Abstract
In Arabidopsis thaliana the CPC-like MYB transcription factors [CAPRICE (CPC), TRIPTYCHON (TRY), ENHANCER OF TRY AND CPC 1, 2, 3/CPC-LIKE MYB 3 (ETC1, ETC2, ETC3/CPL3), TRICHOMELESS 1, 2/CPC-LIKE MYB 4 (TCL1, TCL2/CPL4)] and the bHLH transcription factors [GLABRA3 (GL3) and ENHANCER OF GLABRA 3 (EGL3)] are central regulators of trichome initiation and root-hair differentiation. By transforming the tomato orthologous genes SlTRY (TRY) and SlGL3 (GL3) into Arabidopsis, we demonstrated that SlTRY inhibited trichome initiation and enhanced root-hair differentiation. These results suggest that tomato and Arabidopsis partially use similar transcription factors for epidermal cell differentiation, and that a CPC-like R3 MYB may be a key common regulator of plant trichome and root-hair development. CPC and GL3 are also known to be involved in anthocyanin biosynthesis. Here, we show that anthocyanin accumulation was repressed in the CPC::SlTRY and GL3::SlGL3 transgenic plants, suggesting that SlTRY and SlGL3 can influence anthocyanin pigment synthesis.
Keywords: tomato, Arabidopsis, R3 MYB, CPC, SlTRY, anthocyanin
The differentiation of plant epidermal cells into trichomes and root hairs has proven to be a very useful paradigm for investigating early events in plant cell development. In Arabidopsis thaliana, the CAPRICE (CPC) gene, a member of the R3-type MYB transcription factor family, is an important regulatory factor for trichome initiation and root hair formation.1 Genes closely related to CPC in the Arabidopsis genome include: TRYPTICHON, TRY; ENHANCER OF TRY AND CPC1, ETC1; ENHANCER OF TRY AND CPC2, ETC2; ENHANCER OF TRY AND CPC3/CPC-LIKE MYB3, ETC3/CPL3; TRICHOMELESS1, TCL1 and TRICHOMELESS2/CPC-LIKE MYB4, TCL2/CPL4.2-10 The TRY protein functions primarily during trichome initiation.2,11 Another type of transcription factor in Arabidopsis, the bHLH transcription factor GLABRA3 (GL3), also controls trichome initiation and root hair formation.12 Previously, we reported that the CPC, TRY, ETC1, ETC2 and CPL3 proteins interact with GL3 in yeast and might function together as a transcription factor complex.7
Recently, we identified tomato SlTRY and SlGL3 genes as orthologs of the Arabidopsis TRY and GL3 genes, respectively.13 The CPCp::SlTRY transgenic plants produced more root hairs and had no trichomes, a phenotype analogous to that of the 35S::CPC transgenic plants.13 In contrast, GL3p::SlGL3 transgenic plants closely resembled wild-type. These results suggest that Arabidopsis and tomato have orthologous transcription factors for epidermal cell differentiation including trichome initiation and root-hair differentiation. Furthermore, SlTRY-like R3 MYB appears to function as an important common transcription factor for both trichome initiation and root-hair differentiation.13
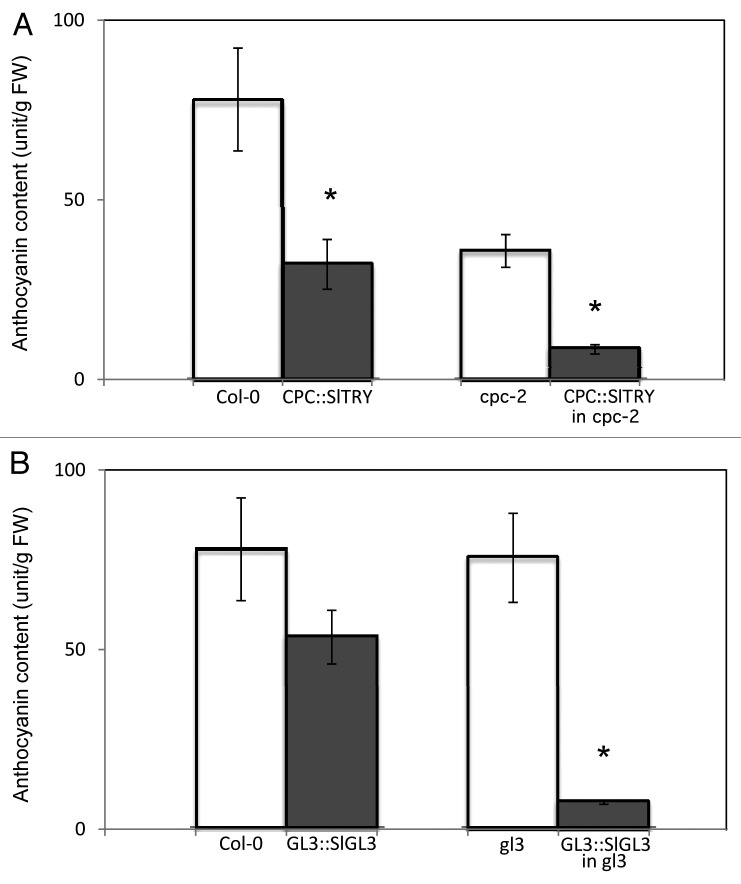
In addition to epidermal cell differentiation, Arabidopsis CPC and GL3 genes are known to regulate anthocyanin biosynthesis.14,15 To verify that tomato SlTRY and SlGL3 also play roles in anthocyanin accumulation, we measured the anthocyanin content of SlTRY and SlGL3 transgenic plants. Total plant pigments were extracted from leaves, and anthocyanin levels were measured according to previously reported protocols.16,17 Compared with the wild-type Col-0, the CPC::SlTRY transgenic plants had significantly reduced levels of anthocyanin accumulation (Fig. 1A). Compared with the cpc-2 mutant, cpc-2 transgenic plants transformed with CPC::SlTRY also accumulated significantly less anthocyanin (Fig. 1A). Anthocyanin accumulation was somewhat reduced in the GL3::SlGL3 transgenic plants compared with wild type Col-0 (Fig. 1B). In comparison with gl3-7454 mutants, gl3-7454 transgenic plants transformed with GL3::SlGL3 also had significantly lower levels of anthocyanin accumulation (Fig. 1B). Therefore, the tomato homologs to the TRY and GL3 genes, SlTRY and SlGL3, also function as repressors of anthocyanin accumulation.
Figure 1. Anthocyanin accumulation is inhibited by SlTRY and SlGL3. (A) Anthocyanin content in wild-type Col-0, CPC::SlTRY, cpc-2 and CPC::SlTRY in cpc-2. Error bars indicate the standard deviations. Bars marked with asterisks indicate a significant difference between the wild-type Col-0 and the CPC::SlTRY transgenic line, or the cpc-2 mutant and the CPC::SlTRY in the cpc-2 transgenic line by Student’s t-test (p < 0.050). (B) Anthocyanin content in wild-type Col-0, GL3::SlGL3, gl3–7454 and GL3::SlGL3 in gl3-7454. Error bars indicate the standard deviations. Bars marked with asterisks indicate a significant difference between the wild-type Col-0 and the GL3::SlGL3 transgenic line, or the gl3-7454 mutant and the GL3::SlGL3 in gl3-7454 transgenic line by Student’s t-test (p < 0.050).
Acknowledgments
This work was financially supported by JSPS KAKENHI Grant Numbers 24658032, 23570057 and by the program “Improvement of Research Environment for Young Researchers” from the Ministry of Education, Culture, Sports, Science and Technology, a grant for Scientific Research on Priority Areas from the University of Miyazaki,
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/24575
References
- 1.Wada T, Tachibana T, Shimura Y, Okada K. Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science. 1997;277:1113–6. doi: 10.1126/science.277.5329.1113. [DOI] [PubMed] [Google Scholar]
- 2.Schellmann S, Schnittger A, Kirik V, Wada T, Okada K, Beermann A, et al. TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 2002;21:5036–46. doi: 10.1093/emboj/cdf524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirik V, Simon M, Huelskamp M, Schiefelbein J. The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev Biol. 2004;268:506–13. doi: 10.1016/j.ydbio.2003.12.037. [DOI] [PubMed] [Google Scholar]
- 4.Kirik V, Simon M, Wester K, Schiefelbein J, Hulskamp M. ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol Biol. 2004;55:389–98. doi: 10.1007/s11103-004-0893-8. [DOI] [PubMed] [Google Scholar]
- 5.Esch JJ, Chen MA, Hillestad M, Marks MD. Comparison of TRY and the closely related At1g01380 gene in controlling Arabidopsis trichome patterning. Plant J. 2004;40:860–9. doi: 10.1111/j.1365-313X.2004.02259.x. [DOI] [PubMed] [Google Scholar]
- 6.Simon M, Lee MM, Lin Y, Gish L, Schiefelbein J. Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev Biol. 2007;311:566–78. doi: 10.1016/j.ydbio.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Tominaga R, Iwata M, Sano R, Inoue K, Okada K, Wada T. Arabidopsis CAPRICE-LIKE MYB 3 (CPL3) controls endoreduplication and flowering development in addition to trichome and root hair formation. Development. 2008;135:1335–45. doi: 10.1242/dev.017947. [DOI] [PubMed] [Google Scholar]
- 8.Wang S, Kwak SH, Zeng Q, Ellis BE, Chen XY, Schiefelbein J, et al. TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development. 2007;134:3873–82. doi: 10.1242/dev.009597. [DOI] [PubMed] [Google Scholar]
- 9.Gan L, Xia K, Chen JG, Wang S. Functional characterization of TRICHOMELESS2, a new single-repeat R3 MYB transcription factor in the regulation of trichome patterning in Arabidopsis. BMC Plant Biol. 2011;11:176. doi: 10.1186/1471-2229-11-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tominaga-Wada R, Nukumizu Y. Expression Analysis of an R3-Type MYB Transcription Factor CPC-LIKE MYB4 (TRICHOMELESS2) and CPL4-Related Transcripts in Arabidopsis. Int J Mol Sci. 2012;13:3478–91. doi: 10.3390/ijms13033478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hülskamp M, Misŕa S, Jürgens G. Genetic dissection of trichome cell development in Arabidopsis. Cell. 1994;76:555–66. doi: 10.1016/0092-8674(94)90118-X. [DOI] [PubMed] [Google Scholar]
- 12.Payne CT, Zhang F, Lloyd AM. GL3 encodes a bHLH protein that regulates trichome development in arabidopsis through interaction with GL1 and TTG1. Genetics. 2000;156:1349–62. doi: 10.1093/genetics/156.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tominaga-Wada R, Nukumizu Y, Sato S, Wada T. Control of plant trichome and root-hair development by a tomato (Solanum lycopersicum) R3 MYB transcription factor. PLoS One. 2013;8:e54019. doi: 10.1371/journal.pone.0054019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu HF, Fitzsimmons K, Khandelwal A, Kranz RG. CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Mol Plant. 2009;2:790–802. doi: 10.1093/mp/ssp030. [DOI] [PubMed] [Google Scholar]
- 15.Feyissa DN, Løvdal T, Olsen KM, Slimestad R, Lillo C. The endogenous GL3, but not EGL3, gene is necessary for anthocyanin accumulation as induced by nitrogen depletion in Arabidopsis rosette stage leaves. Planta. 2009;230:747–54. doi: 10.1007/s00425-009-0978-3. [DOI] [PubMed] [Google Scholar]
- 16.Beggs CJ, Kuhn K, Böcker R, Wellmann E. Phytochrome-induced flavonoid biosynthesis in mustard (<i>Sinapis alba</i> L.) cotyledons. Enzymic control and differential regulation of anthocyanin and quercetin formation. Planta. 1987;172:121–6. doi: 10.1007/BF00403037. [DOI] [PubMed] [Google Scholar]
- 17.Rabino I, Mancinelli AL. Light, temperature, and anthocyanin production. Plant Physiol. 1986;81:922–4. doi: 10.1104/pp.81.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]