Abstract
Post-translational attachment of small ubiquitin-like modifier (SUMO), defined as SUMOylation, has emerged as a new mechanism of protein regulation in plant biology. In plant, SUMOylation has been shown to play crucial roles in a variety of biotic and abiotic stress responses. Recent work using viable mutants with defective SUMOylation have indicated an important role for SUMOylation in a wide range of developmental processes, such as cell division, expansion, survival and differentiation, vegetative growth and reproductive development. This review will summarize the currently emerging information regarding the function of SUMOylation in plant development.
Keywords: SUMOylation, SUMO proteases, AtMMS21, AtSIZ1, development, Arabidopsis
Post-translational modifications of proteins through the reversible covalent attachment of small polypeptide like ubiquitin and ubiquitin-like modifiers plays critical roles in protein stability and biological processes.1 Small ubiquitin-like modifier (SUMO) modification/ SUMOylation has emerged as an important regulatory mechanism in plants.2 SUMOylation of target proteins occurs via a cascade of enzymatic reactions including the sequential action of E1 enzymes for SUMO activation, E2 enzymes for conjugation and E3 enzymes for ligation (Fig. 1). Attachment of SUMO onto substrates is reversible and the SUMO-proteases cleave the SUMO-substrate linkages to recycle free SUMO, as well as involved in generating mature SUMO.3 All of the SUMOylation system components exist in plants and some have been characterized using mutant screening and biochemical approach in Arabidopsis. These approaches demonstrated that SUMO, SUMO-conjugating enzymes and SUMOylation are essential for plant development.4,5 However, the role of SUMOylation in plant development is just beginning to be identified due to the embryonic lethality of the mutations in either E1 (SAE1/2) or E2 (SCE1) or in both the SUMO1 and SUMO2 genes.4
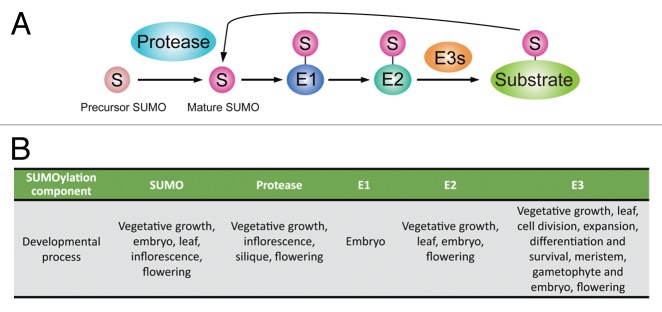
Figure 1. The SUMOylation pathway and its role in plant development. (A) The mechanism of reversible SUMOylation. SUMO is translated into a precursor form and SUMO proteases cleave SUMO into its mature form. Mature SUMO is conjugated to substrate in an ATP-dependent manner in reactions that are catalyzed by three enzymes, E1, E2 and E3, in sequence. SUMOylation is reversible and SUMO deconjugate from the substrate is catalyzed by the SUMO proteases involved in the maturation step. (B) SUMOylation regulates many aspects of developmental processes in plant.
To overcome this embryo lethality and study the role of SUMOylation in plant development, recent work using viable mutants with defect in SUMOylation has improved our understanding of the mechanisms responsible for the plant growth and development. The sumo1 sumo2 knockdown mutant is partially sterile and shows strong developmental phenotypes including dwarfism, leaves crooking, disturbed inflorescence, early senescence and early flowering. Furthermore, SUMO1 and SUMO2 not only act redundantly during embryogenesis, but together regulate many aspects of plant development via the SUMO E3 ligase SIZ1 and the suppression of salicylic acid-dependent signaling.6 Similarly, plants that overexpress a mutant version of SCE with the active site Cys replaced by Ser, show reduced growth, early flowering and changes in the pattern of SUMO conjugates. Also, the effects of overexpression resemble the consequences of defects in SUMO ligase SIZ1 or SUMO protease ESD4.7
It is crucial for SUMO proteases to maintaining cellular balance of SUMOylation.3 Tight regulation of SUMOylation level is important for proper plant development, as shown by the developmental and physiological defects phenotype associated with mutations of SUMO proteases. The esd4 mutants have increased accumulation of SUMOylation and show several phenotypes, including early flowering, dwarf, defective silique and inflorescence development.8,9 Although the absence of other two SUMO proteases OTS1 and OTS2 does not produce any obvious developmental phenotypes under normal growth conditions, overexpressing SUMO1 in the ots1 ots2 double mutants causes a remarkable decrease in plant size.10 Thus, the OTS1 and OTS2 link up plant development and survival under salt stress and the hyper-SUMOylation of key target proteins acts to retard growth to survive stress periods.11 Consistent with the role of SUMOylation level and its components in plant development at a functional level, recent findings reveal that SUMO conjugates accumulate at higher levels in actively growing tissues during plant development.12
In contrast with SUMO proteases deficient mutants, plants without functional SUMO E3 ligase display a reduction in endogenous SUMO conjugate accumulation.13,14 SUMO E3 ligases increase the rate of SUMO conjugation to substrates and influence the substrate specificity of the SUMO conjugation system.15 Although numerous SUMO E3 ligases exist in animals, only two SUMO ligases (AtSIZ1 and AtMMS21) are characterized in Arabidopsis. SUMO ligases knockout mutants siz1 or mms21-1/hpy2 (high ploidy 2) do not compromise plant viability due to the existence of other ligases and cause many obvious phenotypes that offer opportunities to analyze SUMOylation mechanism in plants.14,16 The siz1 mutants exhibit a pleiotropic phenotype and most related to stress responses. SIZ1-dependent SUMOylation is crucial for both abiotic and biotic stress responses, including phosphate starvation, nutrient deficiency, high and low temperature, salt and drought response, copper tolerance and salicylic acid-dependent pathogen defense.2,17,18 Furthermore, mutations in AtSIZ1 lead to dwarf plants with smaller leaves, early flowering, defective female gametophyte and abnormal seed development, indicating that AtSIZ1 also functions in plant development.13,19,20 AtSIZ1 is expressed in almost all plant cells, where it regulates cell expansion and proliferation through SA signaling, as nahG can recovers the defect in cell expansion and cell division caused by the siz1 mutation.19,21 In addition, the phenotypes of siz1 mutant are recovered to wild type phenotypes with the application of exogenous ammonium, which indicates that AtSIZ1 regulates nitrate reduction through its SUMO ligase activity in plant development.17
Another SUMO E3 ligase, AtMMS21/HPY2, an ortholog of MMS21/NSE2-type ligases, was identified independently by two groups.14,22 Similar to siz1 mutants, loss of the AtMMS21 lead to dwarf phenotypes.14,19 However, AtMMS21 and AtSIZ1 are likely to have distinct functions in plant development, as reciprocal expression of AtMMS21 and AtSIZ1 does not complement the single mutant phenotypes.21 The phenotype of siz1 is caused by the accumulation of salicylic acid, while the plants without functional AtMMS21 do not depend on salicylic acid accumulation and exhibit a premature entry into the endocycle.14,21 Mutation of AtMMS21 causes short-root phenotype with impaired expression of the cell division marker (CYCB1, CDKB1, CDKB2) and cytokinin-induced genes, indicating that AtMMS21 regulates meristem development via cell-cycle regulation and cytokinin signaling.14,16 In addition, AtMMS21 is expressed in meristemtic cells and its expression and accumulation are positively regulated by PLT (PLETHORA), which is key transcription factors that mediate the patterning of the root stem cell niche.14,23 Interestingly, our recent data demonstrated that the protein levels of PLT1 and PLT2 are severely reduced in the mms21-1 roots, suggesting a feedback loop might exists between AtMMS21 and PLT in the maintenance of root stem cell niche.24 The involvement of AtMMS21 in the stem cell niche maintenance are demonstrated by the irregular QC organization, the mitotic activation of QC cells, the aberrant expression of QC-specific markers and stem cell transcription factor genes, as well as the appearance of starch granules in the region of the QC and columella stem cells in mms21-1.24 Taken together, our data indicate that AtMMS21 acts in stem cell niches to regulate correct root meristem pattering and function, both during embryogenesis and post-embryonic stages.24
Stem cell niches are dynamic microenvironments that balance stem cell renewal, differentiation and the engagement of programs in response to stress.25 The protection of stem cells against DNA damage is crucial for normal development, but mechanism underlying these processes remains poorly understood.26 A recent study from our laboratory details how AtMMS21 defines the stem cell niche through a reduction in DNA damage.24 This work shows that AtMMS21 maintains the normal cellular organization of the root stem cell niche by preventing DSB-induced cell death. Mutation of AtMMS21 upregulates DSBs and DSB-inducible gene transcription, suggesting that AtMMS21 is involved in DNA damage responses during root development. We further demonstrate that AtMMS21 acts as a component of structural maintenance of chromosomes (SMC) 5/6 complex through its interaction with the SMC5, thus revealing critical roles of AtMMS21 in maintaining genome integrity and root stem cell niche.24 It is noteworthy that it was unknown whether plants possessed SUMOylation mechanism in response to DNA damage, although protein SUMOylation plays important roles in the maintenance of genome integrity from yeast to human. Our work uncovers a novel regulatory framework for the action of SUMO ligase AtMMS21 in correct plant developmental process. Detailed analysis of the genetic interaction between mms21-1 and other mutants defective in the DNA damage response (e.g., atm, atr and sog1 mutants) might help further understanding of AtMMS21 function in the response to DNA damage and stem cell niche maintenance. Additionally, recent proteome-wide screens for SUMO substrates have identified many proteins related to chromatin maintenance/repair processes affected by SUMO conjugation in Arabidopsis, including SMC1, PICKLE, GCN5, ADA2b, NRP1, SYN4, NSE4B, RAD54 (SWI2/SNF2 family) and NUCLEOSOME ASSEMBLY PROTEIN1 (NAP1).27-30 Intriguingly, most of these factors in Arabidopsis have showed involving in stem cell niche maintenance.24 Although Arabidopsis SMC5 and these stem cell regulators/chromatin factors may or may not be a target of AtMMS21, the incorporation of MMS21-mediated SUMOylation into stem cell niche and chromosome maintenance provides several possibilities for how AtMMS21 define the genomic integrity and normal development.
Accumulating evidence suggests that SUMOylation modifies are involved in diverse aspects of developmental processes in Arabidopsis (Fig. 1B). Recent studies have also addressed the function of SUMOylation in vegetative growth and reproductive development by using the monocot rice as models.31,32 It is interesting that the role of SUMOylation for reproductive development remains elusive. Consistently, our unpublished observations showing that gametophyte development and meiosis are processes affected by altered SUMOylation in AtMMS21 deficient plants (Liu et al., submitted). Future identification of SUMO substrates and their functional studies in different spatio-temporal developmental cells will bring new insights into how SUMOylation controls the plant development. It reveals that SUMO system is complex because the downstream pathways and biochemistry properties of SUMOyation components are increasing and diverse. However, the regulation of the SUMOylation system in plants remains poorly understood. Mutant studies have demonstrated that SUMOylation homeostasis is under a tight control and under/over accumulation of SUMOylation cause a misregulation of essential processes. Therefore, studies on the gene expression and enzyme activity of SUMOyation components, as well as the interplay between SUMOylation and other post-translational modifications will be key to understand how SUMO manifests its effects during normal and stress conditions.
Acknowledgments
This work was supported by the National Science Foundation of China (31170269, U1201212) and Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (2010).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/24727
References
- 1.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–56. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 2.Miura K, Hasegawa PM. Sumoylation and other ubiquitin-like post-translational modifications in plants. Trends Cell Biol. 2010;20:223–32. doi: 10.1016/j.tcb.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Hickey CM, Wilson NR, Hochstrasser M. Function and regulation of SUMO proteases. Nat Rev Mol Cell Biol. 2012;13:755–66. doi: 10.1038/nrm3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saracco SA, Miller MJ, Kurepa J, Vierstra RD. Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. 2007;145:119–34. doi: 10.1104/pp.107.102285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park HJ, Kim WY, Park HC, Lee SY, Bohnert HJ, Yun DJ. SUMO and SUMOylation in plants. Mol Cells. 2011;32:305–16. doi: 10.1007/s10059-011-0122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van den Burg HA, Kini RK, Schuurink RC, Takken FL. Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense. Plant Cell. 2010;22:1998–2016. doi: 10.1105/tpc.109.070961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomanov K, Hardtke C, Budhiraja R, Hermkes R, Coupland G, Bachmair A. Small ubiquitin-like modifier conjugating enzyme with active site mutation acts as dominant negative inhibitor of SUMO conjugation in arabidopsis(F) J Integr Plant Biol. 2013;55:75–82. doi: 10.1111/jipb.12016. [DOI] [PubMed] [Google Scholar]
- 8.Murtas G, Reeves PH, Fu YF, Bancroft I, Dean C, Coupland G. A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of SMALL UBIQUITIN-RELATED MODIFIER conjugates. Plant Cell. 2003;15:2308–19. doi: 10.1105/tpc.015487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reeves PH, Murtas G, Dash S, Coupland G. early in short days 4, a mutation in Arabidopsis that causes early flowering and reduces the mRNA abundance of the floral repressor FLC. Development. 2002;129:5349–61. doi: 10.1242/dev.00113. [DOI] [PubMed] [Google Scholar]
- 10.Conti L, Price G, O’Donnell E, Schwessinger B, Dominy P, Sadanandom A. Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell. 2008;20:2894–908. doi: 10.1105/tpc.108.058669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conti L, Kioumourtzoglou D, O’Donnell E, Dominy P, Sadanandom A. OTS1 and OTS2 SUMO proteases link plant development and survival under salt stress. Plant Signal Behav. 2009;4:225–7. doi: 10.4161/psb.4.3.7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaikam V, Karlson DT. Response and transcriptional regulation of rice SUMOylation system during development and stress conditions. BMB Rep. 2010;43:103–9. doi: 10.5483/BMBRep.2010.43.2.103. [DOI] [PubMed] [Google Scholar]
- 13.Catala R, Ouyang J, Abreu IA, Hu Y, Seo H, Zhang X, et al. The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell. 2007;19:2952–66. doi: 10.1105/tpc.106.049981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishida T, Fujiwara S, Miura K, Stacey N, Yoshimura M, Schneider K, et al. SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell. 2009;21:2284–97. doi: 10.1105/tpc.109.068072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol. 2010;11:861–71. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang L, Yang S, Zhang S, Liu M, Lai J, Qi Y, et al. The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant J. 2009;60:666–78. doi: 10.1111/j.1365-313X.2009.03992.x. [DOI] [PubMed] [Google Scholar]
- 17.Park BS, Song JT, Seo HS. Arabidopsis nitrate reductase activity is stimulated by the E3 SUMO ligase AtSIZ1. Nat Commun. 2011;2:400. doi: 10.1038/ncomms1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen CC, Chen YY, Tang IC, Liang HM, Lai CC, Chiou JM, et al. Arabidopsis SUMO E3 ligase SIZ1 is involved in excess copper tolerance. Plant Physiol. 2011;156:2225–34. doi: 10.1104/pp.111.178996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miura K, Lee J, Miura T, Hasegawa PM. SIZ1 controls cell growth and plant development in Arabidopsis through salicylic acid. Plant Cell Physiol. 2010;51:103–13. doi: 10.1093/pcp/pcp171. [DOI] [PubMed] [Google Scholar]
- 20.Ling Y, Zhang C, Chen T, Hao H, Liu P, Bressan RA, et al. Mutation in SUMO E3 ligase, SIZ1, disrupts the mature female gametophyte in Arabidopsis. PLoS ONE. 2012;7:e29470. doi: 10.1371/journal.pone.0029470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishida T, Yoshimura M, Miura K, Sugimoto K. MMS21/HPY2 and SIZ1, two Arabidopsis SUMO E3 ligases, have distinct functions in development. PLoS ONE. 2012;7:e46897. doi: 10.1371/journal.pone.0046897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang S, Qi Y, Yang C. Arabidopsis SUMO E3 ligase AtMMS21 regulates root meristem development. Plant Signal Behav. 2010;5:53–5. doi: 10.4161/psb.5.1.10158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, et al. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell. 2004;119:109–20. doi: 10.1016/j.cell.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 24.Xu P, Yuan D, Liu M, Li C, Liu Y, Zhang S, et al. AtMMS21, an SMC5/6 complex subunit, is involved in stem cell niche maintenance and DNA damage responses in Arabidopsis roots. Plant Physiol. 2013;161:1755–68. doi: 10.1104/pp.112.208942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aichinger E, Kornet N, Friedrich T, Laux T. Plant stem cell niches. Annu Rev Plant Biol. 2012;63:615–36. doi: 10.1146/annurev-arplant-042811-105555. [DOI] [PubMed] [Google Scholar]
- 26.Hashimura Y, Ueguchi C. The Arabidopsis MERISTEM DISORGANIZATION 1 gene is required for the maintenance of stem cells through the reduction of DNA damage. Plant J. 2011;68:657–69. doi: 10.1111/j.1365-313X.2011.04718.x. [DOI] [PubMed] [Google Scholar]
- 27.Budhiraja R, Hermkes R, Müller S, Schmidt J, Colby T, Panigrahi K, et al. Substrates related to chromatin and to RNA-dependent processes are modified by Arabidopsis SUMO isoforms that differ in a conserved residue with influence on desumoylation. Plant Physiol. 2009;149:1529–40. doi: 10.1104/pp.108.135053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elrouby N, Coupland G. Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc Natl Acad Sci USA. 2010;107:17415–20. doi: 10.1073/pnas.1005452107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller MJ, Barrett-Wilt GA, Hua Z, Vierstra RD. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc Natl Acad Sci USA. 2010;107:16512–7. doi: 10.1073/pnas.1004181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park HC, Choi W, Park HJ, Cheong MS, Koo YD, Shin G, et al. Identification and molecular properties of SUMO-binding proteins in Arabidopsis. Mol Cells. 2011;32:143–51. doi: 10.1007/s10059-011-2297-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H, Makeen K, Yan Y, Cao Y, Sun S, Xu G. OsSIZ1 regulates the vegetative growth and reproductive development in rice. Plant Mol Biol Rep. 2011;29:411–7. doi: 10.1007/s11105-010-0232-y. [DOI] [Google Scholar]
- 32.Thangasamy S, Guo CL, Chuang MH, Lai MH, Chen J, Jauh GY. Rice SIZ1, a SUMO E3 ligase, controls spikelet fertility through regulation of anther dehiscence. New Phytol. 2011;189:869–82. doi: 10.1111/j.1469-8137.2010.03538.x. [DOI] [PubMed] [Google Scholar]


