regulator Mypt1 and cofilin (van den Dries et al., 2013a). To validate these proteins as Rock targets in OCs, we blocked the kinase activity of Rock in day-5 BM-OCs for 2 h using an inhibitor, Y-27632, and probed by immunoblotting of total lysates for their canonical phosphorylation sites: cofilin phosphorylation on Ser-3 and Mypt1 phosphorylation on Thr-696 (Supplemental Figure S5). We observed a decrease in p(Ser3)-cofilin but not p(Th696)-Mypt1 ratio normalized to their respective total protein controls. We thus validated cofilin as a downstream target of Rock in mature OCs.
We then investigated whether cofilin phosphorylation was altered in a Rock-dependent manner in RhoE-depleted OCs. We probed for p(Ser3)-cofilin levels in RhoE+/+ and RhoEgt/gt BM-OCs with or without inhibition of Rock activity by Y-27632. In the absence of Y-27632 treatment, p(Ser3)-cofilin was 1.86 times higher in OCs lacking RhoE expression than in wild-type OCs. The addition of Y-27632 reduced cofilin in both situations, whereas Rock-I levels did not vary in any of these conditions (Figure 9B), suggesting that the observed changes in cofilin phosphorylation were independent of the expression level of Rock-I but highly dependent on its kinase activity. Taken together, these experiments revealed that, when expressed in OCs, RhoE partially blocks Rock-mediated cofilin phosphorylation, which consequently leads to maintenance of cofilin in its nonphosphorylated, active form.
Inhibition of cofilin phosphorylation at Ser-3 rescues RhoEgt/gt bone-resorption defect
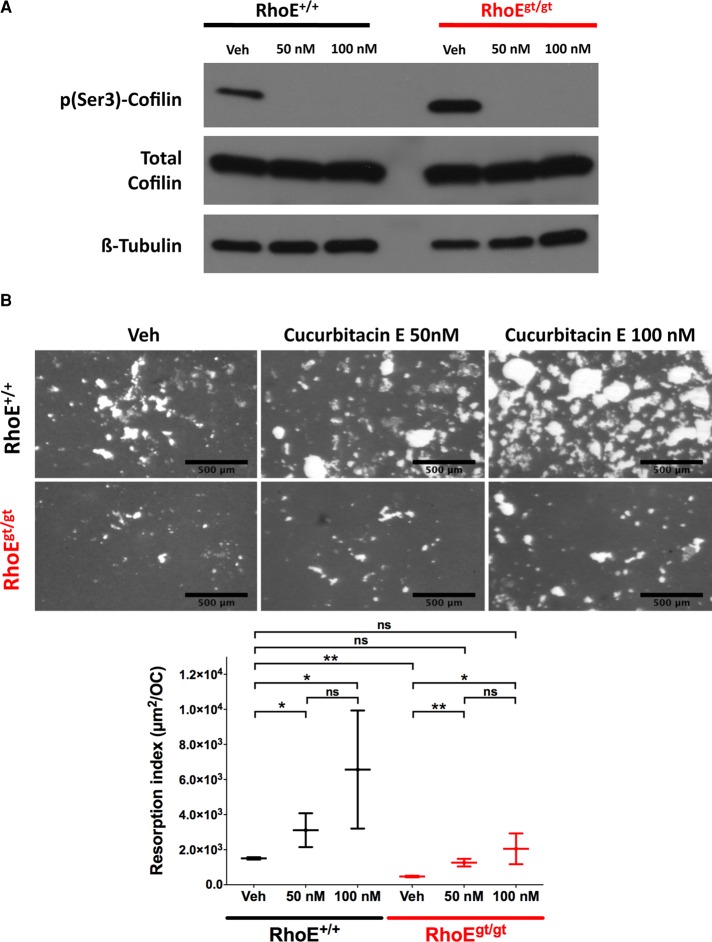
By promoting actin dynamics, cofilin is a regulator of cell migration in cell types other than OCs (Bravo-Cordero et al., 2013). Because cofilin was excessively phosphorylated in RhoE-deficient OCs, we sought to first evaluate the importance of its phosphorylation status to OC function and then explore whether the increased p(Ser3)-cofilin levels observed in RhoEgt/gt OCs could account for their resorption defect. To this end, we assayed the effect of cucurbitacin E, a potent inhibitor of cofilin phosphorylation at Ser-3 (Nakashima et al., 2010), on bone-resorption capacity of RhoE+/+ and RhoEgt/gt OCs. BM precursors from RhoE+/+ and RhoEgt/gt littermates were differentiated directly on bone-mimicking OAS. On day 5 of differentiation, the cultures were treated with 50 and 100 nM cucurbitacin E for 48 h, and the resorption index was determined at day 7. Cucurbitacin E was very efficient in inhibiting cofilin phosphorylation at Ser-3, as no p(Ser3)-Cofilin was detected by immunoblotting starting from a 50 nM concentration of the inhibitor (Figure 10A). In RhoE+/+ and in RhoEgt/gt OCs, the inhibition of cofilin phosphorylation by cucurbitacin E led to a dose-dependent increase in the bone-resorption index (Figure 10B). Of importance, although the resorption index of nontreated RhoEgt/gt OCs was significantly lower that of RhoE+/+ OCs (3.2-fold difference), treatment with 50 nM cucurbitacin E was sufficient for full rescue of the resorption defect resulting from RhoE deficiency (Figure 10B). This result clearly demonstrates a key role of cofilin phosphorylation status in OC-mediated bone resorption. Furthermore, the increased Rock-dependent cofilin phosphorylation resulting from RhoE deficiency is sufficient to account for the resorption defect of RhoEgt/gt OCs.
FIGURE 10:
Inhibition of cofilin phosphorylation at Ser-3 rescues RhoEgt/gt bone-resorption defect. Day-5 BM-OCs initially seeded on bone-mimicking OAS were treated for further 48 h with either vehicle (dimethyl sulfoxide) or 50 or 100 nM cucurbitacin E. (A) Immunoblotting showing inhibition of Ser-3–cofilin phosphorylation by cucurbitacin E at 50 and 100 nM without affecting total cofilin level. (B) Micrographs and plot showing resorption index of OCs seeded for 7 d on OAS, including treatment with cucurbitacin E for the last 48 h. Unresorbed matrix is stained with silver nitrate (gray). Resorbed surface is not stained (white). Scale bar, 500 μm.
DISCUSSION
Actin organization during podosome patterning is central for OC migration and bone resorption. To further decipher the mechanisms involved in actin remodeling in bone-resorbing OC, we took advantage of human monocytic plasticity. Indeed, we were able to generate, from human blood cells, either two types of OCs derived from Mos or DCs or two different types of MGCs derived from DCs (Rivollier et al., 2004; Coury et al., 2008; unpublished data). Characterization of DC-derived MGCs revealed that, despite expressing osteoclastic proteases, they were unable to resorb the mineral matrix (Coury et al., 2008). Moreover, their actin cytoskeleton was organized into circular superstructures reminiscent of SZs. These were named “dysfunctional SZs” because of their inability to ensure resorption of a mineralized matrix in comparison with OC SZs. The existence of bona fide podosomes in DC-derived MGCs is insufficient for SZ formation, and, more important, it suggested that there are yet-unknown SZ-specific molecular regulations that are absent in nonpatterned individual podosomes.
A transcriptomic comparison between bone-resorbing OCs and nonresorbing MGCs allowed us to provide a list of 115 genes that were highly expressed in OCs. Among these, we validated six genes that were highly and exclusively expressed in OCs, namely TM4SF1, AK5, LNX1, MYO1B, RHOE/ARHE/RND3, and PLS3/T-PLASTIN/T-FIMBRIN. Previous studies provided data linking these genes with OCs and/or with actin cytoskeleton. PLS3, a member of the fimbrin family, colocalizes with OC podosomes (Babb et al., 1997). AK5 expression is stimulated by RANKL in osteosarcoma cells (Mori et al., 2007). TM4SF1, a member of the tetraspanin family described for their interaction with integrins at the cell surface, is involved in nanopodia formation and cell migration (Zukauskas et al., 2011). LNX1 is responsible for the ubiquitinylation of Src, a known OC regulator (Weiss et al., 2007). Myo1B, a member of the myosin superfamily, is localized in filopodia in HeLa and Cos-7 cells (Almeida et al., 2011) and regulates Arp2/3 in Golgi-related actin foci. Finally, RhoE, a constitutively active, GTP-bound protein, destabilizes stress fibers and focal adhesions by inhibiting the Rho-Rock pathway. Considering the importance of Rho signaling pathways in OCs (Zou and Teitelbaum, 2010), we focused our attention on RhoE.
RhoE depletion did not affect primary OC differentiation or fusion but greatly decreased bone resorption. RhoEgt/gt OCs formed fewer podosomes but with larger cores compared with RhoE+/+ OCs. FRAP experiments targeting actin in podosomes revealed a significantly slower actin turnover in clusters and rings of RhoE-deficient OCs. Furthermore, deletion of RhoE resulted in smaller SZs in OCs seeded on bone. Besides RhoE, RhoA is another member of the GTPase superfamily that is also required for podosome patterning and SZ spreading (Chellaiah et al., 2003; Destaing et al., 2005). However, in contrast to the results of RhoE knockdown, the chemical inhibition of Rho resulted in the spreading of SZ to a belt at the cell periphery of OCs seeded on ACC (Saltel et al., 2004). The reversed consequences of RhoA and RhoE inhibitions suggest antagonistic functions of these proteins in OCs. In addition to RhoE and RhoA, other members of the GTPase superfamily, such as Rac1/2, Cdc42, ARF6, RhoU, and Rab, are involved in different OC functions (reviewed in Itzstein et al., 2011; Ory et al., 2008). How the role of these GTPases are coordinated within the OC is an open question.
The increase in the number of OCs with rings in the RhoE-depleted OCs led us to investigate ring-driven saltatory migration, which characterizes OC motility. We previously described OC migration as consistent with a long phase of movement in the same direction followed by a short phase consisting of a 90° switch in direction (Hu et al., 2011). In the present study, we provide new quantifiable parameters of this mode of migration, demonstrating that it is considerably altered in RhoEgt/gt OCs, namely by decrease in the velocity of displacement and increase in the frequency of 90° angle turns. Altered RhoEgt/gt OC migration was also characterized by global loss of persistence. These observations make it tempting to speculate that the slow actin turnover rate in RhoEgt/gt OC rings causes their impaired migration and SZ organization.
To find the molecular pathway through which RhoE modulates actin kinetics in OCs, we investigated downstream phosphorylation targets of a well-described RhoE partner, Rock-I (Riento et al., 2003). In contrast to its upstream activator Rho (Chellaiah et al., 2000b; Ory et al., 2008), Rock-I downstream signaling toward podosomes has been poorly studied. However, in nonmyeloid cells, the direct binding of RhoE to Rock-I, but not to Rock-II, in 1:1 complex dimers inhibits the kinase activity of Rock-I (Komander et al., 2008). This Rock-I–RhoE interaction destabilizes actin-based focal adhesions and stress fibers when RhoE is overexpressed (Klein and Aplin, 2009). Here we investigated Rock-I phosphorylation of two targets, Mypt1 and cofilin, in OCs. Mypt1 is the main subunit of the myosin light-chain phosphatase, which activates actomyosin-II contractility (Ito et al., 2004). We asked whether Mypt1 was a Rock-I target in OCs and could explain myosin-II–dependent regulation of podosome size and patterning as suggested in previous studies (Linder and Kopp, 2005; Luxenburg et al., 2012; van den Dries et al., 2013a). The chemical inhibition of Rock activity revealed that Mypt1 phosphorylation status at its primary phosphorylation site, Thr-696, is not Rock dependent in OCs. On the other hand, cofilin is an actin-severing protein that regulates F-actin assembly and disassembly, thus promoting fast actin turnover (Lappalainen and Drubin, 1997; Maekawa et al., 1999). Nonphosphorylated cofilin is the active form, and its phosphorylation on Ser-3 blocks its activity (Abe et al., 1996; Arber et al., 1998). Furthermore, both total and phosphorylated cofilin were shown to localize at podosome clusters and SZ-like structures, but how they regulate these structures was unknown (Blangy et al., 2012; Touaitahuata et al., 2013). We first confirmed that, in OCs, Rock-I phosphorylation of cofilin at Ser-3 is partially inhibited by RhoE. Indeed, when RhoE was not expressed, Rock-I activity could not be blocked, and p(Ser3)-cofilin levels markedly increased. Conversely, in RhoEgt/gt OCs, the chemical inhibition of Rock-I kinase activity mimicked RhoE expression by reducing the increased level of p(Ser3)-cofilin.
Next we validated that nonphosphorylated, active cofilin is needed for bone resorption and that its phosphorylation, that is, inactivation, justifies why RhoEgt/gt OC cannot resorb bone. Indeed, the use of cucurbitacin E, which inhibits cofilin phosphorylation, strongly and dose dependently promotes bone resorption in both RhoE+/+ and RhoEgt/gt OCs. Strikingly, chemical rescue of cofilin activity was sufficient to fully restore resorption activity in the absence of RhoE.
Our findings reveal the RhoE-Rock-cofilin pathway as a new essential mechanism for OC-mediated bone resorption. This pathway ensures the fast actin dynamics needed for podosome organization, collective patterning, ring-driven migration, and sealing zone formation. It also consolidates the emerging model that dissociates podosome dynamics from OC adhesion (Hu et al., 2011) and differentiation (Touaitahuata et al., 2013).
MATERIALS AND METHODS
Ethics statements
All animal procedures were approved by the local ethics committee for animal welfare of the Universidad CEU Cardenal Herrera, ID# CEBA03/2007, and met local guidelines, European regulations (EU Directive 86/609), and Standards for Use of Laboratory Animals (5388-01; National Institutes of Health).
Primary human cell cultures
All of the human cell types used in this study were differentiated directly or indirectly from peripheral monocytes from healthy donor blood (Etablissement Français du Sang, Lyon Gerland, France). All of the human recombinant cytokines used for human cultures were obtained from PeproTech (Rocky Hill, NJ). Briefly, CD14+ Mos were isolated from blood after two successive gradient centrifugations in Leukocyte Separation Medium (CMSMSL01-01; Eurobio, Courtaboeuf, France) and 50% Percoll (Gallois et al., 2010), respectively, followed by negative antibody selection. CD14+ CD16+ Mo were then differentiated into OCs in α-MEM (22561; Life Technologies, Carlsbad, CA) with 10% fetal bovine serum (BioWest, Nuaillé, France), 100 U/ml penicillin, 100 μg/ml streptomycin (15070-063; Life Technologies), 2 mM l-glutamine (25030081; Life Technologies). 50 ng/ml human recombinant M-CSF, and 30 ng/ml human recombinant RANKL. After the first 3 d of culture, medium was changed every 48 h with 25 ng/ml M-CSF and 100 ng/ml RANKL until the end of the culture (5–6 d). Mos were also differentiated in suspension into immature (i.e., CD1a+, CD14−, CD16−) DCs in RPMI with 10% fetal bovine serum, 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 2 mM l-glutamine, 100 ng/ml GM-CSF, and 10 ng/ml IL-4 for 6 d. Experimental details of Mo isolation and differentiation and DC differentiation can be found in Gallois et al. (2010). DCs were then differentiated into either OCs using the same conditions as for Mo-OCs (Rivollier et al., 2004) or into different MGCs such as DC-γ17-MGC (Coury et al., 2008) with or without IFN-γ.
Affymetrix GeneChip assays and comparative analysis
Primary human cultures of the following cell types were performed from several donors: Mos (two), DCs (four), Mo-OCs (two), DC-OCs (two), and DC-derived MGCs with IL-17 (two) or with IL-17 and IFN-γ (three). RNA extraction, antisense cRNA target labeling, array hybridization, and scanning were performed as shown previously (Gallois et al., 2010). Intra-assay and interassay normalizations were performed according to manufacturer's instructions, and gene expression values were averaged per cell type over all donors. When the mean expression value of a gene in a given cell type was superior to the average of all gene means in the same cell type, the gene was considered as expressed. As such, genes commonly expressed in both Mo-OCs and DC-OCs but not expressed in any other cell type used in this study were considered for further investigation.
For the mathematical analysis, after intrachip and interchip normalization, we calculated the mean expression level of a specific RNA in a given cell type (Xc) as follows:
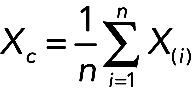 |
where Xc is the mean expression of an RNA X(i) in a cell type c and i is the number of donors for this cell type.
Then we calculated the overall RNA expression per cell type (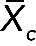 ) as follows:
) as follows:
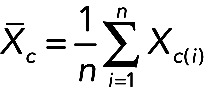 |
where 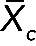 is the average expression of all probed genes Xc(i) in a cell type c and i is the number of probe sets (i = 54,675 for all cell types).
is the average expression of all probed genes Xc(i) in a cell type c and i is the number of probe sets (i = 54,675 for all cell types).
Gene Ontology molecular function enrichment
Molecular functions of 115 genes resulting from the transcriptomic assay were annotated according to Gene Ontology, and their percentages were compared against the human genome using the online software FatiGO (Al-Shahrour et al., 2006). Only statistically overexpressed functions in our specific gene set were represented. Statistical relevance was determined using the Fischer test.
Taqman low-density array
Mos, DCs, Mo-OCs, DC-OCs, and DC-17γ-MGCs triplicate cultures were obtained from three different human blood donors. RNA was extracted as described earlier, quantified with a NanoDrop (ThermoFisher Scientific, Waltham, MA), quality checked with an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA), and finally reverse transcribed using iScript cDNA Synthesis Kit (170-8891; Bio-Rad, Hercules, CA) following kit protocol. Amplification was performed using a made-to-order Taqman Gene Expression Assay (Agilent Technologies) according to manufacturer's instructions. After verification with 18S (Hs99999901_s1) amplification (standard microfluidics technical control for amplification runs), relative cDNA quantities (RQs) calculated based on cycle thresholds (Ct) were normalized with SEC61B (Hs00606455_m1) and TBP (Hs00920494_m1) housekeeping genes. Then the differential expression for each gene in resorbing cells (i.e., Mo-OCs and DC-OCs) was compared with that of nonresorbing cells (i.e., Mos, DCs, and DC-17 γ -MGCs) by using the latter as background in the limma parametric test (Limma package; Affymetrix, Santa Clara, CA) combined with the Benjamini–Hochberg false discovery rate p-value adjustment. Adjusted p < 0.05 was considered significant.
Primary murine osteoclast cultures and transfection
OCs were differentiated from myeloid precursors extracted from RhoE+/+, RhoE+/−, and RhoEgt/gt mice (Mocholi et al., 2011). E15–E18 FL or neonatal BM or spleen (Sp) precursors were isolated and differentiated into OCs following the protocol previously described (Destaing et al., 2003; Chabadel et al., 2007; Schmidt et al., 2011). For each experiment, all cultures were started from the same source tissue (i.e., FL, BM, or Sp) from littermates killed at the same time. Once extracted from their respective tissues by flushing (in the case of BM) or grinding against a 100-μm-pore nylon cell strainer (in the case of FL and Sp), these precursor cells were purified from the gradient interface after centrifugation on a leukocyte separation medium at 1250 × g for 20 min at 20°C. Cells were then washed by centrifugation and put in culture in α-MEM with 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM l-glutamine, 20 ng/ml murine recombinant M-CSF, and murine RANKL (produced by the Production d'analyse des protéines platform, UMS3444, Lyon-Gerland, France). The source of precursors for each experiment is mentioned in the Results. For transfection, day-4 OCs were detached with 0.25 mM EDTA in warm phosphate-buffered saline (PBS) and transfected using the Neon MP-100 electroporator (Life Technologies). Briefly, 0.5 × 106 cells were resuspended with 2 μg of pEGFP-C1-RhoE (23229; Addgene, Cambridge, MA) plasmid DNA in 10 μl of Buffer R (see kit protocol) and then jolted with a 1720-V current in two 10-ms pulses. Electroporated cells were then seeded on coverslips with prewarmed medium and kept overnight. Medium was renewed the following day. For the inhibition of Rock activity with Y-27632 (Sigma-Aldrich, St. Louis, MO), mature OC cultures were serum starved for 2 h and then serum-induced with or without Y-27632 for 2 h. For the inhibition of cofilin phosphorylation at Ser-3, the medium of mature OC cultures was changed with or without addition of cucurbitacin E (Sigma-Aldrich; Nakashima et al., 2010).
Differentiation assays
To quantify differentiation and fusion of FL-derived OCs (FL-OCs) from RhoE+/+, RhoE+/gt, and RhoEgt/gt mice (Mocholi et al., 2011), the same number of precursors (2 × 104 cells/well) from E15.5 littermates was seeded in 96-well, culture-treated plates and on bones slices of the same size. They were then fixed at several time points (days 5, 6, and 8 postseeding) with 4% (wt/vol) paraformaldehyde in PBS for 15 min at room temperature and then stained at 37°C using a TRAP assay kit (Sigma-Aldrich) according to manufacturer's instructions. Stained cells with three or more nuclei were counted as OCs.
Resorption assays
Two methods were used to quantify the resorbed surface per murine OC (i.e., resorption index). For resorption on bovine cortical bone slices, day-8 FL-OCs were assayed for differentiation, and then resorption was measured (Destaing et al., 2003; Harre et al., 2012). For resorption on bone-mimicking Osteo Assay Surface (3988; Corning, Tewksbury, MA), day-4 FL-OCs were detached from plastic wells by flushing after incubation at 37°C for 5 min in PBS plus 0.25 mM EDTA, counted, and then seeded at the same number (2 × 104 cells/well in 96-well plates) in replicate plates and cultured for 4 8h. To measure the total surface of the resorbed matrix, OCs were washed off with distilled water, and then the matrix was stained with a 5% (wt/vol) silver nitrate solution. In parallel wells, the number of OCs was determined by TRAP staining as mentioned previously. Finally, the resorption index was obtained by dividing the total resorbed area per well by the total number of OCs per well.
To determine the resorption index of human DC-OCs and DC-17γ-MGCs on coverslips coated with ACC (Shibutani et al., 2000; Saltel et al., 2004), day-4 DC-OCs and day-14 DC-17γ-MGCs (i.e., stages of differentiation in which both cell types are terminally differentiated; Rivollier et al., 2004; Coury et al., 2008) were detached using 1× Accutase (Sigma-Aldrich) and replated on ACC coverslips with RANKL and M-CSF stimulation to promote resorption. The resorbed area per cell was determined by the absence of matrix crystal complexes subjacent to the cell.
Images of all substrates were captured with an AxioImager microscope (Carl Zeiss, Jena, Germany) topped with a charge-coupled device CoolSnap color camera (Photometrics, Tucson, AZ) and manually quantified with ImageJ (National Institutes of Health, Bethesda, MD).
Random migration assay
Day-4 Sp-OCs were detached and replated at low confluence (5 × 104 cells/well in a 24-well plate) to solicit random (nondirected) OC migration. Cells were given 2 h to adhere and then were imaged in phase contrast on an inverted Axiovert 100M microscope (Carl Zeiss) for 14 h at 37°C with saturated humidity. Images were acquired every 5 min. Consecutive cell positions were determined manually using the MTrackJ plug-in in ImageJ at 20-min intervals, so that displacements were significantly greater than imprecisions due to manual tracking. Small front-to-back polarized OCs that were not in contact with other cells were tracked. In other words, OCs that were fusing or without visible leading and trailing edges were not considered for this assay. Instantaneous velocity (Vi) was defined as the distance traveled between two consecutive points divided by the corresponding time interval (20 min). Instantaneous angular change (Δαi) was defined as the result of the subtraction between the angles of velocity vectors of two consecutive displacements. Persistence of movement (Pi) was defined as the ratio di/Di, with di defined as the direct distance between the position i and the position at the start of the track and Di defined as the length of the track at a given position i.
Fluorescence labeling and confocal microscopy of fixed cells
OCs in culture on coverslips or glass-bottom plates (MatTek Corporation, Ashland, MA) were fixed with 4% paraformaldehyde in PBS for 10 min at 37°C and permeabilized using PBS plus 0.2% Triton X-100 in PBS. All antibody dilutions and washes were thereafter performed with PBS plus 0.1% Triton X-100. Used primary antibodies were anti-vinculin (5 μg/ml final, clone hVIN-1, V9264; Sigma-Aldrich), anti-cortactin (10 μg/ml final, 05-180, clone 4F11; Millipore, Billerica, MA), and anti-paxillin (10 μg/ml final, 610051; BD Biosciences, Franklin Lakes, NJ). Secondary antibodies Alexa Fluor 488 anti-mouse immunoglobulin G (IgG; A11029; Life Technologies) and Alexa Fluor 488 anti-rabbit IgG (A-21245; Life Technologies) were used at 2 μg/ml. Then Alexa Fluor 647–phalloidin (A22287; Life Technologies) was diluted 1:100. Image acquisition was performed with either a DMI4000 microscope (Leica Microsystems, Wetzlar, Germany) equipped with a spinning disk unit CUS22 (Yokogawa, Tokyo, Japan) or an SP7 spectral confocal microscope (Leica Microsystems). For podosome colocalization studies, all images were acquired with the same excitation and detection parameters without reaching signal saturation. To perform pixel-to-pixel multiplication of 16-bit images using ImageJ, nonspecific signal obtained from negative control micrographs (cells with only secondary antibodies) was subtracted from the “green micrographs” on coverslips treated with primary and secondary antibodies. Background noise in “red micrographs” (where Alexa Fluor 647–phalloidin was used) was measured outside the cells and subtracted from each image. For each cell, the green channel image was multiplied with the red channel using the multiplication option of the Image Calculator plug-in in ImageJ, and the result was represented as a 32-bit float image with a ratio lookup table. In the resulting image, the intensity value of a given pixel is therefore the result of the multiplication of the intensity values of the pixels with the same coordinates from the red and green channels. For measurement of SZ sizes on bone slices, contrast minima per field were raised so that bone autofluorescence was no longer observed. For podosome core size measurement, background noise was measured outside of the cells and subtracted from entire images. The podosome core was delimited based on local intensity thresholds using the Find Peaks plug-in in ImageJ with the following parameters: Gaussian blur, 0.72; background method, SD above mean; background parameter, 0; statistics mode, both; background level, 24; search method, half peak value; remove edge maxima, On.
FRAP acquisition and analysis
Day-4 FL-OCs were infected with second-generation packaging viral particles (provided by the Anira-Vectorologie platform, UMS3444, Lyon-Gerland, France) containing a pLVX-LifeAct-mCherry lentiviral construct (obtained by inserting LifeAct coding sequence between EcoR1 and BamH1 sites in pLVX-mCherry-N1 vector [632562, Clontech, Mountain View, CA]) at a multiplicity of infection of 10:1 in 1 ml/well of a six-well plate. OCs were incubated overnight, and medium was changed in the morning. OCs with clusters and rings were fluorescent on day 6 (48 h postinfection) and imaged using a DMI4000 microscope (Leica Microsystems) equipped with a spinning disk unit and a FRAP module piloted by iLas2 software (Roper Scientific, Tucson, AZ). mCherry was excited with a 561-nm laser and bleached with a 473-nm laser at 100% in three iterations. Acquisition was optimized to obtain maximal signal without any saturated pixels and defined in three temporal phases: prebleach phase (five acquisitions, 5-s time interval between consecutive acquisitions), first postbleach phase (20 acquisitions, 3-s interval), and second postbleach phase (11 acquisitions, 30-s interval). The mean relative fluorescence intensity (RFI) of a given region of interest was measured and normalized as mentioned in Negi and Olson (2006). The obtained RFIs were curve fitted and plotted using the formula in Negi and Olson (2006) with consideration of the guidelines in Hardy (2012). Statistical analysis of recovery half-times was calculated from data fitting using the “Do the best fit values of selected parameters differ between datasets differ” function in Prism 6 (GraphPad, La Jolla, CA; Weisswange et al., 2009).
Immunoblotting
Day-5 BM-derived OCs (BM-OCs) were lysed in 2× Laemmli sampling buffer, run on SDS–PAGE, and blotted on polyvinylidene fluoride membranes. Primary antibodies were anti-cofilin (1:1000, clone D3F9, 5157; Cell Signaling, Beverly, MA), anti–phospho(Ser-3)-cofilin (1:1000, clone 77G2, 3313; Cell Signaling), anti–β-tubulin (0.3 mg/ml final, T4026; Sigma-Aldrich), anti–Rock-I (1:1000, clone C8F7, 4036; Cell Signaling), anti-Mypt1 (10 μg/ml, A300-888A; Bethyl Laboratories, Montgomery, TX), and anti-p(696)-Mypt1 (1:1000, STA415; Cell Biolabs, San Diego, CA). Enhanced chemiluminescence detection was performed using Amersham ECL Prime Western Blotting Detection Reagent (RPN2232; GE Healthcare, Piscataway, NJ).
Flow cytometry
Experimental procedures and antibodies are described in Schmidt et al. (2011).
Statistics
Comparison of means was performed with the two-tailed Mann–Whitney nonparametric test in Prism 6 (GraphPad). Unless noted otherwise, values mentioned in the text are mean ± SEM. Confidence level for all tests was 95%. *p < 0.05 is considered significant, **p < 0.01 highly significant, and ***p < 0.001 very highly significant. ns, nonsignificant. Error bars for each sample are defined in the figure legends.
Supplementary Material
Acknowledgments
We acknowledge the contributions of the following from the SFR Biosciences Gerland-Lyon Sud (UMS344/US8) facilities: C. Lionnet, Plateau Technique Imagerie/Microscopie, for valuable input on microscopy; the AniRA-Plateau de Biologie Expérimentale de la Souris for animal care; the AniRA-Vectorologie for lentiviral particle production; and the PAP (Production et Analyse des Proteines) platform for murine RANKL production. D.G. is an Early Stage Researcher at the Tissue Transmigration Training Network (T3-Net) funded by the European Commission under Framework Program 7. D.G. is also funded by the Fondation-ARC (Aides individuelles jeunes chercheurs, ref. DOC20120605264). This project received research grants from the Fondation ARC (Subvention fixes, ref. SFI20101201877), the Fondation Arthritis Courtin, and the PRCEU-UCH22/12 (Projecto Centro de estudios Universitarios de la Universidad Cardenal Herrera numero 22 del 2012).
Abbreviations used:
- BM
bone marrow
- DC
immature dendritic cell
- DC-17γ-MGC
multinucleated giant cell derived from dendritic cell by stimulation with interleukin-17 and interferon-γ
- DC-OC
dendritic cell–derived osteoclast
- FL
fetal liver
- FRAP
fluorescence recovery after photobleaching
- gt
gene trap
- IFN-γ
interferon-γ
- IL-17
interleukin-17
- M-CSF
macrophage colony stimulating factor
- MGC
multinucleated giant cell
- Mo
monocyte
- Mo-OC
monocyte-derived osteoclast
- OAS
Osteo Assay substrate
- OC
osteoclast
- RANKL
receptor activator of nuclear factor κB
- SZ
sealing zone
- TRAP
tartrate-resistant acid phosphatase
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-07-0363) November 27, 2013.
*These authors contributed equally.
REFERENCES
- Abe H, Obinata T, Minamide LS, Bamburg JR. Xenopus laevis actin-depolymerizing factor/cofilin: a phosphorylation-regulated protein essential for development. J Cell Biol. 1996;132:871–885. doi: 10.1083/jcb.132.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida CG, Yamada A, Tenza D, Louvard D, Raposo G, Coudrier E. Myosin 1b promotes the formation of post-Golgi carriers by regulating actin assembly and membrane remodelling at the trans-Golgi network. Nat Cell Biol. 2011;13:779–789. doi: 10.1038/ncb2262. [DOI] [PubMed] [Google Scholar]
- Al-Shahrour F, Minguez P, Tarraga J, Montaner D, Alloza E, Vaquerizas JM, Conde L, Blaschke C, Vera J, Dopazo J. BABELOMICS: a systems biology perspective in the functional annotation of genome-scale experiments. Nucleic Acids Res. 2006;34:W472–W476. doi: 10.1093/nar/gkl172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber S, Barbayannis FA, Hanser H, Schneider C, Stanyon CA, Bernard O, Caroni P. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 1998;393:805–809. doi: 10.1038/31729. [DOI] [PubMed] [Google Scholar]
- Babb SG, Matsudaira P, Sato M, Correia I, Lim SS. Fimbrin in podosomes of monocyte-derived osteoclasts. Cell Motil Cytoskeleton. 1997;37:308–325. doi: 10.1002/(SICI)1097-0169(1997)37:4<308::AID-CM3>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Biosse Duplan M, Zalli D, Stephens S, Zenger S, Neff L, Oelkers JM, Lai FP, Horne W, Rottner K, Baron R. Microtubule dynamic instability controls podosome patterning in osteoclasts through EB1, cortactin and Src. Mol Cell Biol. 2013. doi: 10.1128/MCB.00578-13. [DOI] [PMC free article] [PubMed]
- Blangy A, Touaitahuata H, Cres G, Pawlak G. Cofilin activation during podosome belt formation in osteoclasts. PLoS One. 2012;7:e45909. doi: 10.1371/journal.pone.0045909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce BF. Advances in osteoclast biology reveal potential new drug targets and new roles for osteoclasts. J Bone Mineral Res. 2013;28:711–722. doi: 10.1002/jbmr.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo-Cordero JJ, Magalhaes MA, Eddy RJ, Hodgson L, Condeelis J. Functions of cofilin in cell locomotion and invasion. Nat Rev Mol Cell Biol. 2013;14:405–415. doi: 10.1038/nrm3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzzaniti A, Neff L, Sanjay A, Horne WC, De Camilli P, Baron R. Dynamin forms a Src kinase-sensitive complex with Cbl and regulates podosomes and osteoclast activity. Mol Biol Cell. 2005;16:3301–3313. doi: 10.1091/mbc.E04-12-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabadel A, Banon-Rodriguez I, Cluet D, Rudkin BB, Wehrle-Haller B, Genot E, Jurdic P, Anton IM, Saltel F. CD44 and beta3 integrin organize two functionally distinct actin-based domains in osteoclasts. Mol Biol Cell. 2007;18:4899–4910. doi: 10.1091/mbc.E07-04-0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang EJ, et al. Brain-type creatine kinase has a crucial role in osteoclast-mediated bone resorption. Nat Med. 2008;14:966–972. doi: 10.1038/nm.1860. [DOI] [PubMed] [Google Scholar]
- Chellaiah M, Kizer N, Silva M, Alvarez U, Kwiatkowski D, Hruska KA. Gelsolin deficiency blocks podosome assembly and produces increased bone mass and strength. J Cell Biol. 2000a;148:665–678. doi: 10.1083/jcb.148.4.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellaiah MA, Biswas RS, Rittling SR, Denhardt DT, Hruska KA. Rho-dependent Rho kinase activation increases CD44 surface expression and bone resorption in osteoclasts. J Biol Chem. 2003;278:29086–29097. doi: 10.1074/jbc.M211074200. [DOI] [PubMed] [Google Scholar]
- Chellaiah MA, Soga N, Swanson S, McAllister S, Alvarez U, Wang D, Dowdy SF, Hruska KA. Rho-A is critical for osteoclast podosome organization, motility, and bone resorption. J Biol Chem. 2000b;275:11993–12002. doi: 10.1074/jbc.275.16.11993. [DOI] [PubMed] [Google Scholar]
- Coury F, et al. Langerhans cell histiocytosis reveals a new IL-17A-dependent pathway of dendritic cell fusion. Nat Med. 2008;14:81–87. doi: 10.1038/nm1694. [DOI] [PubMed] [Google Scholar]
- Croke M, Ross FP, Korhonen M, Williams DA, Zou W, Teitelbaum SL. Rac deletion in osteoclasts causes severe osteopetrosis. J Cell Sci. 2011;124:3811–3821. doi: 10.1242/jcs.086280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destaing O, Saltel F, Geminard JC, Jurdic P, Bard F. Podosomes display actin turnover and dynamic self-organization in osteoclasts expressing actin-green fluorescent protein. Mol Biol Cell. 2003;14:407–416. doi: 10.1091/mbc.E02-07-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destaing O, Saltel F, Gilquin B, Chabadel A, Khochbin S, Ory S, Jurdic P. A novel Rho-mDia2-HDAC6 pathway controls podosome patterning through microtubule acetylation in osteoclasts. J Cell Sci. 2005;118:2901–2911. doi: 10.1242/jcs.02425. [DOI] [PubMed] [Google Scholar]
- Destaing O, Sanjay A, Itzstein C, Horne WC, Toomre D, De Camilli P, Baron R. The tyrosine kinase activity of c-Src regulates actin dynamics and organization of podosomes in osteoclasts. Mol Biol Cell. 2008;19:394–404. doi: 10.1091/mbc.E07-03-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong LT, Lakkakorpi PT, Nakamura I, Machwate M, Nagy RM, Rodan GA. PYK2 in osteoclasts is an adhesion kinase, localized in the sealing zone, activated by ligation of alpha(v)beta3 integrin, and phosphorylated by src kinase. J Clin Invest. 1998;102:881–892. doi: 10.1172/JCI3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faccio R, Teitelbaum SL, Fujikawa K, Chappel J, Zallone A, Tybulewicz VL, Ross FP, Swat W. Vav3 regulates osteoclast function and bone mass. Nat Med. 2005;11:284–290. doi: 10.1038/nm1194. [DOI] [PubMed] [Google Scholar]
- Feng S, Deng L, Chen W, Shao J, Xu G, Li YP. Atp6v1c1 is an essential component of the osteoclast proton pump and in F-actin ring formation in osteoclasts. Biochem J. 2009;417:195–203. doi: 10.1042/BJ20081073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallois A, Lachuer J, Yvert G, Wierinckx A, Brunet F, Rabourdin-Combe C, Delprat C, Jurdic P, Mazzorana M. Genome-wide expression analyses establish dendritic cells as a new osteoclast precursor able to generate bone-resorbing cells more efficiently than monocytes. J Bone Miner Res. 2010;25:661–672. doi: 10.1359/jbmr.090829. [DOI] [PubMed] [Google Scholar]
- Guasch RM, Scambler P, Jones GE, Ridley AJ. RhoE regulates actin cytoskeleton organization and cell migration. Mol Cell Biol. 1998;18:4761–4771. doi: 10.1128/mcb.18.8.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy LR. Fluorescence recovery after photobleaching (FRAP) with a focus on F-actin. Curr Protoc Neurosci. 2012 doi: 10.1002/0471142301.ns0217s61. Chapter 2, Unit 2.17. [DOI] [PubMed] [Google Scholar]
- Harre U, et al. Induction of osteoclastogenesis and bone loss by human autoantibodies against citrullinated vimentin. J Clin Invest. 2012;122:1791–1802. doi: 10.1172/JCI60975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Planus E, Georgess D, Place C, Wang X, Albiges-Rizo C, Jurdic P, Geminard JC. Podosome rings generate forces that drive saltatory osteoclast migration. Mol Biol Cell. 2011;22:3120–3126. doi: 10.1091/mbc.E11-01-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Nakano T, Erdodi F, Hartshorne DJ. Myosin phosphatase: structure, regulation and function. Mol Cell Biochem. 2004;259:197–209. doi: 10.1023/b:mcbi.0000021373.14288.00. [DOI] [PubMed] [Google Scholar]
- Itzstein C, Coxon FP, Rogers MJ. The regulation of osteoclast function and bone resorption by small GTPases. Small GTPases. 2011;2:117–130. doi: 10.4161/sgtp.2.3.16453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurdic P, Saltel F, Chabadel A, Destaing O. Podosome and sealing zone: specificity of the osteoclast model. Eur J Cell Biol. 2006;85:195–202. doi: 10.1016/j.ejcb.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Klein RM, Aplin AE. Rnd3 regulation of the actin cytoskeleton promotes melanoma migration and invasive outgrowth in three dimensions. Cancer Res. 2009;69:2224–2233. doi: 10.1158/0008-5472.CAN-08-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Garg R, Wan PT, Ridley AJ, Barford D. Mechanism of multi-site phosphorylation from a ROCK-I:RhoE complex structure. EMBO J. 2008;27:3175–3185. doi: 10.1038/emboj.2008.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen P, Drubin DG. Cofilin promotes rapid actin filament turnover in vivo. Nature. 1997;388:78–82. doi: 10.1038/40418. [DOI] [PubMed] [Google Scholar]
- Linder S, Aepfelbacher M. Podosomes: adhesion hot-spots of invasive cells. Trends Cell Biol. 2003;13:376–385. doi: 10.1016/s0962-8924(03)00128-4. [DOI] [PubMed] [Google Scholar]
- Linder S, Kopp P. Podosomes at a glance. J Cell Sci. 2005;118:2079–2082. doi: 10.1242/jcs.02390. [DOI] [PubMed] [Google Scholar]
- Luxenburg C, Geblinger D, Klein E, Anderson K, Hanein D, Geiger B, Addadi L. The architecture of the adhesive apparatus of cultured osteoclasts: from podosome formation to sealing zone assembly. PLoS One. 2007;2:e179. doi: 10.1371/journal.pone.0000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxenburg C, Parsons JT, Addadi L, Geiger B. Involvement of the Src-cortactin pathway in podosome formation and turnover during polarization of cultured osteoclasts. J Cell Sci. 2006;119:4878–4888. doi: 10.1242/jcs.03271. [DOI] [PubMed] [Google Scholar]
- Luxenburg C, Winograd-Katz S, Addadi L, Geiger B. Involvement of actin polymerization in podosome dynamics. J Cell Sci. 2012;125:1666–1672. doi: 10.1242/jcs.075903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Sadashivaiah K, Madayiputhiya N, Chellaiah MA. Regulation of sealing ring formation by L-plastin and cortactin in osteoclasts. J Biol Chem. 2010;285:29911–29924. doi: 10.1074/jbc.M109.099697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Samanna V, Chellaiah MA. Dramatic inhibition of osteoclast sealing ring formation and bone resorption in vitro by a WASP-peptide containing pTyr294 amino acid. J Mol Signal. 2008;3:4. doi: 10.1186/1750-2187-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa M, Ishizaki T, Boku S, Watanabe N, Fujita A, Iwamatsu A, Obinata T, Ohashi K, Mizuno K, Narumiya S. Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science. 1999;285:895–898. doi: 10.1126/science.285.5429.895. [DOI] [PubMed] [Google Scholar]
- Meddens MB, Rieger B, Figdor CG, Cambi A, van den Dries K. Automated podosome identification and characterization in fluorescence microscopy images. Microsc Microanal. 2013;19:180–189. doi: 10.1017/S1431927612014018. [DOI] [PubMed] [Google Scholar]
- Mocholi E, Ballester-Lurbe B, Arque G, Poch E, Peris B, Guerri C, Dierssen M, Guasch RM, Terrado J, Perez-Roger I. RhoE deficiency produces postnatal lethality, profound motor deficits and neurodevelopmental delay in mice. PLoS One. 2011;6:e19236. doi: 10.1371/journal.pone.0019236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreaux J, Hose D, Kassambara A, Reme T, Moine P, Requirand G, Goldschmidt H, Klein B. Osteoclast-gene expression profiling reveals osteoclast-derived CCR2 chemokines promoting myeloma cell migration. Blood. 2011;117:1280–1290. doi: 10.1182/blood-2010-04-279760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Berreur M, Blanchard F, Chevalier C, Guisle-Marsollier I, Masson M, Redini F, Heymann D. Receptor activator of nuclear factor-kappaB ligand (RANKL) directly modulates the gene expression profile of RANK-positive Saos-2 human osteosarcoma cells. Oncol Rep. 2007;18:1365–1371. [PubMed] [Google Scholar]
- Nakashima S, Matsuda H, Kurume A, Oda Y, Nakamura S, Yamashita M, Yoshikawa M. Cucurbitacin E as a new inhibitor of cofilin phosphorylation in human leukemia U937 cells. Bioorg Med Chem Lett. 2010;20:2994–2997. doi: 10.1016/j.bmcl.2010.02.062. [DOI] [PubMed] [Google Scholar]
- Negi SS, Olson MO. Effects of interphase and mitotic phosphorylation on the mobility and location of nucleolar protein B23. J Cell Sci. 2006;119:3676–3685. doi: 10.1242/jcs.03090. [DOI] [PubMed] [Google Scholar]
- Olsson Akefeldt S, et al. Chemoresistance of human monocyte-derived dendritic cells is regulated by IL-17A. PLoS One. 2013;8:e56865. doi: 10.1371/journal.pone.0056865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ory S, Brazier H, Pawlak G, Blangy A. Rho GTPases in osteoclasts: orchestrators of podosome arrangement. Eur J Cell Biol. 2008;87:469–477. doi: 10.1016/j.ejcb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Ory S, Munari-Silem Y, Fort P, Jurdic P. Rho and Rac exert antagonistic functions on spreading of macrophage-derived multinucleated cells and are not required for actin fiber formation. J Cell Sci. 2000;113(Pt 7):1177–1188. doi: 10.1242/jcs.113.7.1177. [DOI] [PubMed] [Google Scholar]
- Riento K, Guasch RM, Garg R, Jin B, Ridley AJ. RhoE binds to ROCK I and inhibits downstream signaling. Mol Cell Biol. 2003;23:4219–4229. doi: 10.1128/MCB.23.12.4219-4229.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riento K, Villalonga P, Garg R, Ridley A. Function and regulation of RhoE. Biochem Soc Trans. 2005;33:649–651. doi: 10.1042/BST0330649. [DOI] [PubMed] [Google Scholar]
- Rivollier A, Mazzorana M, Tebib J, Piperno M, Aitsiselmi T, Rabourdin-Combe C, Jurdic P, Servet-Delprat C. Immature dendritic cell transdifferentiation into osteoclasts: a novel pathway sustained by the rheumatoid arthritis microenvironment. Blood. 2004;104:4029–4037. doi: 10.1182/blood-2004-01-0041. [DOI] [PubMed] [Google Scholar]
- Saltel F, Destaing O, Bard F, Eichert D, Jurdic P. Apatite-mediated actin dynamics in resorbing osteoclasts. Mol Biol Cell. 2004;15:5231–5241. doi: 10.1091/mbc.E04-06-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S, Nakchbandi I, Ruppert R, Kawelke N, Hess MW, Pfaller K, Jurdic P, Fassler R, Moser M. Kindlin-3-mediated signaling from multiple integrin classes is required for osteoclast-mediated bone resorption. J Cell Biol. 2011;192:883–897. doi: 10.1083/jcb.201007141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibutani T, Iwanaga H, Imai K, Kitago M, Doi Y, Iwayama Y. Use of glass slides coated with apatite-collagen complexes for measurement of osteoclastic resorption activity. J Biomed Mater Res. 2000;50:153–159. doi: 10.1002/(sici)1097-4636(200005)50:2<153::aid-jbm9>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Speziani C, et al. Murine dendritic cell transdifferentiation into osteoclasts is differentially regulated by innate and adaptive cytokines. Eur J Immunol. 2007;37:747–757. doi: 10.1002/eji.200636534. [DOI] [PubMed] [Google Scholar]
- Takayanagi H. The role of NFAT in osteoclast formation. Ann NY Acad Sci. 2007;1116:227–237. doi: 10.1196/annals.1402.071. [DOI] [PubMed] [Google Scholar]
- Takayanagi H, et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell. 2002;3:889–901. doi: 10.1016/s1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- Touaitahuata H, Planus E, Albiges-Rizo C, Blangy A, Pawlak G. Podosomes are dispensable for osteoclast differentiation and migration. Eur J Cell Biol. 2013;92:139–149. doi: 10.1016/j.ejcb.2013.03.001. [DOI] [PubMed] [Google Scholar]
- van den Dries K, Meddens MB, de Keijzer S, Shekhar S, Subramaniam V, Figdor CG, Cambi A. Interplay between myosin IIA-mediated contractility and actin network integrity orchestrates podosome composition and oscillations. Nat Commun. 2013a;4:1412. doi: 10.1038/ncomms2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Dries K, Schwartz SL, Byars J, Meddens MB, Bolomini-Vittori M, Lidke DS, Figdor CG, Lidke KA, Cambi A. Dual color super-resolution microscopy reveals nanoscale organization of mechanosensory podosomes. Mol Biol Cell. 2013b;24:2112–2123. doi: 10.1091/mbc.E12-12-0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives V, Laurin M, Cres G, Larrousse P, Morichaud Z, Noel D, Cote JF, Blangy A. The Rac1 exchange factor Dock5 is essential for bone resorption by osteoclasts. J Bone Miner Res. 2011;26:1099–1110. doi: 10.1002/jbmr.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A, Baumgartner M, Radziwill G, Dennler J, Moelling K. c-Src is a PDZ interaction partner and substrate of the E3 ubiquitin ligase Ligand-of-Numb protein X1. FEBS Lett. 2007;581:5131–5136. doi: 10.1016/j.febslet.2007.09.062. [DOI] [PubMed] [Google Scholar]
- Weisswange I, Newsome TP, Schleich S, Way M. The rate of N-WASP exchange limits the extent of ARP2/3-complex-dependent actin-based motility. Nature. 2009;458:87–91. doi: 10.1038/nature07773. [DOI] [PubMed] [Google Scholar]
- Zambonin-Zallone A, Teti A, Carano A, Marchisio PC. The distribution of podosomes in osteoclasts cultured on bone laminae: effect of retinol. J Bone Miner Res. 1988;3:517–523. doi: 10.1002/jbmr.5650030507. [DOI] [PubMed] [Google Scholar]
- Zou W, Teitelbaum SL. Integrins, growth factors, and the osteoclast cytoskeleton. Ann NY Acad Sci. 2010;1192:27–31. doi: 10.1111/j.1749-6632.2009.05245.x. [DOI] [PubMed] [Google Scholar]
- Zukauskas A, Merley A, Li D, Ang LH, Sciuto TE, Salman S, Dvorak AM, Dvorak HF, Jaminet SC. TM4SF1: a tetraspanin-like protein necessary for nanopodia formation and endothelial cell migration. Angiogenesis. 2011;14:345–354. doi: 10.1007/s10456-011-9218-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.