Abstract
Transmission electron microscopy and associated methods such as single particle analysis, 2-D crystallography, helical reconstruction and tomography, are highly data-intensive experimental sciences, which also have substantial variability in experimental technique. Object-oriented databases present an attractive alternative to traditional relational databases for situations where the experiments themselves are continually evolving. We present EMEN2, an easy to use object-oriented database with a highly flexible infrastructure originally targeted for transmission electron microscopy and tomography, which has been extended to be adaptable for use in virtually any experimental science. It is a pure object-oriented database designed for easy adoption in diverse laboratory environments, and does not require professional database administration. It includes a full featured, dynamic web interface in addition to APIs for programmatic access. EMEN2 installations currently support roughly 800 scientists worldwide with over 1/2 million experimental records and over 20 TB of experimental data. The software is freely available with complete source.
Keywords: TEM, cryo-EM, database, electronic notebook, software, web interface
Introduction
Every scientist is expected to maintain a complete and accurate record of the experiments they perform. While we have seen an explosion in computational tools that make it easier for us to analyze our data, do literature research, and even write manuscripts, electronic lab notebook (ELN) systems designed to replace traditional paper lab notebooks have not seen widespread adoption. Part of the problem is that ELN systems must solve a nearly impossible problem. How does one store information flexibly enough such that any possible experiment performed in a lab can be recorded, while still organizing that information such that it can be searched and/or mined in a useful way, with enough detail that similar experiments between different scientists can be reasonably compared? In addition, any such system must be at least as easy as a paper lab notebook to use, or else scientists will not have a compelling reason to adopt the electronic system. While groups in many scientific disciplines have developed ontologies to describe the various experiments, classification systems, and terms used in a particular field, the question of how to accurately represent the diverse specific experimental protocols used in different labs, while still maintaining some standardization, remains a difficult question.
A number of projects have attempted to solve the ELN dilemma, often in very different ways. Some microscopy database projects, like the OME (Open Microscopy Environment) (Goldberg et al., 2005; Allan et al., 2012) and Leginon / Appion (Suloway et al., 2005; Lander et al., 2009) focus on accurately recording the sequence of operations performed on images after data collection, since application of a specific image processing algorithm from a specific version of a specific software package is unambiguous. A similar approach is taken by Galaxy (Blankenberg et al., 2010; Goecks et al., 2010; Giardine et al., 2005), which integrates many domain-specific programs and toolkits into a common environment. On the other end of the spectrum, projects like OpenWetWare (http://openwetware.org) are built on a free-form wiki approach with open sharing of protocols and data, but with less organization and searchability. In the middle ground between these approaches are projects like iLAP (Stocker et al., 2009) which aims to create a flexible system for describing experimental protocols, and Bisque (Kvilekval et al., 2010) which uses a user-defined data model and incorporates many architectural ideas from cloud computing.
In this manuscript, we describe the Electron Microscopy Electronic Notebook (EMEN2), a system which attempts to bridge these two disparate goals using a simple, yet highly flexible design. EMEN2 has been under development at the National Center for Macromolecular Imaging for the past 8 years, and in active use for the last 5. While our primary goal was to develop a system useful for the cryo-EM and structural biology communities, we designed it with sufficient flexibility to make it potentially useful in virtually any scientific discipline.
EMEN2 is a pure object-oriented database (OODB) designed for storage and mining of scientific data in collaborative environments. The goal of EMEN2 is to provide an environment where the end-user scientist is free to express their experiment with great flexibility, but also subtly encouraged to define their data in terms of existing definitions whenever possible. This is handled as a social engineering problem where adhering to standards is the path of least resistance, and each additional level of flexibility, and thus deviation from standards, requires slightly more effort. This encourages the scientist to deviate from standards only when required to accurately record their experiment. In addition, since correct attribution is critical in science, all recorded information has complete user attribution, with a full history of any changes made. Numerous other issues are also considered, such as ease of use, limited maintenance requirements, scalability, security, storage of large amounts of raw data, and a mechanism for directly interacting with computerized lab equipment to reduce user effort and improve data reliability.
Results and Discussion
EMEN development began over a decade ago, as a mechanism for archiving collected electron microscopy data and sharing with collaborators for a single cryo-EM facility. This original version of EMEN (Ludtke et al., 2003) was in active use from 2001 through 2007. As the amount of data generated began to exceed EMEN’s original design specifications, we began development of EMEN2, a new system able to handle tens of millions of records. It was also designed using a more flexible model, incorporating newly developed technologies such as asynchronous “Web 2.0” interfaces and embracing several of the ideas behind the Semantic Web (Berners-Lee et al., 2001; Shadbolt et al., 2006). We believed that this new system could be an attractive choice for many scientific disciplines, so we ensured that any laboratory could easily adopt, and adapt it to their own internal practices and standards without the need to hire a dedicated database administrator. Despite its ease of use and administration, EMEN2 provides capabilities and performance that can rival or surpass expensive dedicated relational databases in appropriate environments. Key EMEN2 concepts and features will be introduced in the following case study of EMEN2’s use at the National Center for Macromolecular Imaging (NCMI), with more detailed explanations of each concept in subsequent sections.
EMEN2 from the end-user’s perspective
When a user logs in to EMEN2, they are immediately presented with a summary “dashboard” showing projects they have permissions to view and any recent activity in those projects. At the NCMI, recent project activity would include records such as CCD frames, microscopy sessions, grid preparations, and so on, with links to the complete record details and thumbnail images where appropriate. Projects are designed to support multi-scientist collaborations. At any given moment in time, one or more specific users are designated as responsible for the next stage of the research in each project, making it easier for users and collaborators to know who to contact for more information. As different types of users have different needs from an ELN system, we consider the user experience of four representative users as examples of how EMEN2 is currently being used.
First, a microscopist, preparing for a microscope session, might log in to the web interface, select the project they are working on, and add a new entry describing a set of grids they have just vitrified. In addition to linking to the selected project, this vitrification record would also link to specific aliquot of the purified specimen, the source of the grids, and the specific freezing apparatus used in the vitrification. This is also a point at which the power of EMEN2’s flexible data model could potentially come into play. Consider a specific experiment where the pH of the specimen is adjusted at the time of freezing, with a specific time delay between pH change and vitrification. We would like to record both the original and new pH as well as the time delay for each grid, even though these parameters would not normally be part of a standard vitrification protocol. EMEN2 permits such additional parameters to be stored within each individual grid record, and remain fully searchable within the query system, even when the parameters were not part of the original freezing protocol.
As the microscopist begins the session and cools down the microscope, they launch an EMEN2 client program called EMDash on the CCD camera computer to help manage their session and upload data. After requesting login, project, and grid details, EMDash runs passively in the background and monitors a specified directory for new images and metadata files. As these files appear, they are uploaded automatically to the EMEN2 server, never interrupting the flow of data collection. By default EMDash operates independently of any vendor-specific software, although a plugin interface is available for integrating with specific systems. While the imaging takes place, the microscopist is given the option to assign images a quality rating and take notes. Newly collected data is immediately available to collaborators through the web interface, while the imaging session is still underway. If the database goes down or becomes unavailable for some reason, EMDash will wait for it to become available again, and then continue the upload process.
As our second example, we consider the viewpoint of an outside collaborator working with the NCMI, who may be off-site, or even international. A collaborator preparing to ship a new specimen for imaging would log in to the web interface, browse to the project, and then create records describing the purification and specific aliquots being shipped. Note that a typical collaborator with only one or two collaborative projects would see only those projects, and there is no mechanism by which they could gain any information about any other projects in the system without explicitly being granted access. Alternatively, the collaborator might send the specimen details on paper, and have it entered on-site by the microscopist. When an imaging session begins, a collaborator logged in to the database will see the images appearing in real-time as they are being collected, complete with a “Google Maps”-style interface for quickly screening recorded image files. The collaborator is also able to add comments to the images, which are immediately visible to the microscopist. Collaborators may even begin downloading images and processing data while the microscopy session is still underway, giving the microscopist real-time feedback on data quality.
Third, we consider users performing image processing. While some simple image viewing and evaluation tasks are available in EMEN2, and it would certainly be possible to incorporate portions of the image processing pipeline directly into EMEN2, we currently view tasks such as 3-D reconstruction to be external processes. The typical image processor would log in to the database, locate a project and grid imaging session of interest, and then download some or all of the images in the session. For users of our related EMAN2 software package (Tang et al., 2007), the user can browse an EMEN2 database directly from the file selection dialog and download data directly into their processing workflow. However, for users in general, this can be handled via the web interface without any specialized software. Selected images can be downloaded individually, or all together in a single tar format archive file. These archives are created by the database server on-the-fly, avoiding annoying delays for the user. After downloading the data, the user performs processing using software of their choice. Again, for users who happen to be using EMAN2, we provide capabilities for automatically uploading selected image processing metadata and results back to the EMEN2 server, into the same project where the image data originated. Users of other software would create records of their reconstructions interactively, and could similarly attach any images, reconstructed volumes, annotations, and other files. While EMEN2 will accept files in any format, some file types are specially recognized via a plugin system that provides additional functionality (such as the previously mentioned “Google Maps”-style browser, which supports virtually any standard cryo-EM format via an EMAN2 plug-in).
Finally, we consider EMEN2 from the lab director’s perspective. A lab director will generally have access to all of the projects in their lab, with the goal of keeping sensible track of how they are progressing, and who to consult if they are not progressing. While most professors can simply recite this information from memory, it is worth mentioning that the NCMI currently has over 200 active projects (with over 1000 specific biological subprojects) in all stages of their life-cycle, from newly started to complete. EMEN2 provides a convenient way to organize and summarize this information. Immediately upon logging in, the lab director is presented with an “eagle’s eye” view of the entire lab, and a detailed recent history of any specific project or subproject is only a few clicks away. In addition to being highly useful to the manager, this also provides subtle encouragement to lab members to actually record what they are doing in the database, as a perceived lack of recorded activity may result in direct questioning about a specific project.
In addition, extensions can be readily added to EMEN2 to dramatically reduce faculty workloads. For example, as an NIH / NIGMS P41 sponsored center, the NCMI is required to submit a detailed annual report of its collaborative and service research activities. We have created an EMEN2 extension to automatically query all active projects in the database and build an XML file, representing most of the content required for this report, that is uploaded to the NIGMS website. While some organizational effort is required via EMEN2 to identify which specific projects to include and to update annual progress summaries, a process that previously required manually assembling multiple copies of a 100+ page report is now handled electronically, with a dramatic reduction in man-hours.
Database Structure
Now that we have considered how EMEN2 appears to typical end users, we must examine how the database is structured, and how this provides the promised interdisciplinary flexibility. EMEN2 is an object-oriented database built around two fundamental concepts: protocols and parameters. Parameters are simply values that might be recorded during an experiment, such as “Buffer pH” or “Vitrified Grid Temperature”. Each parameter is defined by a description, data type (float, string, etc.) and permitted units, when appropriate (Table 1).
Table 1.
Built-in data types. Every parameter in EMEN2 has an associated data type and a flag indicating if the parameter value is a scalar or an array. Additional data types can be added by extending one of the built-in types and providing a validation method.
| Data Type | Constraints |
|---|---|
|
| |
| text | Long-format text |
| choice | One value from a list of choices |
| int | Integer |
| oat | Floating Point |
| coordinates | Array of x, y coordinates |
| boolean | True / False |
| datetime | yyyy/mm/dd HH:MM:SS |
| uri | Remote URI |
| user | Valid username |
| recorddef | Valid Protocol |
| record | Valid Record ID |
| binary | Reference to managed file on disk |
A protocol is a definition of a single specific type of record to be stored in the database, such as vitrifying a grid, running a column for protein purification, collecting an image on the microscope, or any other experimental procedure. The protocol itself is a plain-text description, as might be written in a lab notebook, with an embedded list of experimental parameters which are expected to be recorded during the experiment marked in the text (Figure 1). Note that the protocol is not the record of the actual experiment, but is a definition of how a recurring experiment is performed. The experimental values unique to each specific experiment are stored in records, which are tied to a specific protocol, which gives the values a context.
Figure 1.

Example EMEN2 experimental protocol with embedded parameters. The protocol has been truncated for display in this figure; full protocols may be viewed on our site. The experimental protocol is written in plain text, as would appear in a lab notebook, with the different parameters to be recorded marked with “$$”. The parameter’s description can be inserted using “$#”. Both HTML and Markdown formatting are supported. Beyond the main protocol definition, several additional “views” of the data may be provided using the same syntax, including a which columns to display in tables, and a “summary” view with key parameters.
For example, a cryo-EM vitrification protocol would describe the sequence of experimental steps during vitrification, with parameters to be recorded at each step such as specimen concentration, blotting time, humidity, and temperature. Parameters may be shared between protocols when the contextual meaning is the same. While the freezing protocol for an FEI Vitrobot is different than for a Gatan plunger, the meaning of blotting time is the same in both protocols, and they would share a common parameter definition. New parameters need only be added when no parameter already exists with the appropriate meaning. Similarly, protocols need only be added when a new experiment is designed or an old experiment undergoes a substantial change of methodology.
A third fundamental concept in EMEN2 is the use of hierarchical relationships between parameters and protocols to create an ontology that provides unique organizational and descriptive power (Figure 2 and Supplementary Figures 1 and 2). This concept is particularly useful for data mining. Extending the example above, a vitrification protocol might have specialized child protocols for each type of freezing apparatus. One could pose a broad question, “find all vitrification experiments where the buffer temperature was between 25 and 30°C,” or more narrowly, “find all Mark IV Vitrobot experiments where the buffer temperature was between 25 and 30°C,” which would return a subset of the first query. Likewise, the temperature parameter could be specific such as “buffer temperature”, “room temperature,” or a very general “any temperature measurement”. Records are also organized hierarchically, with very few constraints on the user and frequent use of multiple-parent relationships (Figure 2c). Queries can combine several types of relationships, e.g. “find all images in the GroEL project acquired with the JEOL 2010F microscope.” This would return any imaging records (CCD frame, scanned micrograph, tomogram, etc.) that are children of both the given project and microscope. Although currently only references to protocols and parameters in the local system are supported, there is planned support for cross-referencing terms in other ontologies such as the Gene Ontology (http://www.geneontology.org), the National Cancer Institute Thesaurus (http://ncit.nci.nih.gov), or any of the hundreds of ontologies available through the National Center for Biomedical Ontologies’ BioPortal site (http://bioportal.bioontology.org).
Figure 2.
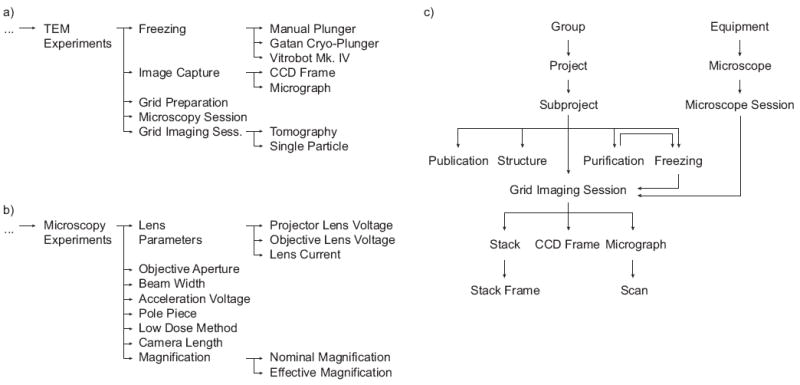
Representative example of ontologies for (a) cryo-EM protocols and (b) parameters. The default EMEN2 installation provides a number of protocols and parameters for common cryo-EM equipment, procedures, and experimental data. Only a few items are shown here; the full ontologies can be viewed in Supplementary Figures 1 and 2. New protocols and parameters can be created and shared between EMEN2 installations. Records are organized in a similar hierarchy. While there are no restrictions on how records may be organized, EMEN2 offers suggestions based on common patterns (c). Multiple parents are commonly used to provide context without duplication of data.
While these may appear to be fairly simple concepts, they provide a solution to one of the largest problems in electronic notebooks: variation of experimental protocols over time and between different investigators. Protocols and parameters in EMEN2 can be extended by any user to define new experiments, at any time, without limiting the search flexibility of the system. While such a system could be implemented in a relational database, the ever expanding pool of tables would require continual updating of SQL scripts, and professional database administration would be a virtual necessity.
Once a protocol has been used, the definition should never change (other than cosmetically), as that would effectively change the contextual definition of existing data. Instead, if a protocol requires an update, a new protocol is created, using the original as a template. This ensures that the historical meaning of all records is preserved. EMEN2 uses this strategy to anticipate the natural evolution of experimental protocols that occurs over time and to encourage self-documentation.
In many experimental settings, many changes to an experimental protocol are tried and abandoned before settling on a standard protocol. In these cases, documenting every attempted experiment with a new protocol is inefficient and time consuming. To combat proliferation of protocols, and the time it would require to create them, an alternative mechanism is provided for reporting trials of variations of existing protocols: Individual records may be annotated with text and parameter values not defined in the protocol. For example, in an particular freezing session where the sample’s pH was being controlled, the user could opt to make a normal freezing record and add an annotation that the pH was controlled along with the parameter value for the specific pH. Unlike many traditional databases, where such parameters would likely be textual notes, such parameters are fully searchable in EMEN2. A query on the database for all records with pH would return this record, along with any records where pH was an explicit part of the protocol.
This gives the end-users of the system complete flexibility, while subtly encouraging them to keep the database definition as simple as is reasonable to do. If the user plans on performing a specific variation of a protocol only once or twice, it is undoubtedly easier for them to use an existing protocol and make an annotation. However, if this is a permanent change to the protocol, the user would soon tire of adding all the annotations, and adding a new protocol becomes the path of least-resistance. Thus, while an EMEN2 database will certainly have a much richer and more complicated schema than a traditional database, it does so while preserving full search capabilities, and more importantly, preserving a complete scientific historical record of experiments. The EMEN2 distribution includes a complete schema for cryo-EM as performed in a typical cryo-EM facility with multiple instruments and a variety of different biological projects and experimental protocols. As the schema defines the experimental practices in a specific lab, the schema must be, and is, easily customized directly via the web interface. However, as most labs will likely start with the existing schema, databases in different labs should share a vast majority of common parameters and root protocols, making information exchange among labs still feasible.
Automatic Data Archiving
In automation-oriented systems such as Leginon / Appion (Suloway et al., 2005; Lander et al., 2009), the data acquisition and processing software tightly integrates with the database. EMEN2, however, uses a modular approach for greater portability to different software, data acquisition systems and equipment manufacturers. One aspect of this approach is implemented in EMDash, a platform-independent client program which runs on the image acquisition computer (Figure 3). As mentioned above, rather than requiring specific knowledge of any particular image acquisition system, EMDash operates passively in the background by monitoring a directory and waiting for new files to appear. New files are added to the upload queue automatically, and copying to the database takes place as a low priority background process as not to disrupt data collection. In addition, EMDash permits the user to optionally interactively assess each image in the queue, before, during or after uploading, and stores the assessment along with the image. While EMDash generally runs uncoupled from the acquisition system, a plugin interface is provided for automatically collecting additional metadata from explicitly recognized systems. Currently supported systems include Gatan DigitalMicrograph, DirectElectron, JADAS (Zhang et al., 2009) and SerialEM (Mastronarde, 2005). Adding enhanced metadata support for additional systems is quite straightforward as EMDash, like EMEN2, is written in Python, with a fully documented API and example code.
Figure 3.
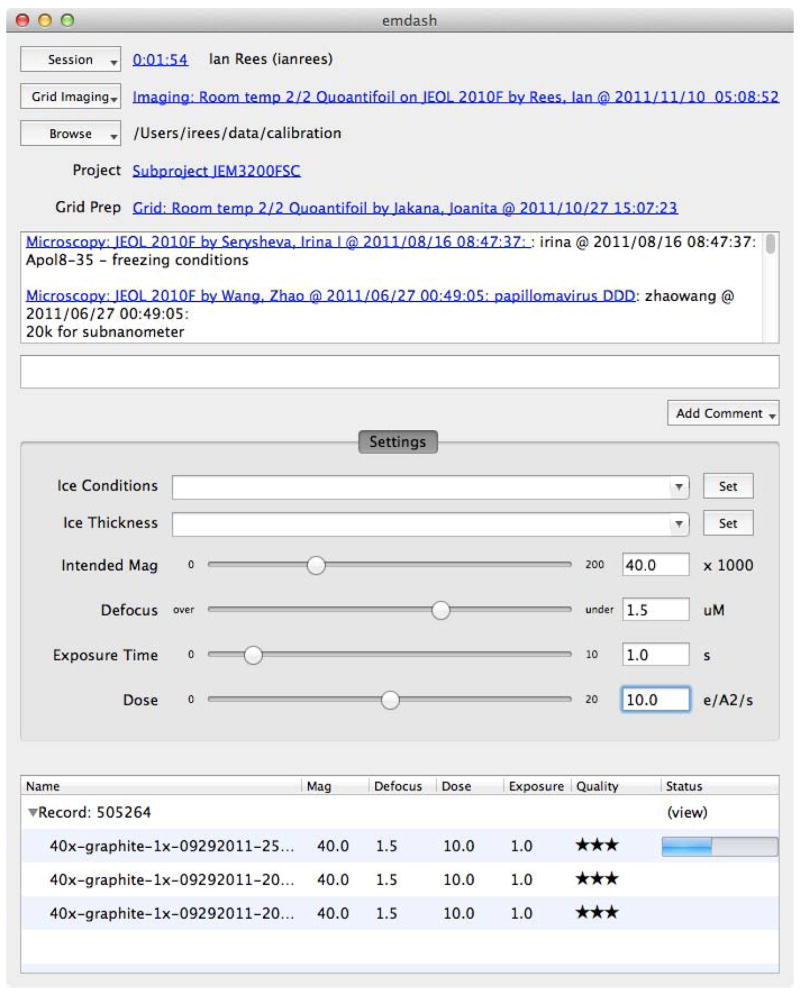
Screenshot of EMDash microscope client. EMDash is started at the beginning of a microscopy session and continues to upload new images in the background during data collection. Several “wizards” are provided to accomplish common tasks and lower barriers for new users. Uploaded data is immediately available via the web interface.
Beyond automatic archival of the raw data itself, EMDash’s session management provides valuable microscope usage accounting details. This data can be queried and viewed in several ways (calendar, date histogram, grouped by user or project, etc.) directly on the EMEN2 web server. The standard data model also separates microscope details (“Microscopy” protocol) from data collection on a specific specimen (“Grid Imaging” protocol). Default security is such that all users can browse the project-independent microscopy sessions where the user can record information about microscope performance or maintenance issues. The grid imaging sessions, however, contain specimen-specific information and are secured based on site policy. This ensures that users can always check previous sessions to be aware of any ongoing problems, but will only have access to biological data on projects where they have been assigned permissions.
EMDash also provides tools for uploading image processing and reconstruction metadata from EMAN2 (Tang et al., 2007), including CTF parameters, coordinates of boxed particles, and image quality metrics such as number of particles per frame. This data can be plotted vs. other database parameters to look for trends (Figure 5 and Supplementary Figure 3). Further integration with EMAN2 is underway to provide a fully integrated experience where data can be downloaded directly into an EMAN2 project, and information about processing is automatically uploaded during processing.
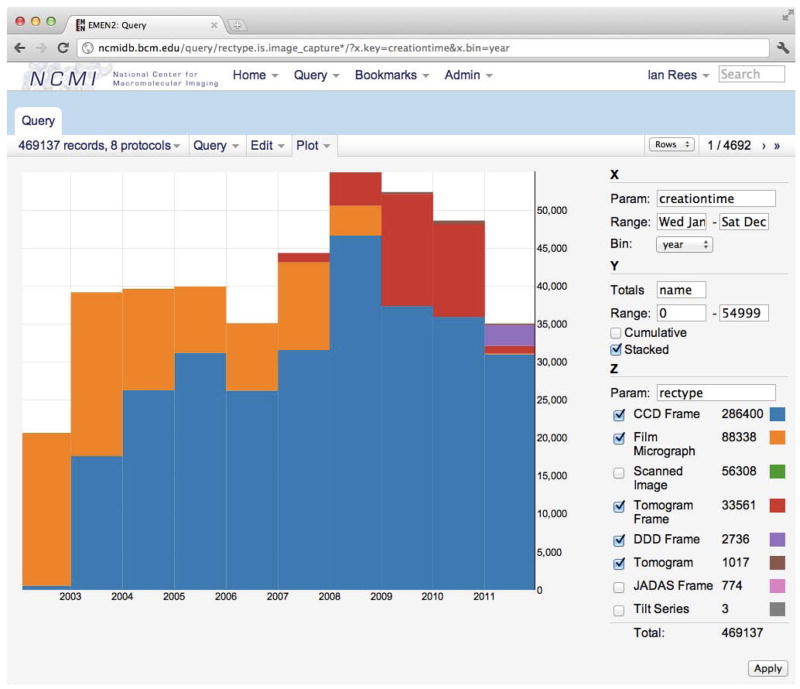
Figure 5.
Plot showing images collected at one facility over the past decade, illustrating the migration from film to digital images. The web interface includes an interactive visualization tool for examining relationships between parameters or trends in data. Several plotting modes (scatter plot, histogram, stacked bars, etc.) are supported. Additional examples are provided in Supplementary Figure 3. Query results can also be exported in CSV or JSON format for additional analysis.
Security and Logging
While the fundamental principles of science make it, almost by definition, an open enterprise, situations still frequently arise where security and privacy are, at least temporarily, important. In EMEN2, each record has an access control list with users assigned to one of four increasing layers of privilege: read-only, read & comment, full editing, and record ownership. A user without read permission on a record will not be able to even detect the record’s existence via queries or any other mechanism. Groups are implemented to simplify permissions management and provide role-based access (administrator, authenticated users, anonymous access, etc.). This fine-grained access control system allows for a wide variety of security designs. Some parts of a project may be private, while others, such as published data sets, available for unrestricted dissemination.
Since EMEN2 is intended as a replacement for paper laboratory notebooks, change tracking is a critical factor. In a paper lab notebook, all changes are recorded with initials, date, and a comment describing the reason for the change. This is implemented in EMEN2 transparently to the user, with all changes written to a read-only log documenting who made the change, the time of the change, and the parameter value prior to the change. This log is integral to the record and is available whenever the record is viewed. Users are also encouraged to add a comment to the record explaining the cause for the change. While it would be possible for an administrator with unrestricted physical access to the computer to alter these logs, there is no mechanism within the database to circumvent logging, regardless of permission levels. To provide true verifiable traceability, database snapshots can be cryptographically signed, dated, and maintained on WORM (write once, read many) media.
Binary Data
With microscopy or any other imaging science, binary images represent the majority of the contents of the database. In EMEN2, as with most databases, uploaded images and other binary files are simply stored in a managed set of folders in the file system, with programmable naming conventions. Files are treated as attachments to records, with record permissions governing access via the network.
Any type of file can be uploaded and stored by EMEN2. At the most basic level, the file is simply archived without any additional processing. However, EMEN2 also provides a plugin interface for handling specific file types, with abstract methods that can be implemented to read file metadata, calculate statistics, generate thumbnail images, etc. Plugins may also implement a full “Google Maps” style image browser for exploring images in detail without downloading the entire image (Figure 4); this works by caching tiled arrays of 256 × 256 pixel images at various scales. There are currently plugins for most common cryo-EM image formats, including TIFF, HDF5, MRC, and DM3; in most cases, both 2D and 3D (volume and stack) data is supported. These plugins use the EMAN2 library to read and manipulate images, and most EMAN2 image attributes are 1:1 mapped to EMEN2 parameters. Users can also create new plugins by subclassing an existing plugin. Support for additional file types, including multi-channel images, is planned. Custom image processing and analytical tools, such as an online particle picker, can also be added using the extension API.
Figure 4.
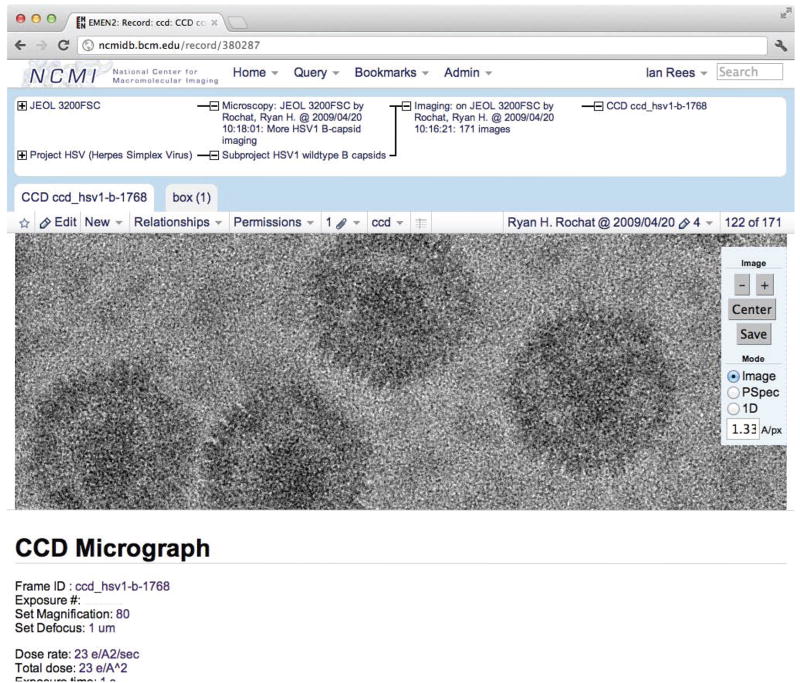
Screenshot of EMEN2 web interface showing a CCD frame record. Parent relationships are shown as a tree with paths back to the root record, and child records are shown as tabs. Common data formats can be previewed inside the browser with a “Google Maps” style interface, as well as a 2D FFT of the image. Particle coordinates can be viewed and edited with an in-browser tool.
Database Implementation
EMEN2 is a pure object-oriented database implemented in Python on top of a BerkeleyDB (Oracle, Inc.) back end. BerkeleyDB is a high-performance open-source embedded database (Olson et al., 1999) written in C, and completely unrelated to Oracle’s commercial relational database products. It is available at no cost for use in open-source projects, and essentially provides a high performance mechanism for storing and retrieving sets of key/value pairs. Despite the simple key/value API, BerkeleyDB provides a robust ACID (atomicity, consistency, isolation, durability) environment to ensure data integrity and robust failure recovery. Berkeley DB also supports clustering using a single master, multiple replica system; this is not currently enabled in EMEN2, but could be used in computationally demanding or high-availability scenarios. Most image processing operations are run as subprocesses in a LIFO (last in, first out) queue, with a maximum number executing at any given time.
EMEN2’s “Web 2.0” interface uses open standards (XHTML, CSS, JSON) and asynchronous queries to provide a highly interactive experience. The web server is built using established technologies such as Twisted.Web (http://twistedmatrix.com), Mako Templates (http://makotemplates.org), and the jQuery javascript library (http://jquery.com). The web interface is very modular, and defines a simple mechanism for creating custom plug-ins and customizations (Figures 4-6, Supplementary Figure 4). EMEN2 also provides a public API that is available to client programs both directly on the server (Python) through a web-service using XML-RPC (Winer, 1999) or JSON-RPC (Crockford, 2006).
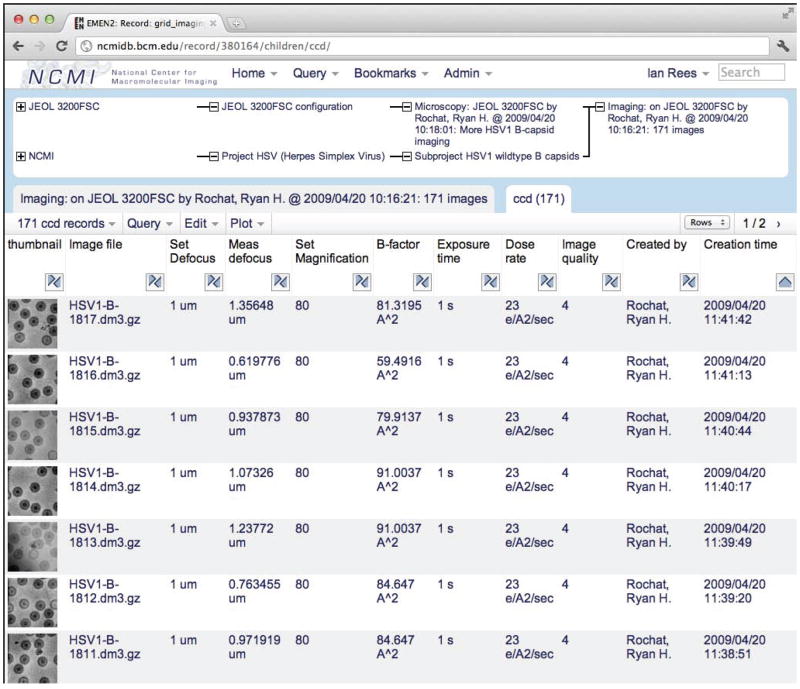
Figure 6.
A table showing CCD frames collected during a microscopy session. The table widget includes controls for batch editing and sorting, as well as drop-down menus for refining the query, displaying statistics, generating plots, and downloading the table as a spreadsheet.
EMEN2 is designed to run as a production system for long periods with minimal maintenance. To minimize the risk that a critical component will become deprecated or abandoned in the long term, a conscious effort was made to select dependencies that are scalable, mature, and widely used. Likewise, to reduce the risk of software “lock in,” the entire contents and schema of an EMEN2 database can be dumped to a flat file in either XML or JSON format.
Conclusions
EMEN2 has been in production use in our laboratory for 5 years with nearly 800 users participating in more than 300 projects, and has also been installed in several other independent labs. As of July 2012, we have over 520,000 records, including more than 20TB of associated image data. The infrastructure has been tested for scalability to many millions of records. Apart from the storage systems, the hardware requirements are modest. We have also exploited EMEN2’s flexibility as a platform to build a number of additional systems, including a grant-reporting tool that harvests data from active projects for annual reporting, the website for the 2010 Cryo-EM Modeling Challenge (http://ncmi.bcm.edu/ncmi/events/workshops/workshops_115_test) and a “mirror” of our in-house database containing numerous published raw data sets for public access (http://ncmi.bcm.edu/publicdata/db/home).
EMEN2 is freely available under a BSD-compatible license. Documentation and installation instructions are provided at our site: http://blake.grid.bcm.edu/emanwiki/EMEN2. EMEN2 is also a registered PyPI package (http://pypi.python.org).
Supplementary Material
Tree view of the complete included EMEN2 parameter ontology. In addition to generic parameters common to any scientific endeavor, this includes common data types and parameters used in cryo-EM projects, including metadata from image headers, microscope and freezing apparatus settings, reconstruction metadata, etc.
Tree view of the included EMEN2 protocol ontology. Protocols for common cryo-EM experiments and data types are included. Users can customize these protocols by creating modified versions. For non-cryo-EM use, the cryo-EM portion of the ontology can be easily removed.
Additional plotting examples. This scatterplot of CTF B-Factor vs. measured defocus, grouped by the microscope used shows differences in the quality of data acquired with each instrument. The plot could alternatively be grouped by specimen, microscopist, date, etc., to see if the differences are spurious or something that needs to be seriously investigated. The plot is highly interactive and updates dynamically.
Screenshot tour of additional database features. The main home page features user administration tools, a table of recently created records, and a list of top-level projects.
Interactive online particle picker. This tool uses EMAN2 on the server to render the micrographs and boxed particles as tiles that are sent to the browser in real time. The micrograph may be panned and zoomed, and particles are added with a click and removed with a shift-click. The boxed particles appear in the right sidebar. Particle coordinates can be imported and exported from EMAN1 and EMAN2.
Screenshot of the basic query interface for finding records by protocol, user, project, etc.
Acknowledgments
We would like to thank NCMI users and collaborators for feedback to drive improvements and development of EMEN2. Support for EMEN2 development was provided by NIH grants P41GM103832 (previously P41RR02250) and PN2EY016525, R01GM080139 for development of EMAN2 related features, and Ian Rees was supported by NIH training grant T15LM007093 through the Gulf Coast Consortia.
References
- Allan C, Burel JM, Moore J, Blackburn C, Linkert M, Loynton S, Macdonald D, Moore WJ, Neves C, Patterson A, Porter M, Tarkowska A, Loranger B, Avondo J, Lagerstedt I, Lianas L, Leo S, Hands K, Hay RT, Patwardhan A, Best C, Kleywegt GJ, Zanetti G, Swedlow JR. OMERO: flexible, model-driven data management for experimental biology. Nat Methods. 2012;9:245–253. doi: 10.1038/nmeth.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berners-Lee T, Hendler J, Lassila O. The semantic web. Scientific American. 2001;284:28–37. [Google Scholar]
- Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol. 2010;Chapter 19:Unit 19.10.1–Unit 19.1021. doi: 10.1002/0471142727.mb1910s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crockford D. The application/json Media Type for JavaScript Object Notation (JSON) IETF Request for Comments. 2006;4627 [Google Scholar]
- Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, Miller W, Kent WJ, Nekrutenko A. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J Galaxy Team. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg I, Allan C, Burel JM, Creager D, Falconi A, Hochheiser H, Johnston J, Mellen J, Sorger P, Swedlow J. The Open Microscopy Environment (OME) Data Model and XML file: open tools for informatics and quantitative analysis in biological imaging. Genome Biology. 2005;6:R47. doi: 10.1186/gb-2005-6-5-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvilekval K, Fedorov D, Obara B, Singh A, Manjunath BS. Bisque: a platform for bioimage analysis and management. Bioinformatics. 2010;26:544–552. doi: 10.1093/bioinformatics/btp699. [DOI] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, et al. Appion: an integrated, databasedriven pipeline to facilitate EM image processing. Journal of structural biology. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Nason L, Tu H, Peng L, Chiu W. Object oriented database and electronic notebook for transmission electron microscopy. Microsc Microanal. 2003;9:556–565. doi: 10.1017/S1431927603030575. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Olson MA, Bostic K, Seltzer M. Berkeley DB. Proceedings of the FREENIX Track: 1999 USENIX Annual Technical Conference; Monterey, CA. 1999. pp. 183–192. USENIX Association. [Google Scholar]
- Shadbolt N, Hall W, Berners-Lee T. The semantic web revisited. Intelligent Systems, IEEE. 2006;21:96–101. [Google Scholar]
- Stocker G, Fischer M, Rieder D, Bindea G, Kainz S, Oberstolz M, McNally JG, Trajanoski Z. iLAP: a workflow-driven software for experimental protocol development, data acquisition and analysis. BMC Bioinformatics. 2009;10:390. doi: 10.1186/1471-2105-10-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Winer D. [19 November 2011];XML-RPC Specification. 1999 http://xmlrpc.com.
- Zhang J, Nakamura N, Shimizu Y, Liang N, Liu X, Jakana J, Marsh MP, Booth CR, Shinkawa T, Nakata M, Chiu W. JADAS: a customizable automated data acquisition system and its application to ice-embedded single particles. J Struct Biol. 2009;165:1–9. doi: 10.1016/j.jsb.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tree view of the complete included EMEN2 parameter ontology. In addition to generic parameters common to any scientific endeavor, this includes common data types and parameters used in cryo-EM projects, including metadata from image headers, microscope and freezing apparatus settings, reconstruction metadata, etc.
Tree view of the included EMEN2 protocol ontology. Protocols for common cryo-EM experiments and data types are included. Users can customize these protocols by creating modified versions. For non-cryo-EM use, the cryo-EM portion of the ontology can be easily removed.
Additional plotting examples. This scatterplot of CTF B-Factor vs. measured defocus, grouped by the microscope used shows differences in the quality of data acquired with each instrument. The plot could alternatively be grouped by specimen, microscopist, date, etc., to see if the differences are spurious or something that needs to be seriously investigated. The plot is highly interactive and updates dynamically.
Screenshot tour of additional database features. The main home page features user administration tools, a table of recently created records, and a list of top-level projects.
Interactive online particle picker. This tool uses EMAN2 on the server to render the micrographs and boxed particles as tiles that are sent to the browser in real time. The micrograph may be panned and zoomed, and particles are added with a click and removed with a shift-click. The boxed particles appear in the right sidebar. Particle coordinates can be imported and exported from EMAN1 and EMAN2.
Screenshot of the basic query interface for finding records by protocol, user, project, etc.