Abstract
Autophagy is a major pathway for the delivery of proteins or organelles to be degraded in the vacuole and recycled. It can be induced by abiotic stresses, senescence, and pathogen infection. Recent research has shown that autophagy is activated by ER stress. Here we review the major progress that has been made in the study of autophagy and ER stress in plants, and describe the links between ER stress and autophagy to guide further study on how autophagy is regulated in response to ER stress.
Keywords: autophagy, ER stress, unfolded protein response, TOR, IRE1
Introduction
Autophagy is a major pathway for delivery of proteins and organelles to lysosomes in mammalian cells or vacuoles in plant cells, where they are degraded and recycled. Three major types of autophagy have been described based on their mechanism; macroautophagy,1 microautophagy2 and chaperone-mediated autophagy.3 Both macroautophagy and microautophagy can be either non-selective or selective, while chaperone-mediate autophagy is highly selective.4,5 To date, macroautophagy and microautophagy have been described in plants,6,7 and in this review autophagy refers to macroautophagy. When autophagy is induced, the material that needs to be degraded begins to be surrounded by a double-membrane cup-shaped structure called a phagophore which is completed to form a double-membrane vesicle, the autophagosome. Upon delivery to the vacuole, the outer membrane of the autophagosome fuses with the vacuole membrane, and the inner membrane with the cargo is degraded by vacuolar hydrolases.8 Three types of markers have been used commonly to monitor autophagosomes in plants; monodansylcadaverine staining, an acidotropic dye that stains acidic membrane compartments;9 LysoTracker Red, another acidotropic dye that has been used most frequently in animal cells10 and green fluorescent protein (GFP)-ATG8 fusion proteins, since ATG8 is required for autophagosome formation via association with the autophagosome membrane.9,11,12 These markers have allowed the identification of conditions under which autophagy is activated in plants, most recently upon encountering endoplasmic reticulum (ER) stress.56 In this review we describe the pathways known to regulate autophagy and ER stress in plants and the relationships between them that are beginning to be revealed (Fig. 1).
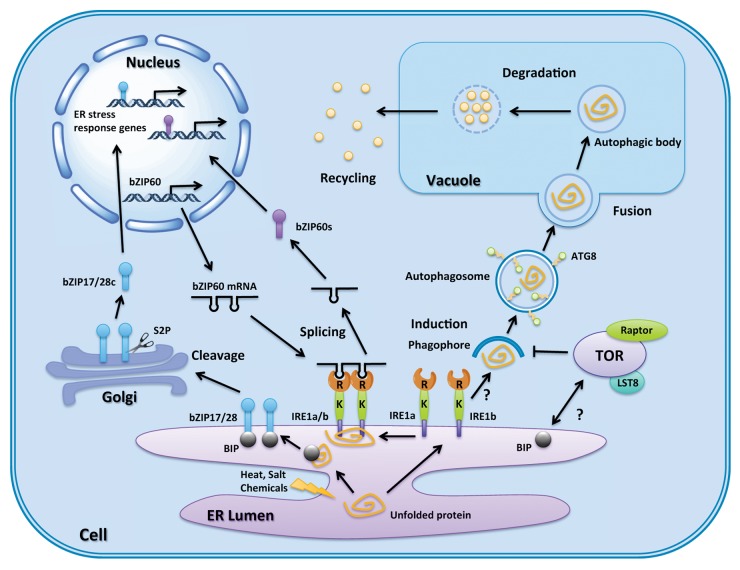
Figure 1. Autophagy and ER stress regulation pathways in plants. A general schematic of autophagy and the UPR under ER stress is shown. ER stress is caused by disturbance of protein folding, and two branches of the UPR are induced by unfolded or misfolded protein aggregates in the ER lumen. The first pathway starts when BIP recognizes and binds to unfolded or misfolded proteins, which activates bZIP17 or bZIP28 to translocate to the Golgi and be cleaved by S2P. The cleaved bZIP17 or bZIP28 enters the nucleus and initiates ER stress response gene transcription. The second pathway begins with oligomerization of IRE1 and its binding to unfolded or misfolded protein. The activated IRE1 splices bZIP60 mRNA. Spliced bZIP60 mRNA is translated and enters the nucleus to begin transcription of ER stress response genes. IRE1b is also required for autophagy induction, although how it is involve in the activation process is still unknown. ATG8 is a key protein for autophagosome formation, and is also used as a marker to monitor autophagy. Autophagosomes then carry cargo to the vacuole where the outer membrane fuses with the vacuole membrane, and the inner membrane and cargo are degraded in the vacuole. Breakdown products are exported into the cytosol for reuse. bZIP17/28c, cleaved form of bZIP17 or bZIP28; bZIP60s, translated protein from IRE1-spliced bZIP60; R, ribonuclease domain of IRE1; K, kinase domain of IRE1; S2P, golgi-associated site 2 protease.
Function and Regulation of the Autophagy Pathway in Plants
The molecular mechanism of autophagy was initially studied in yeast and animals, facilitating the discovery of components in the plant autophagy pathway. To date, more than 30 genes that function in autophagy have been identified in yeast genetic screens, and many of them have also been identified in plants. The identified genes can be divided into several functional groups:8,13 the ATG9 cycling system, which is proposed to be responsible for the initiation of autophagosomes;14-17 an ATG1-ATG13 kinase group, which functions in ATG9 movement and autophagy induction;12,18-20 two ubiquitin-like conjugation systems, that are responsible for the conjugation of ATG8 to the membrane lipid phosphatidylethanolamine (PE), with this ATG8-PE conjugation system being required for the complete formation of autophagosomes;1,11 and a phosphatidylinositol 3-kinase (PtdIns3K) complex, which is required for the initiation of autophagosome formation in yeast and animals,1,8 but is still poorly studied in terms of its function in plant autophagy.
A key pathway for autophagy regulation is the TOR (target of rapamycin) signaling pathway. TOR is a PtdIns3K-related kinase that functions as a serine/threonine protein kinase, and it works as a component of larger complexes. In yeast and animals, there are two types of TOR complexes, mTORC1 and mTORC2, differentiated by distinct binding partners and functions.21,22 In plants, only homologs of mTORC1 components have been identified,23,24 including RAPTOR, which recruits substrates and presents them to TOR for phosphorylation,25-27 and LST8, which stabilizes the complex.28,29 Previous studies indicated that TOR is a negative regulator of autophagy in plants,30,31 and it signals through phosphorylation of downstream substrates.32,33 In plants, ribosomal p70 S6 kinase has been identified as a substrate of TOR,34,35 and other potential TOR substrates include Arabidopsis Mei2-like1, based on its in vitro interaction with RAPTOR;36 ErbB-3 epidermal growth factor receptor binding protein, based on the correlation of its expression and TOR37,38 and Tap46, which is phosphorylated by TOR and interacts with protein phosphatase type 2A, a regulator of autophagy in yeast.39
Autophagy has been extensively studied in humans and animals due to its important role in cancer, aging, immunity and inflammatory responses.13,40-42 In plants, autophagy is involved in the response to abiotic stresses and pathogen infection, in protein degradation during senescence, with a basal level of autophagy under normal conditions functioning as a housekeeping process.7,16,43-46 Under stress conditions, autophagy is induced,47,48 and these stresses include nutrient deprivation,49-52 salt and drought stress53 and oxidative stress.54,55 A recent study shows that autophagy can also be induced by ER stress.56,57
The ER Stress Response
The ER is the organelle in which many membrane and secretory proteins are synthesized and folded. The ER folding machinery consists of a variety of molecular chaperones and other factors that assist in correctly folding polypeptides to ensure the final structure and function of proteins. One of the molecular chaperones in the ER is binding protein (BIP), a heat shock protein 70 that assists protein folding in the ER lumen by binding to nascent proteins when they enter the ER.58 If proteins are not correctly folded, an ER quality control system is able to detect these unfolded or misfolded proteins and they can be degraded by an ER-associated degradation system. However, if the amount of unfolded or misfolded proteins exceeds the capacity of the folding and degradation system, it results in ER stress.59,60 ER stress can be triggered by environmental conditions such as drought and salt stresses, pathogen attack, and heat stress or biotic agents,61-63 leading to a homeostatic response called the unfolded protein response (UPR). The UPR aids proper folding and degradation of unfolded or misfolded proteins by upregulation of UPR-related genes, including BIP, calnexin, calreticulin, and protein disulfide isomerase.64,65 In the laboratory, the UPR can be induced by chemicals such as tunicamycin, which interferes with N-linked glycosylation of glycoproteins, and dithiothreitol (DTT), a reducing agent that inhibits the proper formation of disulfide bonds.66
ER Stress Sensor System
ER stress sensors located on the ER membrane initiate the UPR signaling pathway. Three branches of the UPR signaling pathway have been identified from work in yeast and mammals, initiated by three classes of ER sensor/transducers.67 The first ER stress sensor, inositol-requiring enzyme-1 (IRE1), was identified in yeast, and functions as both a kinase and a ribonuclease.68,69 Another two ER stress sensors were later discovered in mammals, activating transcription factor 6 (ATF6), a membrane-associated transcription factor that upon ER stress translocates to the nucleus to upregulate UPR genes,70 and protein kinase RNA-like endoplasmic reticulum kinase (PERK), a membrane-associated protein kinase that affects translation by phosphorylating a translation initiation factor, eukaryotic initiation factor-2α.71 However, in plants, only two branches of the UPR signaling pathway, those initiated by IRE1 and ATF6, have been identified so far.72,73
IRE1 is a highly conserved ER stress sensor that has been identified in yeast, nematodes, fruit flies, zebrafish and mammals. ER stress activates IRE1 via its oligomerization and autophosphorylation,74 and IRE1 splices a membrane-associated basic leucine zipper (bZIP) transcription factor mRNA, HAC1 in yeast or XBP1 in mammals.75 Both HAC1 and XBP1 mRNA have a highly conserved secondary structure that is recognized by IRE1.76,77 Spliced HAC1 and XBP1 mRNA are then translated and the proteins translocate to the nucleus, activating the transcription of UPR genes.68,69 In Arabidopsis thaliana, two IRE1 homologs have been identified, IRE1a and IRE1b, both of which are located in the perinuclear ER.72 In an analogous manner to yeast and mammals, Arabidopsis IRE1 splices the mRNA encoding the bZIP60 transcription factor, upon which the bZIP60 protein is synthesized and translocates into the nucleus to upregulate UPR genes such as BIP.78-80 IRE1a and IRE1b have been suggested to have partially redundant functions in the splicing of bZIP60 mRNA in seedlings, though IRE1b seems to have a predominant role.63,78,79
The IRE1/bZIP60 system seems to be present in other plants in addition to Arabidopsis. IRE1 has a rice (Oryza sativa) homolog, OsIRE181 and genes similar to bZIP60 that can be spliced by IRE1 have also been found recently in rice, OsbZIP74 and OsbZIP50,82,83 and in maize (Zea mays), ZmbZIP60.
Another membrane-associated ER transducer that is well studied in mammalian cells is ATF6, a type II membrane protein with a single-pass transmembrane domain, a bZIP domain facing the cytosol, and a C-terminal tail facing the ER lumen under unstressed conditions. Under ER stress, ATF6 is transported from the ER to the Golgi, where its C-terminal tail is proteolytically cleaved by a Golgi-associated protease, site 1 protease, and its membrane domain is cleaved by site 2 protease. The cleaved ATF6 is activated and translocates to the nucleus to upregulate target genes.66,84,85
In Arabidopsis, two homologs of ATF6 have been identified and studied, bZIP17 and bZIP28. Both of them localize to the ER membrane in normal conditions. Under ER stress, they are proteolytically cleaved by site 1 and site 2 proteases and translocate to the nucleus, where they upregulate the target genes for the UPR, including BIP3.73,86
Interestingly, evidence exists suggesting that the bZIP17/bZIP28 and IRE1/bZIP60 pathways might converge for UPR gene regulation. bZIP60 seems to function by heterodimerization with bZIP17 and bZIP28,87 and BIP3 is suggested to be upregulated by both bZIP28 and bZIP60 under ER stress in Arabidopsis.87,88
Relationship Between ER Stress and Autophagy
Autophagy was first found to be induced by ER stress in mammalian cells, with IRE1 as a link between the two processes, and the kinase function of IRE1, instead of its ribonuclease function, required for the induction of autophagy.89 The c-Jun N-terminal kinase pathway is activated by IRE1 and is required for autophagy induction upon ER stress.66
In plants, autophagy was also recently discovered to be induced by ER stress.56 In Arabidopsis, the ER stress agents DTT and tunicamycin cause activation of autophagy as evidenced by the formation of autophagosomes containing ER membrane.56 As in mammalian cells, IRE1 is suggested to be a link between autophagy and ER stress in plants. While disruption of IRE1a seems to have no effect on autophagy, no autophagosomes are observed in an ire1b knockout mutant when treated with ER stress agents, suggesting that IRE1b is required for the induction of autophagy. In Arabidopsis, the only target mRNA identified so far that is spliced by IRE1 is bZIP60. However, a knockout mutant in the bZIP60 gene has no effect on autophagy induction by ER stress. In animals, the c-Jun N-terminal kinase pathway functions downstream of IRE1 in autophagy activation, but there is no evidence that this pathway exists in plants, suggesting that the components downstream of IRE1 in plants must be different from those in animals. Therefore, we suggest that either additional as yet unidentified factors can be spliced by IRE1b in plants, leading to autophagy activation or that the kinase function of IRE1 in plants is responsible for autophagy via an alternative downstream pathway.
As mentioned above, TOR is a negative regulator of autophagy in plants.30,31 Recent research has shown that although it has been generally accepted for a number of years that Arabidopsis is resistant to the TOR inhibitor rapamycin, it is actually sensitive to rapamycin when used at a higher concentration than in yeast and animals24,35 and the model green alga Chlamydomonas reinhardtii shows high sensitivity to rapamycin, indicating that this inhibitor may be useful in studying photosynthetic organisms.90 Chlamydomonas reinhardtii is a unicellular organism in which the interaction between TOR and LST8 was first shown,28 and TOR also negatively regulates autophagy induced by stresses in Chlamydomonas reinhardtii. Recent studies in this model alga identify BIP as a link between TOR signaling and ER stress.91,92 Inhibition of TOR by rapamycin causes repression of BIP activity by phosphorylation, to the same extent as inhibition of protein synthesis with cycloheximide, while ER stress induced by tunicamycin or heat shock results in BIP activation by dephosphorylation.91 Though this research suggests that BIP modification might be regulated by TOR signaling via control of protein synthesis,91 the function of TOR in autophagy raises the interesting possibility that BIP is also a link between autophagy regulation and ER stress.
Conclusions and Perspectives
Although a major pathway for protein degradation and recycling under stress, autophagy is still poorly understood in plants. Recent studies have shown that autophagy is involved in the response to ER stress in Arabidopsis.56 A variety of future challenges remain in understanding how autophagy functions and is regulated under ER stress.
How autophagy is involved in the response to ER stress in plants is still not clear. As a major pathway for protein and organelle vacuolar delivery and degradation, autophagy may play a role in reducing the amount of misfolded and unfolded proteins in the ER by aiding the breakdown and recycling of ER components. During ER stress, pieces of the ER membrane and its contents are engulfed by autophagosomes and delivered to the vacuole.57 Whether this engulfment is selective in unclear; one possibility is that a receptor protein specifically recognizes components within the ER membrane when ER stress responses are activated and causes their incorporation into autophagosomes. Alternatively, ER stress may cause some fragmentation of the ER, enabling its engulfment by autophagosomes. Selective autophagy has been described in plants,93-95 but the mechanism of degradation of organelles by autophagy is still not well understood.5
As noted above, IRE1b has been suggested to be required for the induction of autophagy by ER stress in Arabidopsis. IRE1b can function as both a kinase and a ribonuclease since it has a kinase domain and a ribonuclease domain facing the cytosol.68,69 Though IRE1b has been studied primarily as a non-conventional splicing factor of bZIP60, its function in splicing bZIP60 mRNA does not seem to be involved in autophagy induction.56 This suggests that either other components exist that can be spliced by IRE1b and function in autophagy pathway activation, or that other functions of IRE1b activate autophagy, such as its kinase activity. Future research is needed to distinguish between these possibilities and to identify the downstream regulators of the IRE1b pathway in autophagy signaling.
Different species of plants shows slight differences in components functioning in autophagy signaling in response to ER stress. In rice, only one IRE1 homolog has been identified.81 Studies of the model green alga, Chlamydomonas reinhardtii, indicated BIP as a link between ER stress and TOR signaling, which is a key pathway in autophagy regulation, but it is not known whether this is also true for higher plant species. Even in Arabidopsis, IRE1a, as a highly conserved homolog of IRE1b, seems to have no effect on autophagy induction.56 Although it is possible that the reason for this difference is the low expression of IRE1a in some tissues, IRE1a and IRE1b seem to have only partial functional overlap.56,63 Therefore, future work is needed in various plant species to determine the commonalities and differences in the factors that link autophagy and ER stress responses.
In mammalian cells, a third ER stress response pathway is initiated by PERK, a type I transmembrane protein kinase that phosphorylates eukaryotic initiation factor-2α in response to ER stress and leads to the inhibition of general mRNA translation to reduce the generation of unfolded proteins.67 However, none of the components of the PERK pathway have been discovered in plants. It is possible that functional homologs of some components in the PERK pathway are present in plants, but with little sequence homology to the animal components. Even if the PERK pathway is absent, whether there is another branch of the ER stress sensing and regulation pathway in plants is an open question. Note to Author: Please provide in-text citation for Figure 1
Acknowledgments
This work was supported by grant no. MCB-1051818 from the National Science Foundation to D.C.B.
Glossary
Abbreviations:
- GFP
green fluorescent protein
- PE
phosphatidylethanolamine
- PtdIns3K
phosphatidylinositol 3-kinase
- TOR
target of rapamycin
- ER
endoplasmic reticulum
- BIP
Binding protein
- UPR
unfolded protein response
- DTT
dithiothreitol
- IRE1
inositol-requiring enzyme-1
- ATF6
activating transcription factor 6
- PERK
protein kinase RNA-like endoplasmic reticulum kinase
- bZIP
membrane-associated basic leucine zipper
- R
ribonuclease domain of IRE1
- K
kinase domain of IRE1
- S2P
golgi-associated site 2 protease
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/24297
References
- 1.Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol. 2009;335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mijaljica D, Prescott M, Devenish RJ. Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy. 2011;7:673–82. doi: 10.4161/auto.7.7.14733. [DOI] [PubMed] [Google Scholar]
- 3.Orenstein SJ, Cuervo AM. Chaperone-mediated autophagy: molecular mechanisms and physiological relevance. Semin Cell Dev Biol. 2010;21:719–26. doi: 10.1016/j.semcdb.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li WW, Li J, Bao JK. Microautophagy: lesser-known self-eating. Cell Mol Life Sci. 2012;69:1125–36. doi: 10.1007/s00018-011-0865-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Floyd BE, Morriss SC, Macintosh GC, Bassham DC. What to eat: evidence for selective autophagy in plants. J Integr Plant Biol. 2012;54:907–20. doi: 10.1111/j.1744-7909.2012.01178.x. [DOI] [PubMed] [Google Scholar]
- 6.Van der Wilden W, Herman EM, Chrispeels MJ. Protein bodies of mung bean cotyledons as autophagic organelles. Proc Natl Acad Sci USA. 1980;77:428–32. doi: 10.1073/pnas.77.1.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bassham DC. Plant autophagy--more than a starvation response. Curr Opin Plant Biol. 2007;10:587–93. doi: 10.1016/j.pbi.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Bassham DC. Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol. 2012;63:215–37. doi: 10.1146/annurev-arplant-042811-105441. [DOI] [PubMed] [Google Scholar]
- 9.Contento AL, Xiong Y, Bassham DC. Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J. 2005;42:598–608. doi: 10.1111/j.1365-313X.2005.02396.x. [DOI] [PubMed] [Google Scholar]
- 10.Swanson SJ, Bethke PC, Jones RL. Barley aleurone cells contain two types of vacuoles. Characterization Of lytic organelles by use of fluorescent probes. Plant Cell. 1998;10:685–98. doi: 10.1105/tpc.10.5.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, et al. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 2004;16:2967–83. doi: 10.1105/tpc.104.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li F, Vierstra RD. Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 2012;17:526–37. doi: 10.1016/j.tplants.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–22. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He C, Klionsky DJ. Atg9 trafficking in autophagy-related pathways. Autophagy. 2007;3:271–4. doi: 10.4161/auto.3912. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198:219–33. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, et al. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 2002;129:1181–93. doi: 10.1104/pp.011024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, et al. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell. 2009;21:2914–27. doi: 10.1105/tpc.109.068635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–9. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 19.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–13. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vigani G. Discovering the role of mitochondria in the iron deficiency-induced metabolic responses of plants. J Plant Physiol. 2012;169(1):1–11. doi: 10.1016/j.jplph.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 21.Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–68. doi: 10.1016/S1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 22.Wedaman KP, Reinke A, Anderson S, Yates J, 3rd, McCaffery JM, Powers T. Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:1204–20. doi: 10.1091/mbc.E02-09-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Díaz-Troya S, Pérez-Pérez ME, Florencio FJ, Crespo JL. The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy. 2008;4:851–65. doi: 10.4161/auto.6555. [DOI] [PubMed] [Google Scholar]
- 24.Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, et al. Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci USA. 2002;99:6422–7. doi: 10.1073/pnas.092141899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–89. doi: 10.1016/S0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 26.Anderson GH, Veit B, Hanson MR. The Arabidopsis AtRaptor genes are essential for post-embryonic plant growth. BMC Biol. 2005;3:12. doi: 10.1186/1741-7007-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deprost D, Truong HN, Robaglia C, Meyer C. An Arabidopsis homolog of RAPTOR/KOG1 is essential for early embryo development. Biochem Biophys Res Commun. 2005;326:844–50. doi: 10.1016/j.bbrc.2004.11.117. [DOI] [PubMed] [Google Scholar]
- 28.Díaz-Troya S, Florencio FJ, Crespo JL. Target of rapamycin and LST8 proteins associate with membranes from the endoplasmic reticulum in the unicellular green alga Chlamydomonas reinhardtii. Eukaryot Cell. 2008;7:212–22. doi: 10.1128/EC.00361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moreau M, Azzopardi M, Clément G, Dobrenel T, Marchive C, Renne C, et al. Mutations in the Arabidopsis homolog of LST8/GβL, a partner of the target of Rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. Plant Cell. 2012;24:463–81. doi: 10.1105/tpc.111.091306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Bassham DC. TOR is a negative regulator of autophagy in Arabidopsis thaliana. PLoS ONE. 2010;5:e11883. doi: 10.1371/journal.pone.0011883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pérez-Pérez ME, Florencio FJ, Crespo JL. Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol. 2010;152:1874–88. doi: 10.1104/pp.109.152520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raught B, Gingras AC, Sonenberg N. The target of rapamycin (TOR) proteins. Proc Natl Acad Sci USA. 2001;98:7037–44. doi: 10.1073/pnas.121145898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren M, Qiu S, Venglat P, Xiang D, Feng L, Selvaraj G, et al. Target of rapamycin regulates development and ribosomal RNA expression through kinase domain in Arabidopsis. Plant Physiol. 2011;155:1367–82. doi: 10.1104/pp.110.169045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahfouz MM, Kim S, Delauney AJ, Verma DP. Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell. 2006;18:477–90. doi: 10.1105/tpc.105.035931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiong Y, Sheen J. Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. J Biol Chem. 2012;287:2836–42. doi: 10.1074/jbc.M111.300749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson GH, Hanson MR. The Arabidopsis Mei2 homologue AML1 binds AtRaptor1B, the plant homologue of a major regulator of eukaryotic cell growth. BMC Plant Biol. 2005;5:2. doi: 10.1186/1471-2229-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolaï M, et al. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 2007;8:864–70. doi: 10.1038/sj.embor.7401043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horváth BM, Magyar Z, Zhang Y, Hamburger AW, Bakó L, Visser RG, et al. EBP1 regulates organ size through cell growth and proliferation in plants. EMBO J. 2006;25:4909–20. doi: 10.1038/sj.emboj.7601362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahn CS, Han JA, Lee HS, Lee S, Pai HS. The PP2A regulatory subunit Tap46, a component of the TOR signaling pathway, modulates growth and metabolism in plants. Plant Cell. 2011;23:185–209. doi: 10.1105/tpc.110.074005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones SA, Mills KH, Harris J. Autophagy and inflammatory diseases. Immunol Cell Biol. 2013;91:250–8. doi: 10.1038/icb.2012.82. [DOI] [PubMed] [Google Scholar]
- 41.Liu G, Bi Y, Wang R, Wang X. Self-eating and self-defense: autophagy controls innate immunity and adaptive immunity. J Leukoc Biol. 2012;93(4):511–9. doi: 10.1189/jlb.0812389. [DOI] [PubMed] [Google Scholar]
- 42.Quy PN, Mizushima N. [Aging and Bio-motor function. Aging and autophagy] Clin Calcium. 2013;23:39–44. [PubMed] [Google Scholar]
- 43.Bassham DC. Function and regulation of macroautophagy in plants. Biochim Biophys Acta. 2009;1793:1397–403. doi: 10.1016/j.bbamcr.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 44.Hayward AP, Dinesh-Kumar SP. What can plant autophagy do for an innate immune response? Annu Rev Phytopathol. 2011;49:557–76. doi: 10.1146/annurev-phyto-072910-095333. [DOI] [PubMed] [Google Scholar]
- 45.Thomas H. Senescence, ageing and death of the whole plant. New Phytol. 2013;197:696–711. doi: 10.1111/nph.12047. [DOI] [PubMed] [Google Scholar]
- 46.Liu Y, Schiff M, Czymmek K, Tallóczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121:567–77. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 47.Sláviková S, Shy G, Yao Y, Glozman R, Levanony H, Pietrokovski S, et al. The autophagy-associated Atg8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. J Exp Bot. 2005;56:2839–49. doi: 10.1093/jxb/eri276. [DOI] [PubMed] [Google Scholar]
- 48.Xiong Y, Contento AL, Bassham DC. Disruption of autophagy results in constitutive oxidative stress in Arabidopsis. Autophagy. 2007;3:257–8. doi: 10.4161/auto.3847. [DOI] [PubMed] [Google Scholar]
- 49.Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem. 2002;277:33105–14. doi: 10.1074/jbc.M204630200. [DOI] [PubMed] [Google Scholar]
- 50.Phillips AR, Suttangkakul A, Vierstra RD. The ATG12-conjugating enzyme ATG10 Is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics. 2008;178:1339–53. doi: 10.1534/genetics.107.086199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 2005;138:2097–110. doi: 10.1104/pp.105.060673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiong Y, Contento AL, Bassham DC. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 2005;42:535–46. doi: 10.1111/j.1365-313X.2005.02397.x. [DOI] [PubMed] [Google Scholar]
- 53.Liu Y, Xiong Y, Bassham DC. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy. 2009;5:954–964. doi: 10.4161/auto.5.7.9290. [DOI] [PubMed] [Google Scholar]
- 54.Xiong Y, Contento AL, Nguyen PQ, Bassham DC. Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 2006;143:291–9. doi: 10.1104/pp.106.092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiong Y, Contento AL, Bassham DC. Disruption of autophagy results in constitutive oxidative stress in Arabidopsis. Autophagy. 2007;3:257–8. doi: 10.4161/auto.3847. [DOI] [PubMed] [Google Scholar]
- 56.Liu Y, Burgos JS, Deng Y, Srivastava R, Howell SH, Bassham DC. Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell. 2012;24:4635–51. doi: 10.1105/tpc.112.101535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Y, Bassham DC. Degradation of the endoplasmic reticulum by autophagy in plants. Autophagy. 2013;9:622–3. doi: 10.4161/auto.23559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Otero JH, Lizák B, Hendershot LM. Life and death of a BiP substrate. Semin Cell Dev Biol. 2010;21:472–8. doi: 10.1016/j.semcdb.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–29. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 60.Vitale A, Boston RS. Endoplasmic reticulum quality control and the unfolded protein response: insights from plants. Traffic. 2008;9:1581–8. doi: 10.1111/j.1600-0854.2008.00780.x. [DOI] [PubMed] [Google Scholar]
- 61.Liu JX, Howell SH. Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell. 2010;22:2930–42. doi: 10.1105/tpc.110.078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ye C, Dickman MB, Whitham SA, Payton M, Verchot J. The unfolded protein response is triggered by a plant viral movement protein. Plant Physiol. 2011;156:741–55. doi: 10.1104/pp.111.174110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moreno AA, Mukhtar MS, Blanco F, Boatwright JL, Moreno I, Jordan MR, et al. IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS ONE. 2012;7:e31944. doi: 10.1371/journal.pone.0031944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kamauchi S, Nakatani H, Nakano C, Urade R. Gene expression in response to endoplasmic reticulum stress in Arabidopsis thaliana. FEBS J. 2005;272:3461–76. doi: 10.1111/j.1742-4658.2005.04770.x. [DOI] [PubMed] [Google Scholar]
- 65.Martínez IM, Chrispeels MJ. Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell. 2003;15:561–76. doi: 10.1105/tpc.007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Howell SH. Endoplasmic Reticulum Stress Responses in Plants. Annu Rev Plant Biol. 2013 doi: 10.1146/annurev-arplant-050312-120053. [DOI] [PubMed] [Google Scholar]
- 67.Bravo R, Parra V, Gatica D, Rodriguez AE, Torrealba N, Paredes F, et al. Endoplasmic reticulum and the unfolded protein response: dynamics and metabolic integration. Int Rev Cell Mol Biol. 2013;301:215–90. doi: 10.1016/B978-0-12-407704-1.00005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mori K, Kawahara T, Yoshida H, Yanagi H, Yura T. Signalling from endoplasmic reticulum to nucleus: transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells. 1996;1:803–17. doi: 10.1046/j.1365-2443.1996.d01-274.x. [DOI] [PubMed] [Google Scholar]
- 69.Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/S0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 70.Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J Biol Chem. 1998;273:33741–9. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- 71.Shi Y, Vattem KM, Sood R, An J, Liang J, Stramm L, et al. Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Mol Cell Biol. 1998;18:7499–509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koizumi N, Martinez IM, Kimata Y, Kohno K, Sano H, Chrispeels MJ. Molecular characterization of two Arabidopsis Ire1 homologs, endoplasmic reticulum-located transmembrane protein kinases. Plant Physiol. 2001;127:949–62. doi: 10.1104/pp.010636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu JX, Srivastava R, Che P, Howell SH. An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell. 2007;19:4111–9. doi: 10.1105/tpc.106.050021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Korennykh AV, Egea PF, Korostelev AA, Finer-Moore J, Zhang C, Shokat KM, et al. The unfolded protein response signals through high-order assembly of Ire1. Nature. 2009;457:687–93. doi: 10.1038/nature07661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gonzalez TN, Sidrauski C, Dörfler S, Walter P. Mechanism of non-spliceosomal mRNA splicing in the unfolded protein response pathway. EMBO J. 1999;18:3119–32. doi: 10.1093/emboj/18.11.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–91. doi: 10.1016/S0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 77.Oikawa D, Tokuda M, Hosoda A, Iwawaki T. Identification of a consensus element recognized and cleaved by IRE1 alpha. Nucleic Acids Res. 2010;38:6265–73. doi: 10.1093/nar/gkq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Deng Y, Humbert S, Liu JX, Srivastava R, Rothstein SJ, Howell SH. Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc Natl Acad Sci USA. 2011;108:7247–52. doi: 10.1073/pnas.1102117108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nagashima Y, Mishiba K, Suzuki E, Shimada Y, Iwata Y, Koizumi N. Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci Rep. 2011;1:29. doi: 10.1038/srep00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Iwata Y, Koizumi N. An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc Natl Acad Sci USA. 2005;102:5280–5. doi: 10.1073/pnas.0408941102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Okushima Y, Koizumi N, Yamaguchi Y, Kimata Y, Kohno K, Sano H. Isolation and characterization of a putative transducer of endoplasmic reticulum stress in Oryza sativa. Plant Cell Physiol. 2002;43:532–9. doi: 10.1093/pcp/pcf063. [DOI] [PubMed] [Google Scholar]
- 82.Hayashi S, Wakasa Y, Takahashi H, Kawakatsu T, Takaiwa F. Signal transduction by IRE1-mediated splicing of bZIP50 and other stress sensors in the endoplasmic reticulum stress response of rice. Plant J. 2012;69:946–56. doi: 10.1111/j.1365-313X.2011.04844.x. [DOI] [PubMed] [Google Scholar]
- 83.Lu SJ, Yang ZT, Sun L, Sun L, Song ZT, Liu JX. Conservation of IRE1-regulated bZIP74 mRNA unconventional splicing in rice (Oryza sativa L.) involved in ER stress responses. Mol Plant. 2012;5:504–14. doi: 10.1093/mp/ssr115. [DOI] [PubMed] [Google Scholar]
- 84.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–99. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ye J, Rawson RB, Komuro R, Chen X, Davé UP, Prywes R, et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–64. doi: 10.1016/S1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 86.Liu JX, Srivastava R, Che P, Howell SH. Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. 2007;51:897–909. doi: 10.1111/j.1365-313X.2007.03195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu JX, Howell SH. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell. 2010;22:782–96. doi: 10.1105/tpc.109.072173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Iwata Y, Fedoroff NV, Koizumi N. Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell. 2008;20:3107–21. doi: 10.1105/tpc.108.061002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–31. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Crespo JL, Díaz-Troya S, Florencio FJ. Inhibition of target of rapamycin signaling by rapamycin in the unicellular green alga Chlamydomonas reinhardtii. Plant Physiol. 2005;139:1736–49. doi: 10.1104/pp.105.070847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Díaz-Troya S, Pérez-Pérez ME, Pérez-Martín M, Moes S, Jeno P, Florencio FJ, et al. Inhibition of protein synthesis by TOR inactivation revealed a conserved regulatory mechanism of the BiP chaperone in Chlamydomonas. Plant Physiol. 2011;157:730–41. doi: 10.1104/pp.111.179861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Crespo JL. BiP links TOR signaling to ER stress in Chlamydomonas. Plant Signal Behav. 2012;7:273–5. doi: 10.4161/psb.18767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Svenning S, Lamark T, Krause K, Johansen T. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy. 2011;7:993–1010. doi: 10.4161/auto.7.9.16389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zientara-Rytter K, Lukomska J, Moniuszko G, Gwozdecki R, Surowiecki P, Lewandowska M, et al. Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy. 2011;7:1145–58. doi: 10.4161/auto.7.10.16617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhou J, Wang J, Cheng Y, Chi YJ, Fan B, Yu JQ, et al. NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 2013;9:e1003196. doi: 10.1371/journal.pgen.1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]