Abstract
Background
Previous studies on the association of X-ray repair cross-complementing group 1 (XRCC1) Arg194Trp, Arg399Gln, and Arg280His polymorphisms with head and neck cancer (HNC) have produced inconsistent results. The aim of the present study was to evaluate the effects of these three polymorphic variants on HNC risk.
Methods
The PubMed and EMBASE databases were searched for genetic association studies on the XRCC1 Arg194Trp, Arg399Gln, and Arg280His polymorphisms and HNC risk. (The most recent search was conducted on 20 August, 2013.) Twenty-six studies were identified and meta-analysis was performed to evaluate the association between the polymorphism and HNC by calculating combined odds ratios and 95% confidence intervals.
Results
No significant association was found under the allelic, homozygous, heterozygote, and dominant genetic models in the overall comparison. Further, no significant association between the XRCC1 Arg399Gln and Arg280His polymorphisms and HNC risk was detected under the four genetic models in subgroup analyses based on ethnicity, cancer site, and whether or not the studies had been adjusted for cigarette smoking and alcohol. However, in stratified analyses based on cancer site, a significant association was found between the XRCC1 Arg194Trp polymorphism and oral cancer under the allelic, heterozygote, and dominant models. The XRCC1 Arg194Trp polymorphism was significantly associated with HNC risk in studies that were adjusted for smoking and alcohol under the homozygous and heterozygote models.
Conclusion
The meta-analysis results suggest that the XRCC1 Arg399Gln and Arg280His polymorphisms are probably not associated with the risk of HNC, but the XRCC1 Arg194Trp polymorphism was associated with increased risk of HNC in the subgroup analysis of studies adjusted for smoking and alcohol and with increased risk of oral cancer in the stratified analyses based on cancer site. Further studies with larger samples are needed to confirm these findings.
Introduction
Head and neck cancer (HNC), including cancers in the oral cavity, pharynx (other than nasopharynx), and larynx, is the sixth most common cancer in the world [1]. Approximately 540,000 new cases and 271,000 deaths are reported annually worldwide, indicating a mortality of approximately 50% [2]. HNC is considered to be a complex disease because both genetic and environmental risk factors contribute to its etiology [3]. The principal risk factors for HNC include tobacco and alcohol use, and exposure to the human papillomavirus (HPV), which together contribute to the development of at least 90% of squamous cell carcinoma of the head and neck cases [1]. Furthermore, many recent studies have provided evidence that genetic factors including family history [4] and polymorphisms in genes [5]–[8] play important roles in the development of HNC.
Recent evidence indicates that DNA repair genes may determine individual susceptibility to HNC [9], [10]. Polymorphisms in the repair genes that encode enzymes may increase or decrease DNA repair capacity. The DNA repair pathway involves the direct reversal pathway, the excision repair pathway, and the post-replication/bypass pathway. The excision repair pathway includes base excision repair (BER), nucleotide excision repair, and mismatch repair [11]. X-ray repair cross-complementing group 1 (XRCC1) is an important DNA repair protein in the BER pathway [12]. In vitro and vivo studies have shown that XRCC1 plays a role either directly during the repair of single-strand breaks or indirectly during BER. Loss of XRCC1 activity resulted in decreased genetic stability, including the increased frequency of spontaneous and/or induced chromosome translocations and deletions [13]–[16]. Although more than 200 single nucleotide polymorphisms (SNPs) have been identified in XRCC1, only three common SNPs have been widely investigated in cancer risk. They are Arg194Trp (rs1799782), Arg280His (rs25489), and Arg399Gln (rs25487), located in exons 6, 9, and 10, respectively, of the XRCC1 gene [3]. In HapMap (http://snp.cshl.org/cgi-perl/gbrowse/hapmap24_B36/), the minor allele frequency (MAF) of Arg194Trp is 0.09 for Caucasians and 0.26 for Asians; the MAF of Arg399Gln is 0.37 for Caucasians and 0.26 for Asians; and the MAF of Arg280His is 0.04 for Caucasians and 0.09 for Asians. Some studies reported the association of the Arg194Trp [8], [17], Arg280His [17], [18], and Arg399Gln [19], [20] polymorphisms with risk of various cancers.
Lately, a number of studies have reported the association between XRCC1 polymorphisms and HNC risk, but the results are inconsistent. Tea et al. [9] and Ramachandran et al. [10] found that the Arg194Trp polymorphism might increase the HNC risk, while Matullo et al. [21] reported the opposite finding. The function of the Arg280His polymorphism is still not fully understood. Chuang et al. [22] conducted a pooled analysis and found that rare XRCC1 Arg280His homozygotes were associated with HNC risk after adjustment for cigarette smoking and alcohol consumption, while, in other studies, no such association was found [9], [23]–[25]. For the Arg399Gln polymorphism, the results are still controversial [10], [11], [24]. Therefore, we performed a meta-analysis to assess the association between the XRCC1 Arg194Trp, Arg280His and Arg399Gln polymorphisms and HNC risk.
Materials and Methods
Search Strategy
We searched PubMed and EMBASE databases for all genetic association studies on XRCC1 and HNC risk. (The most recent search was conducted on 20 August, 2013). Various combinations of the following terms were used in the search: “head and neck cancer”, “oral cancer”, “oropharyngeal cancer”, “laryngeal cancer”, “pharyngeal cancer”, “XRCC1”, “X-ray cross-complementing group 1”, “base excision repair”, “BER”, “SNP”, “single nucleotide polymorphism”, “polymorphism” and “variant”. Only English language papers were included in the search. The references cited in the original studies or review articles concerning the relevant topic were retrieved to potentially broaden the search for additional relevant publications.
Inclusion and Exclusion Criteria
The following criteria were used to select the articles for the meta-analysis: (a) case-control study or cohort study methodology was used; (b) association of HNC with the XRCC1 Arg194Trp or Arg399Gln or Arg280His polymorphisms was explored; and (c) sufficient data of genotypes presented with estimated odds ratios (ORs) and 95% confidence intervals (CIs) were available. The exclusion criteria used were: (a) the control population included malignant tumor patients or the study had no controls; (b) insufficient information was available about genotype frequency or number; (c) duplicate publications or publications that contained overlapping data.
Data Extraction
Information was extracted from all eligible publications carefully and independently by two investigators (Wei Wu and Lu Liu) using a standard protocol and data-collecting form based on the inclusion criteria. The original extraction data were checked by another investigator (Zhihua Yin), and disagreements were resolved by discussion among the three investigators. The following data were extracted: name of first author, year of publication, ethnicity of studied populations, site of cancer, genotyping method, source of controls, matching criteria, adjusted variables, and cases and controls with different genotypes.
Statistical Analysis
The Hardy-Weinberg equilibrium (HWE) test [26] was conducted on the control groups to evaluate the genetic equilibrium of each study. A P value>0.05 was taken to indicate no significant disequilibrium. To avoid the inclusion of unknown heterogeneities, studies in which the distribution of the genotypes of the XRCC1 gene polymorphisms in the control groups not consistent with the HWE were excluded in the subsequent analysis. The MAF was computed in the control groups. MAF is an estimate of the frequency at which the less common allele occurs in a given population. The strength of the association between an XRCC1 polymorphism and HNC risk was assessed by combined odds ratios (ORs) with 95% confidence interval (CIs). The significance of the combined ORs was determined by a Z test and two-sided P values<0.05 were considered significant. The chi-square-based Q statistical test was used for heterogeneity analysis [27]. In this study, P values<0.05 were taken to indicate significant heterogeneity among studies. The random-effects model was used when heterogeneity was significant [28]; otherwise, the fixed-effects model was used [29]. Heterogeneity across studies was detected using an I2 test. I2 values of <25% were considered low, I2 values of 25% to 75% were considered moderate, and I2 values of >75% were considered high [30]. We calculated the OR using four different genetic models: allelic model (B vs. A), homozygous model (BB vs. AA), heterozygote model (AB vs. AA), and dominant model (BB+AB vs. AA), where A represents the major allele and B represents the minor allele. Stratified analyses of each study by ethnicity, cancer site, and whether the data had been adjusted for smoking and alcohol were also conducted using the four genetic models, to identify the relationship between the XRCC1 polymorphism and HNC risk. Whenever possible, adjusted ORs in a logistic model were used to compute combined OR and 95% CI for studies adjusted for smoking and alcohol. Furthermore, sensitivity analyses were conducted to confirm the stability and reliability of our results [31]. Visual inspection of Begg's funnel plot and Egger's test were used to evaluate the publication bias in the meta-analysis and P values<0.05 were considered statistically significant [32], [33]. All statistical tests were performed with the software Stata version 12.0 (Stata Corporation, College Station, TX, USA).
Results
Study Characteristics
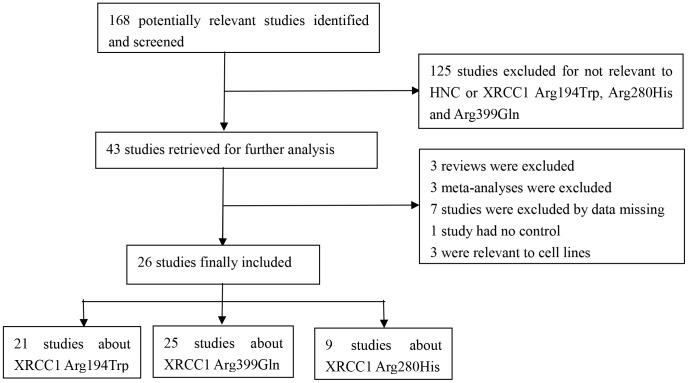
A total of 168 potentially relevant studies were retrieved after a comprehensive search of the PubMed and EMBASE databases (Figure 1), and 125 of these studies were excluded as not relevant to HNC or the XRCC1 Arg194Trp, Arg280His, and Arg399Gln polymorphisms. A further 17 studies were excluded including three reviews, three meta-analyses, seven studies with missing data, one study with no controls, and three studies relevant to cell lines. Consequently, 26 studies [9]–[11], [21], [23]–[25], [34]–[52] of the association of the XRCC1 Arg194Trp, Arg280His, and Arg399Gln polymorphisms with the risk of HNC were included in the meta-analysis. Twenty-one of the studies were about XRCC1 Arg194Trp, 25 were about XRCC1 Arg399Gln, and nine were about XRCC1 Arg280His (Figure 1).
Figure 1. Flowchart of the process used for selection of eligible studies.
The characteristics of the 26 eligible studies, including year of publication, ethnicity of studied populations, site of cancer, method of genotyping, source of controls, matching criteria, adjusted variables, cases and controls with different genotypes, HWE in controls, and MAF in controls for the XRCC1 Arg194Trp, Arg399Gln, and Arg280His polymorphisms are listed in Tables 1–3 respectively. All the studies were published between 1999 and 2013. The most commonly used genotyping method was polymerase chain reaction (PCR)-restriction fragment length polymorphism. The distribution of the genotypes of the XRCC1 Arg194Trp, Arg280His, and Arg399Gln polymorphisms in the control groups was consistent with the HWE, except in three of the studies. Two of these studies were related to the Arg194Trp polymorphism [37], [43], and one was related to the Arg399Gln polymorphism [51]. These studies were excluded from the subsequent analyses. Finally, to analyze the association of the three XRCC1 polymorphisms with the risk of HNC, 19 studies were selected for Arg194Trp, 24 studies were selected for Arg399Gln, and nine studies were selected for Arg280His.
Table 1. Characteristics of the Studies about the XRCC1 Arg194Trp polymorphism (rs1799782) Included in the Meta-analysis.
| Author | Year | Ethnicity | Cancer Site | Genotyping Method | Control Source | Matching Criteria | Adjusted Variables | Case | Control | HWEd | MAFe | ||||||
| N | AA | AB | BB | N | AA | AB | BB | ||||||||||
| Dos Reis | 2013 | Mixed | Oral | PCR-RFLP | PBa | Age, sex, race, smoke | None | 150 | 127 | 23 | 0 | 150 | 123 | 24 | 3 | 0.174 | 0.100 |
| Yen | 2008 | Asian | Oral | PCR-RFLP | PB | None | Age, sex, alcohol, smoke, betel nut chewing | 103 | 48 | 40 | 15 | 98 | 54 | 35 | 9 | 0.348 | 0.270 |
| Sturgis | 1999 | Mixed | Head&neck | PCR-RFLP | HBb | Age, sex, race | Age, sex, ethnicity, smoke, alcohol | 203 | 180 | 22 | 1 | 424 | 363 | 61 | 0 | 0.110 | 0.072 |
| Majumder | 2005 | Asian | Oral | PCR-RFLP | HB | None | None | 310 | 249 | 58 | 3 | 348 | 285 | 57 | 6 | 0.122 | 0.099 |
| Harth | 2008 | Caucasian | Head&neck | RT-PCR | HB | None | Age, sex | 258 | 217 | 40 | 1 | 300 | 259 | 39 | 2 | 0.690 | 0.072 |
| Applebaum | 2009 | Mixed | Head&neck | PCR-RFLP | PB | Age, sex | Age, sex, race, education, smoke, alcohol, HPV16 | 484 | 427 | 55 | 2 | 549 | 485 | 61 | 3 | 0.476 | 0.061 |
| Majumder | 2007 | Asian | Oral | PCR-RFLP | HB | None | Age, sex, smoke | 309 | 248 | 58 | 3 | 387 | 317 | 62 | 8 | 0.022 | 0.101 |
| Tae | 2004 | Asian | Head&neck | MAPA | HB | None | Age, smoke, alcohol | 120 | 59 | 52 | 9 | 145 | 101 | 39 | 5 | 0.611 | 0.169 |
| Varzim | 2003 | Caucasian | Larynx | PCR-RFLP | PB | None | None | 88 | 80 | 8 | 0 | 178 | 160 | 18 | 0 | 0.477 | 0.051 |
| Csejtei | 2009 | Caucasian | Head&neck | PCR-RFLP | HB | Age, sex | None | 108 | 96 | 11 | 1 | 102 | 85 | 15 | 2 | 0.191 | 0.093 |
| Kowalski | 2009 | Caucasian | Head&neck | PCR-RFLP | HB | Age | None | 92 | 71 | 21 | 0 | 124 | 102 | 22 | 0 | 0.278 | 0.089 |
| Demokan | 2005 | Caucasian | Head&neck | PCR-RFLP | PB | None | None | 95 | 78 | 14 | 3 | 98 | 88 | 8 | 2 | 0.004 | 0.061 |
| Ramachandran | 2006 | Asian | Oral | PCR-RFLP | PB | Age, sex | Age, sex, smoke, alcohol, betel quid chewing | 110 | 66 | 37 | 7 | 110 | 90 | 19 | 1 | 0.998 | 0.095 |
| Olshan | 2002 | Caucasian | Head&neck | PCR-RFLP | HB | Age, sex | Age, sex | 98 | 82 | 16 | 0 | 161 | 135 | 26 | 0 | 0.265 | 0.081 |
| Kietthubthew | 2006 | Asian | Oral | PCR-RFLP | PB | Age, sex, smoke, alcohol, betel chewing, religion | Other genes, betel quid chewing | 106 | 40 | 50 | 16 | 164 | 77 | 67 | 20 | 0.365 | 0.326 |
| Kumar | 2012 | Asian | Head&neck | PCR-RFLP | PB | Age, sex, habits | None | 278 | 144 | 111 | 23 | 278 | 121 | 131 | 26 | 0.264 | 0.329 |
| Gugatschka | 2011 | Caucasian | Head&neck | TaqMan | PB | None | None | 168 | 148 | 20 | 0 | 463 | 397 | 63 | 3 | 0.772 | 0.075 |
| Ho | 2007 | Mixed | Oral | PCR-RFLP | HB | None | Age, sex, ethnicity, family history, smoke, alcohol, radiation exposure | 137 | 108 | 29 | 0 | 503 | 433 | 69 | 1 | 0.306 | 0.071 |
| Rydzanicz | 2005 | Caucasian | Head&neck | PCR-RFLP | PB | Smoke, occupational exposure | None | 182 | 165 | 16 | 1 | 143 | 129 | 14 | 0 | 0.538 | 0.049 |
| Gajecka | 2005 | Caucasian | Larynx | PCR-RFLP | PB | None | None | 290 | 262 | 27 | 1 | 325 | 291 | 33 | 1 | 0.950 | 0.054 |
| Matullo | 2006 | Caucasian | Head&neck | TaqMan | UKc | None | None | 82 | 78 | 4 | 0 | 1094 | 951 | 141 | 2 | 0.171 | 0.066 |
PB: Population based;
HB: Hospital based;
UK: Unknown or unstated;
HWE: Hardy-Weinberg equilibrium in controls;
MAF: Minor allele frequency in controls.
Table 3. Characteristics of the Studies about the XRCC1 Arg280His polymorphism (rs25489) Included in the Meta-analysis.
| Author | Year | Ethnicity | Cancer Site | Genotyping Method | Control Source | Matching Criteria | Adjusted Variables | Case | Control | HWEc | MAFd | ||||||
| N | AA | AB | BB | N | AA | AB | BB | ||||||||||
| Majumder | 2005 | Asian | Oral | PCR-RFLP | HBa | None | None | 310 | 228 | 79 | 3 | 348 | 264 | 81 | 3 | 0.232 | 0.125 |
| Harth | 2008 | Caucasian | Head&neck | RT-PCR | HB | None | Age, sex | 312 | 283 | 28 | 1 | 300 | 270 | 30 | 0 | 0.362 | 0.050 |
| Applebaum | 2009 | Mixed | Head&neck | PCR-RFLP | PBb | Age, sex | Age, sex, race, education, smoke, alcohol, HPV16 | 484 | 437 | 46 | 1 | 548 | 492 | 52 | 4 | 0.052 | 0.055 |
| Majumder | 2007 | Asian | Oral | PCR-RFLP | HB | None | Age, sex, smoke | 307 | 225 | 79 | 3 | 387 | 297 | 87 | 3 | 0.213 | 0.120 |
| Tae | 2004 | Asian | Head&neck | MAPA | HB | None | Age, smoke, alcohol | 135 | 113 | 21 | 1 | 168 | 139 | 29 | 0 | 0.221 | 0.086 |
| Ramachandran | 2006 | Asian | Oral | PCR-RFLP | PB | Age, sex | Age, sex, smoke, alcohol, betel quid chewing | 110 | 77 | 31 | 2 | 110 | 83 | 26 | 1 | 0.502 | 0.127 |
| Kumar | 2012 | Asian | Head&neck | PCR-RFLP | PB | Age, sex, habits | None | 278 | 129 | 123 | 26 | 278 | 142 | 116 | 20 | 0.575 | 0.281 |
| Gugatschka | 2011 | Caucasian | Head&neck | TaqMan | PB | None | None | 168 | 159 | 9 | 0 | 463 | 430 | 32 | 1 | 0.621 | 0.037 |
| Ho | 2007 | Mixed | Oral | PCR-RFLP | HB | None | Age, sex, ethnicity, family history, smoke, alcohol, radiation exposure | 138 | 125 | 13 | 0 | 503 | 453 | 50 | 0 | 0.241 | 0.050 |
HB: Hospital based;
PB: Population based;
HWE: Hardy-Weinberg equilibrium in controls;
MAF: Minor allele frequency in controls.
Table 2. Characteristics of the Studies about the XRCC1 Arg399Gln polymorphism (rs25478) Included in the Meta-analysis.
| Author | Year | Ethnicity | Cancer Site | Genotyping Method | Control Source | Matching Criteria | Adjusted Variables | Case | Control | HWEd | MAFe | ||||||
| N | AA | AB | BB | N | AA | AB | BB | ||||||||||
| Dos Reis | 2013 | Mixed | Oral | PCR-RFLP | PBa | Age, sex, race, smoke | None | 150 | 64 | 62 | 24 | 150 | 62 | 54 | 34 | 0.002 | 0.407 |
| Yuan | 2012 | Asian | Head&neck | TaqMan | PB | Age, sex | Age, sex, smoke, alcohol | 390 | 221 | 146 | 23 | 886 | 481 | 339 | 66 | 0.558 | 0.266 |
| Sturgis | 1999 | Mixed | Head&neck | PCR-RFLP | HBb | Age, sex, race | None | 203 | 94 | 77 | 32 | 424 | 181 | 197 | 46 | 0.483 | 0.341 |
| Majumder | 2005 | Asian | Oral | PCR-RFLP | HB | None | None | 310 | 135 | 143 | 32 | 348 | 158 | 163 | 27 | 0.088 | 0.312 |
| Harth | 2008 | Caucasian | Head&neck | RT-PCR | HB | None | Age, sex | 310 | 114 | 166 | 30 | 300 | 143 | 121 | 36 | 0.189 | 0.322 |
| Applebaum | 2009 | Mixed | Head&neck | PCR-RFLP | PB | Age, sex | Age, sex, race, education, smoke, alcohol, HPV16 | 483 | 192 | 229 | 62 | 547 | 232 | 246 | 69 | 0.763 | 0.351 |
| Li | 2007 | Caucasian | Head&neck | PCR-RFLP | PB | Age, sex, race | Age, sex, smoke, alcohol | 830 | 335 | 374 | 121 | 854 | 360 | 385 | 109 | 0.702 | 0.353 |
| Majumder | 2007 | Asian | Oral | PCR-RFLP | HB | None | Age, sex, smoke | 309 | 134 | 143 | 32 | 385 | 170 | 179 | 36 | 0.255 | 0.326 |
| Tae | 2004 | Asian | Head&neck | MAPA | HB | None | Age, smoke, alcohol | 129 | 69 | 51 | 9 | 157 | 86 | 64 | 7 | 0.251 | 0.248 |
| Varzim | 2003 | Caucasian | Larynx | PCR-RFLP | PB | None | None | 88 | 37 | 40 | 11 | 178 | 80 | 80 | 18 | 0.759 | 0.326 |
| Csejtei | 2009 | Caucasian | Head&neck | PCR-RFLP | HB | Age, sex | None | 108 | 50 | 47 | 11 | 102 | 53 | 41 | 8 | 0.986 | 0.279 |
| Kowalski | 2009 | Caucasian | Head&neck | PCR-RFLP | HB | Age | None | 92 | 37 | 44 | 11 | 124 | 49 | 53 | 22 | 0.253 | 0.391 |
| Demokan | 2005 | Caucasian | Head&neck | PCR-RFLP | PB | None | None | 95 | 42 | 41 | 12 | 98 | 39 | 46 | 13 | 0.922 | 0.367 |
| Krupa | 2011 | Caucasian | Larynx | PCR-RFLP | HB | Age, sex | None | 253 | 93 | 111 | 49 | 253 | 105 | 113 | 35 | 0.603 | 0.362 |
| Kostrzewska-Poczekaj | 2013 | Caucasian | Head&neck | PCR-RFLP | PB | None | None | 290 | 110 | 154 | 26 | 158 | 50 | 81 | 27 | 0.550 | 0.427 |
| Jelonek | 2010 | Caucasian | Head&neck | PCR-RFLP | PB | Age, sex | None | 104 | 47 | 50 | 7 | 110 | 35 | 62 | 13 | 0.068 | 0.400 |
| Ramachandran | 2006 | Asian | Oral | PCR-RFLP | PB | Age, sex | Age, sex, smoke, alcohol, betel quid chewing | 110 | 46 | 48 | 16 | 110 | 73 | 33 | 4 | 0.910 | 0.186 |
| Olshan | 2002 | Caucasian | Head&neck | PCR-RFLP | HB | Age, sex | Age, sex | 98 | 45 | 50 | 3 | 161 | 62 | 82 | 17 | 0.183 | 0.360 |
| Kietthubthew | 2006 | Asian | Oral | PCR-RFLP | PB | Age, sex, smoke, alcohol, betel chewing, religion | None | 106 | 55 | 45 | 6 | 164 | 67 | 74 | 23 | 0.724 | 0.366 |
| Kumar | 2012 | Asian | Head&neck | PCR-RFLP | PB | Age, sex, habits | None | 278 | 128 | 124 | 26 | 278 | 98 | 144 | 36 | 0.133 | 0.388 |
| Gugatschka | 2011 | Caucasian | Head&neck | TaqMan | PB | None | None | 168 | 70 | 74 | 24 | 463 | 204 | 198 | 61 | 0.241 | 0.346 |
| Ho | 2007 | Mixed | Oral | PCR-RFLP | HB | None | Age, sex, ethnicity, family history, smoke, alcohol, radiation exposure | 138 | 61 | 62 | 15 | 503 | 220 | 216 | 67 | 0.229 | 0.348 |
| Rydzanicz | 2005 | Caucasian | Head&neck | PCR-RFLP | PB | Smoke, occupational exposure | None | 182 | 63 | 98 | 21 | 143 | 59 | 63 | 21 | 0.535 | 0.367 |
| Gajecka | 2005 | Caucasian | Larynx | PCR-RFLP | PB | None | None | 293 | 106 | 153 | 34 | 319 | 124 | 145 | 50 | 0.484 | 0.384 |
| Matullo | 2006 | Caucasian | Head&neck | TaqMan | UKc | None | None | 82 | 34 | 38 | 10 | 1094 | 484 | 482 | 128 | 0.632 | 0.337 |
PB: Population based;
HB: Hospital based;
UK: Unknown or unstated;
HWE: Hardy-Weinberg equilibrium in controls;
MAF: Minor allele frequency in controls.
Quantitative Data Synthesis
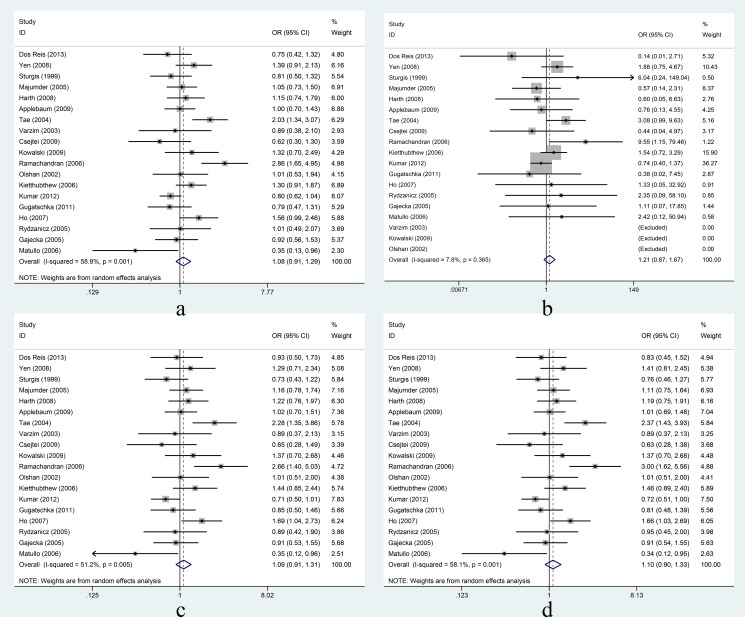
XRCC1 Arg194Trp: In the overall comparison, the Arg194Trp polymorphism was not significantly associated with HNC risk under the four different genetic models (Figure 2). Further, in the subgroup analyses based on ethnicity, the Arg194Trp polymorphism was found to be a risk factor in Asians while it was a protective factor in Caucasians under all genetic models; however, the association of the Arg194Trp polymorphism with HNC risk in Asians and in Caucasians was not significant (Table 4). In the stratified analyses based on cancer site, the Arg194Trp polymorphism was significantly associated with oral cancer using the allelic, heterozygote, and dominant models (allelic model: OR = 1.35, 95%CI = 1.00–1.82, I2 = 63.5%, Pheterogeneity = 0.02; heterozygote model: OR = 1.40, 95%CI = 1.13–1.73, I2 = 28.5%, Pheterogeneity = 0.22; dominant model: OR = 1.40, 95%CI = 1.14–1.72, I2 = 53.1%, Pheterogeneity = 0.06), but not significantly associated with oral cancer using the homozygous model. The Arg194Trp polymorphism was not significantly associated with larynx cancer under all four genetic models (Table 4). In the analyses of studies adjusted for smoking and alcohol, the Arg194Trp polymorphism was significantly associated with HNC risk under the homozygous and heterozygote models (homozygous model: OR = 2.21, 95%CI = 1.44–3.38, I2 = 0.0%, Pheterogeneity = 0.50; heterozygote model: OR = 1.65, 95%CI = 1.15–2.38, I2 = 50.0%, Pheterogeneity = 0.01), but the association was not significant under the dominant model. When the studies were not adjusted for smoking and alcohol, the Arg194Trp polymorphism was not significantly associated with HNC risk using any of the four genetic models (Table 4).
Figure 2. Association between XRCC1 Arg194Trp and risk of head and neck cancer under four genetic models.
Forest plots for a: Trp vs. Arg; b: TrpTrp vs. ArgArg; c: ArgTrp vs. ArgArg; d: TrpTrp+ ArgTrp vs. ArgArg. Random-effects models were used for a, c, and d; a fixed-effects model was used for b. Squares and horizontal lines represent the study-specific OR and 95% CI respectively; diamond indicates the summary OR and 95% CI.
Table 4. Stratified analyses of the association of the XRCC1 Arg194Trp (rs1799782), XRCC1 Arg399Gln (rs25487), and XRCC1 Arg280His (rs25489) polymorphisms with HNC risk.
| XRCC1(polymorphism) | Variables | Na | B vs. A | BB vs. AA | AB vs. AA | BB+AB vs. AA | ||||||||
| OR(95%CI) | P b | I2 (%) | OR(95%CI) | P | I2 (%) | OR(95%CI) | P | I2 (%) | OR(95%CI) | P | I2 (%) | |||
| XRCC1 Arg194Trp | Ethnicity | |||||||||||||
| (rs1799782) | Asian | 6 | 1.39c(0.97–1.98) | 0.00 | 81.2 | 1.34(0.93–1.92) | 0.052 | 54.5 | 1.40c(0.93–2.10) | 0.00 | 75.9 | 1.46c(0.95–2.24) | 0.00 | 80.4 |
| Caucasian | 9 | 0.90(0.74–1.11) | 0.50 | 0.0 | 0.78(0.27–2.30) | 0.91 | 0.0 | 0.92(0.74–1.14) | 0.52 | 0.0 | 0.91(0.74–1.12) | 0.49 | 0.0 | |
| Cancer Site | ||||||||||||||
| Oral | 6 | 1.35c(1.00–1.82)* | 0.02 | 63.5 | 1.48(0.92–2.38) | 0.19 | 33.0 | 1.40(1.13–1.73)* | 0.22 | 28.5 | 1.40(1.14–1.72)* | 0.06 | 53.1 | |
| Larynx | 2 | 0.92(0.59–1.42) | 0.95 | 0.0 | 1.11(0.07–17.85) | N/A | N/A | 0.90(0.57–1.43) | 0.97 | 0.0 | 0.98(0.58–1.43) | 0.96 | 0.0 | |
| Smoking and alcohol | ||||||||||||||
| Adjusted | 6 | N/A | N/A | N/A | 2.21(1.44–3.38)* | 0.50 | 0.0 | 1.65c(1.15–2.38)* | 0.01 | 50.0 | 1.41c(0.42–4.71) | 0.00 | 90.4 | |
| Unadjusted | 13 | 0.93(0.81–1.06) | 0.37 | 7.5 | 0.92(0.61–1.38) | 0.77 | 0.0 | 0.93(0.80–1.08) | 0.39 | 5.2 | 1.07c(0.89–1.29) | 0.01 | 49.0 | |
| XRCC1 Arg399Gln | Ethnicity | |||||||||||||
| (rs25487) | Asian | 7 | 1.02c(0.80–1.29) | 0.00 | 80.3 | 1.03c(0.62–1.71) | 0.00 | 73.6 | 0.98c(0.78–1.22) | 0.03 | 58.5 | 1.00c(0.76–1.30) | 0.00 | 74.3 |
| Caucasian | 14 | 1.02(0.95–1.10) | 0.09 | 35.9 | 0.98(0.84–1.15) | 0.06 | 40.4 | 1.10(0.99–1.23) | 0.24 | 19.6 | 1.07(0.98–1.19) | 0.16 | 27.2 | |
| Cancer Site | ||||||||||||||
| Oral | 5 | 1.09c(0.80–1.49) | 0.00 | 82.3 | 1.14c(0.60–2.19) | 0.00 | 76.1 | 1.06(0.89–1.27) | 0.06 | 55.5 | 1.11c(0.79–1.57) | 0.00 | 74.7 | |
| Larynx | 3 | 1.09(0.94–1.28) | 0.37 | 0.2 | 1.14(0.82–1.59) | 0.17 | 44.4 | 1.16(0.92–1.46) | 0.89 | 0.0 | 1.16(0.93–1.44) | 0.94 | 0.0 | |
| Smoking and alcohol | ||||||||||||||
| Adjusted | 6 | N/A | N/A | N/A | 1.13c(0.81–1.56) | 0.04 | 56.5 | 1.07(0.94–1.21) | 0.13 | 41.2 | 1.01(0.88–1.16) | 0.72 | 0.0 | |
| Unadjusted | 18 | 0.96c(0.88–1.06) | 0.02 | 46.4 | 0.90c(0.72–1.12) | 0.02 | 46.7 | 1.01c(0.88–1.15) | 0.04 | 39.7 | 1.03c(0.90–1.18) | 0.00 | 55.8 | |
| XRCC1 Arg280His | Ethnicity | |||||||||||||
| (rs25489) | Asian | 5 | 1.16(0.99–1.35) | 0.97 | 0.0 | 1.46(0.86–2.47) | 0.97 | 0.0 | 1.15(0.96–1.38) | 0.93 | 0.0 | 1.17(0.98–1.40) | 0.94 | 0.0 |
| Caucasian | 2 | 0.87(0.57–1.33) | 0.54 | 0.0 | 1.66(0.20–13.63) | 0.62 | 0.0 | 0.84(0.54–1.31) | 0.74 | 0.0 | 0.85(0.55–1.32) | 0.64 | 0.0 | |
| Cancer Site | ||||||||||||||
| Oral | 4 | 1.14(0.94–1.39) | 0.89 | 0.0 | 1.38(0.49–3.83) | 0.91 | 0.0 | 1.15(0.93–1.43) | 0.91 | 0.0 | 1.16(0.93–1.43) | 0.89 | 0.0 | |
| Smoking and alcohol | ||||||||||||||
| Adjusted | 4 | N/A | N/A | N/A | 1.59(0.08–32.17) | 0.02 | 82.5 | 1.22(0.91–1.63) | 0.10 | 51.7 | 0.98(0.52–1.86) | N/A | N/A | |
| Unadjusted | 5 | 1.11(0.95–1.30) | 0.75 | 0.0 | 1.43(0.84–2.43) | 0.98 | 0.0 | 1.10(0.92–1.33) | 0.76 | 0.0 | 1.09(0.93–1.27) | 0.86 | 0.0 | |
Number of comparisons;
P-value for Q-test;
The random-effects model was used when the P-value for the Q-test for heterogeneity was <0.05, otherwise the fixed-effects model was used.
Statistically significant, P<0.05.
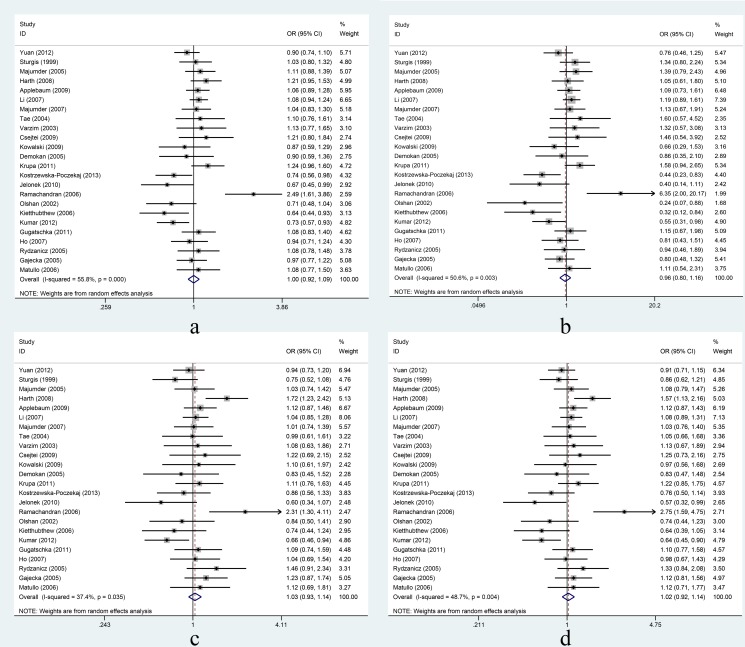
XRCC1 Arg399Gln: In the overall comparison, the Arg399Gln polymorphism was not significantly associated with HNC risk under the four different genetic models (Figure 3). Further, in the subgroup analyses based on ethnicity, the Arg399Gln polymorphism was not significantly associated with HNC risk in Asians or Caucasians (Table 4). In the stratified analyses based on cancer site, the Arg399Gln polymorphism was not significantly associated with oral cancer or larynx cancer (Table 4). When studies either adjusted or unadjusted for smoking and alcohol were analyzed, the Arg399Gln polymorphism was not significantly associated with HNC risk (Table 4).
Figure 3. Association between XRCC1 Arg399Gln and risk of head and neck cancer under four genetic models.
Forest plots for a: Gln vs. Arg; b: GlnGln vs. ArgArg; c: ArgGln vs. ArgArg; d: GlnGln+ ArgGln vs. ArgArg. Random-effects models were used for c and d; fixed-effects models were used for a and b. Squares and horizontal lines represent the study-specific OR and 95% CI respectively; diamond indicates the summary OR and 95% CI.
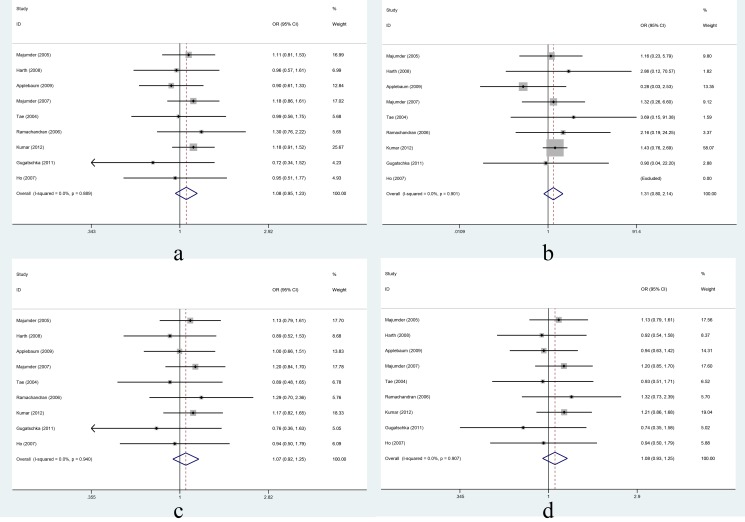
XRCC1 Arg280His: In the overall comparison, the Arg280His polymorphism was not significantly associated with HNC risk under the four different genetic models (Figure 4). Further, in the subgroup analyses based on ethnicity, the Arg280His polymorphism was not significantly associated with HNC risk in Asians or Caucasians (Table 4). In the stratified analyses based on cancer site, the Arg280His polymorphism was not significantly associated with oral cancer (Table 4). When studies either adjusted or unadjusted for smoking and alcohol were analyzed, the Arg280His polymorphism was not significantly associated with HNC risk (Table 4).
Figure 4. Association between XRCC1 Arg280His and risk of head and neck cancer under four genetic models.
Forest plots for a: His vs. Arg; b: HisHis vs. ArgArg; c: ArgHis vs. ArgArg; d: HisHis+ ArgHis vs. ArgArg. Fixed-effects models were used for a, b, c, and d. Squares and horizontal lines represent the study-specific OR and 95% CI respectively; diamond indicates the summary OR and 95% CI.
Heterogeneity Analysis
Evidence of heterogeneity between studies in this meta-analysis was detected for XRCC1 Arg194Trp, and Arg399Gln, but the reasons for the heterogeneity were unclear. In the subgroup analyses, significant heterogeneity was found in the studies that used Asian populations, but not in the studies that used Caucasians, indicating that the publications that used Asians were probably the main source of heterogeneity in our study. In addition, significant heterogeneity was found in studies among oral cancers but not larynx cancers, indicating that the publications that focused on oral cancers were another probable source of heterogeneity. HNC includes cancers from different sites and risk factors for these cancers are different. Therefore, further studies with larger sample sizes and different tumor sites are needed to investigate the possible sources of the heterogeneity.
Sensitivity Analysis
Sensitivity analyses were conducted to assess the influence of individual studies on the combined ORs by omitting each study in turn. For all three polymorphisms under all four genetic models, the significance of the combined ORs was not materially altered by the exclusion of any individual study (data not shown). This result indicated that our results were statistically robust. Figure S1 shows the sensitivity analysis of XRCC1 Arg194Trp obtained under the allelic model by deleting of one study at a time.
Publication Bias
Begg's funnel plot and Egger's test were used to estimate the publication bias in the literature. For all three polymorphisms, the shapes of the Begg's funnel plots under all four genetic models showed no obvious asymmetry. Figure S2 shows the shape of the Begg's funnel plot for XRCC1 Arg399Gln under the dominant model. Egger's test also did not reveal significant evidence of publication bias for the three polymorphisms under all four genetic models (data not shown); the one exception was for XRCC1 Arg280His under the heterozygote model (t = −2.56, P = 0.037). Nevertheless, we found no significant difference between the corrected OR and uncorrected OR in the trim and fill analysis, which supported the robustness of our findings.
Discussion
In the overall comparison, the meta-analysis detected no significant association between the XRCC1 Arg194Trp, Arg399Gln, and Arg280His polymorphisms and HNC risk under all four genetic models. Further, in the subgroup analyses based on ethnicity, cancer site, and whether adjusted or unadjusted for smoking and alcohol, no significant association was found between the XRCC1 Arg399Gln, and Arg280His polymorphisms and HNC risk under the four genetic models. Nevertheless, in the stratified analyses based on cancer site, significant association was found between the XRCC1 Arg194Trp polymorphism and oral cancer under the allelic, heterozygote, and dominant models. When the studies adjusted for smoking and alcohol were analyzed, significant association was found between the XRCC1 Arg194Trp polymorphism and HNC risk under the homozygous and heterozygote models. Our results indicated that, while the XRCC1 Arg399Gln and Arg280His polymorphisms may not increase or decrease the risk of HNC, when cigarette smoking and alcohol consumption were taken into account, the XRCC1 Arg194Trp polymorphism was associated with increased risk of HNC and also may modulate genetic susceptibility to oral cancer.
The XRCC1Arg194Trp polymorphism is located in the region of the protein that separates the DNA polymerase-b and poly (ADP-ribose) polymerase-interacting domains. Tae et al. reported a highly significant association under the dominant genetic model of XRCC1 Arg194Trp with increased risk of squamous cell carcinoma of the head and neck among Korean patients and normal controls [9]. However, most other studies have found no association of XRCC1 Arg194Trp with HNC risk [11], [24], [25], [34], [35], [40], [46], [48]. In the present study, an intriguing finding was that the Arg194Trp polymorphism was a risk factor in Asians and a protective factor in Caucasians under all four genetic models; however, these associations were not statistically significant. This finding may have happened by chance, or may have resulted from different gene frequencies in the different populations; the MAF of XRCC1 Arg194Trp is 0.26 for Asians but only 0.09 for Caucasians. A number of studies have reported the association between the XRCC1 Arg194Trp polymorphism and oral cancer risk [10], [39], [41], [43], [44], [51]. It has been reported that the XRCC1 Arg194Trp polymorphism may result in decreased repair efficiency of DNA damage, and the repair deficit may eventually increase an individual's susceptibility to oral cancer [53], [54]. In our subgroup meta-analysis, we also detected the association of XRCC1 Arg194Trp with oral cancer risk. We found that under the allelic model. Trp allele carriers had a higher risk of oral cancer than Arg allele carriers. We also found that individuals with the Arg/Trp genotype had a higher risk of developing oral cancer under the heterozygote model; however, this association was not detected under the homozygous model. We speculate that the main reason for this finding may be the low occurrence of the Trp/Trp genotype in the study populations; indeed, in several studies, the number of Trp/Trp genotype was reported to be zero. The low occurrence of the Trp/Trp genotype will lead to poor statistical power. Under the dominant model, when the Trp/Trp and Arg/Trp genotypes were analyzed together, the association of the Arg/Trp genotype with oral cancer was still statistically significant. This finding is in accordance with our speculation that the heterozygote and homozygote models gave different results because of the low occurrence of the Trp/Trp genotype in the study population. However, this hypothesis needs to be tested with larger sample sizes in future studies.
The XRCC1 Arg399Gln polymorphism is located in the zinc finger domain area (PARP binding site) of the protein that detects DNA strand breaks [55]. The carriers of this variant were shown to have a higher level of DNA adducts [56] and tobacco-related DNA damage [46], [57]–[59]. XRCC1 Arg399Gln has been reported to be significantly associated with risks of gastric [60], lung [61], and colorectal [20] cancers. Ramachandran et al. found that the XRCC1 Arg399Gln polymorphism was associated with increased risk of oral cancer in an Indian population [10], while Kostrzewska-Poczekaj et al. found that XRCC1 Arg399Gln was a protective factor for squamous cell carcinoma of the head and neck in young adults [52]. Most other studies have found no significant association of XRCC1 Arg399Gln with HNC risk [9], [25], [34], . In the present study, we also found no association between XRCC1 Arg399Gln and HNC risk under all four genetic models.
The XRCC1 Arg280His polymorphism is located in the proliferating cell nuclear antigen binding region [62] in the apurinic/apyrimidinic endonuclease (APE)-binding domain of the protein [54], [63]. The Arg280His polymorphism could potentially alter the structure of XRCC1 and affect its ability to interact with APE [54], [64]. In a functional study, the XRCC1 protein carrying His 280 failed to rescue the single-strand break repair deficiency of mutant cells when human XRCC1 variant proteins were introduced into XRCC1 mutant Chinese hamster ovary cells [65]. Although functional studies revealed a possible mechanism for the association of the XRCC1 Arg280His polymorphism with cancer risk, our meta-analysis did not detect a significant association between XRCC1 Arg280His and HNC risk. This null result may be because of the limited number of studies that were included in our analyses. Clearly, larger sample sizes are needed to clarify the association of the XRCC1 Arg280His polymorphism with HNC risk.
Although we conducted a comprehensive analysis, our study has a number of limitations. First, only a limited number of eligible studies were found and so the sample size was relatively small. Therefore, especially in the stratified analyses, the association detected in our study may have occurred by chance. Second, because almost all the studies that were selected for meta-analysis were case-control studies, the patients were cancer survivors and patients who did not survive were not included. As a result, selection/survival bias could not be avoided.
In conclusion, the meta-analysis detected no association between the XRCC1 Arg399Gln and Arg280His polymorphisms and risk of HNC. However, in the subgroup analyses of studies adjusted for smoking and alcohol, the XRCC1 Arg194Trp polymorphism was associated with increased risk of HNC and, in the stratified analyses based on cancer site, XRCC1 Arg194Trp was associated with increased risk of oral cancer. Further studies with larger samples are needed to further evaluate the association between XRCC1 polymorphisms and HNC risk.
Supporting Information
Sensitivity analysis of XRCC1 Arg194Trp using the allelic model.
(DOC)
Begg's funnel plot of publication bias test for XRCC1 Arg399Gln using the dominant model.
(DOC)
PRISMA Checklist.
(DOC)
PRISMA Flowchart.
(DOC)
Funding Statement
This study was supported by grant no. 81272293 from National Natural Science Foundation of China, grant no. 81102194 from National Natural Science Foundation of China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Parkin DM, Bray F, Ferlay J, Pisani P (2005) Global cancer statistics, 2002. CA Cancer J Clin 55: 74–108. [DOI] [PubMed] [Google Scholar]
- 2. Szymanska K, Levi JE, Menezes A, Wunsch-Filho V, Eluf-Neto J, et al. (2010) TP53 and EGFR mutations in combination with lifestyle risk factors in tumours of the upper aerodigestive tract from South America. Carcinogenesis 31: 1054–1059. [DOI] [PubMed] [Google Scholar]
- 3. Wang M, Chu H, Zhang Z, Wei Q (2013) Molecular epidemiology of DNA repair gene polymorphisms and head and neck cancer. J Biomed Res 27: 179–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Negri E, Boffetta P, Berthiller J, Castellsague X, Curado MP, et al. (2009) Family history of cancer: pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. Int J Cancer 124: 394–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hiyama T, Yoshihara M, Tanaka S, Chayama K (2008) Genetic polymorphisms and head and neck cancer risk (Review). Int J Oncol 32: 945–973. [PubMed] [Google Scholar]
- 6. Flores-Obando RE, Gollin SM, Ragin CC (2010) Polymorphisms in DNA damage response genes and head and neck cancer risk. Biomarkers 15: 379–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vaezi A, Feldman CH, Niedernhofer LJ (2011) ERCC1 and XRCC1 as biomarkers for lung and head and neck cancer. Pharmgenomics Pers Med 4: 47–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang Y, Wang Y, Wu J, Li LJ (2013) XRCC1 Arg194Trp polymorphism is associated with oral cancer risk: evidence from a meta-analysis. Tumour Biol 34: 2321–2327. [DOI] [PubMed] [Google Scholar]
- 9. Tae K, Lee HS, Park BJ, Park CW, Kim KR, et al. (2004) Association of DNA repair gene XRCC1 polymorphisms with head and neck cancer in Korean population. Int J Cancer 111: 805–808. [DOI] [PubMed] [Google Scholar]
- 10. Ramachandran S, Ramadas K, Hariharan R, Rejnish Kumar R, Radhakrishna Pillai M (2006) Single nucleotide polymorphisms of DNA repair genes XRCC1 and XPD and its molecular mapping in Indian oral cancer. Oral Oncol 42: 350–362. [DOI] [PubMed] [Google Scholar]
- 11. Kumar A, Pant MC, Singh HS, Khandelwal S (2012) Associated risk of XRCC1 and XPD cross talk and life style factors in progression of head and neck cancer in north Indian population. Mutat Res 729: 24–34. [DOI] [PubMed] [Google Scholar]
- 12. Matullo G, Palli D, Peluso M, Guarrera S, Carturan S, et al. (2001) XRCC1, XRCC3, XPD gene polymorphisms, smoking and (32)P-DNA adducts in a sample of healthy subjects. Carcinogenesis 22: 1437–1445. [DOI] [PubMed] [Google Scholar]
- 13. Thompson LH, Brookman KW, Dillehay LE, Mooney CL, Carrano AV (1982) Hypersensitivity to mutation and sister-chromatid-exchange induction in CHO cell mutants defective in incising DNA containing UV lesions. Somatic Cell Genet 8: 759–773. [DOI] [PubMed] [Google Scholar]
- 14. Carrano AV, Minkler JL, Dillehay LE, Thompson LH (1986) Incorporated bromodeoxyuridine enhances the sister-chromatid exchange and chromosomal aberration frequencies in an EMS-sensitive Chinese hamster cell line. Mutat Res 162: 233–239. [DOI] [PubMed] [Google Scholar]
- 15. Op het Veld CW, Jansen J, Zdzienicka MZ, Vrieling H, van Zeeland AA (1998) Methyl methanesulfonate-induced hprt mutation spectra in the Chinese hamster cell line CHO9 and its xrcc1-deficient derivative EM-C11. Mutat Res 398: 83–92. [DOI] [PubMed] [Google Scholar]
- 16. Rouse J, Jackson SP (2002) Interfaces between the detection, signaling, and repair of DNA damage. Science 297: 547–551. [DOI] [PubMed] [Google Scholar]
- 17. Fang Z, Chen F, Wang X, Yi S, Chen W, et al. (2013) XRCC1 Arg194Trp and Arg280His polymorphisms increase bladder cancer risk in asian population: evidence from a meta-analysis. PLoS One 8: e64001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang K, Zhou B, Wang Y, Rao L, Zhang L (2012) The XRCC1 Arg280His polymorphism contributes to cancer susceptibility: an update by meta-analysis of 53 individual studies. Gene 510: 93–101. [DOI] [PubMed] [Google Scholar]
- 19. Nissar S, Lone TA, Banday MZ, Rasool R, Chowdri NA, et al. (2013) Arg399Gln polymorphism of XRCC1 gene and risk of colorectal cancer in Kashmir: A case control study. Oncol Lett 5: 959–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tian Z, Li YL, Liu JG (2013) XRCC1 Arg399Gln polymorphism contributes to increased risk of colorectal cancer in Chinese population. Mol Biol Rep 40: 4147–4151. [DOI] [PubMed] [Google Scholar]
- 21. Matullo G, Dunning AM, Guarrera S, Baynes C, Polidoro S, et al. (2006) DNA repair polymorphisms and cancer risk in non-smokers in a cohort study. Carcinogenesis 27: 997–1007. [DOI] [PubMed] [Google Scholar]
- 22. Chuang SC, Agudo A, Ahrens W, Anantharaman D, Benhamou S, et al. (2011) Sequence Variants and the Risk of Head and Neck Cancer: Pooled Analysis in the INHANCE Consortium. Front Oncol 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ho T, Li G, Lu J, Zhao C, Wei Q, et al. (2007) X-ray repair cross-complementing group 1 (XRCC1) single-nucleotide polymorphisms and the risk of salivary gland carcinomas. Cancer 110: 318–325. [DOI] [PubMed] [Google Scholar]
- 24. Harth V, Schafer M, Abel J, Maintz L, Neuhaus T, et al. (2008) Head and neck squamous-cell cancer and its association with polymorphic enzymes of xenobiotic metabolism and repair. J Toxicol Environ Health A 71: 887–897. [DOI] [PubMed] [Google Scholar]
- 25. Applebaum KM, McClean MD, Nelson HH, Marsit CJ, Christensen BC, et al. (2009) Smoking modifies the relationship between XRCC1 haplotypes and HPV16-negative head and neck squamous cell carcinoma. Int J Cancer 124: 2690–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wittke-Thompson JK, Pluzhnikov A, Cox NJ (2005) Rational inferences about departures from Hardy-Weinberg equilibrium. Am J Hum Genet 76: 967–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cochran WG (1950) The comparison of percentages in matched samples. Biometrika 37: 256–266. [PubMed] [Google Scholar]
- 28. DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7: 177–188. [DOI] [PubMed] [Google Scholar]
- 29. Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22: 719–748. [PubMed] [Google Scholar]
- 30. Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. Bmj 327: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Attia J, Thakkinstian A, D'Este C (2003) Meta-analyses of molecular association studies: methodologic lessons for genetic epidemiology. J Clin Epidemiol 56: 297–303. [DOI] [PubMed] [Google Scholar]
- 32. Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. Bmj 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stuck AE, Rubenstein LZ, Wieland D (1998) Bias in meta-analysis detected by a simple, graphical test. Asymmetry detected in funnel plot was probably due to true heterogeneity. Bmj 316: 469 author reply 470-461. [PMC free article] [PubMed] [Google Scholar]
- 34. Sturgis EM, Castillo EJ, Li L, Zheng R, Eicher SA, et al. (1999) Polymorphisms of DNA repair gene XRCC1 in squamous cell carcinoma of the head and neck. Carcinogenesis 20: 2125–2129. [DOI] [PubMed] [Google Scholar]
- 35. Olshan AF, Watson MA, Weissler MC, Bell DA (2002) XRCC1 polymorphisms and head and neck cancer. Cancer Lett 178: 181–186. [DOI] [PubMed] [Google Scholar]
- 36. Varzim G, Monteiro E, Silva RA, Fernandes J, Lopes C (2003) CYP1A1 and XRCC1 gene polymorphisms in SCC of the larynx. Eur J Cancer Prev 12: 495–499. [DOI] [PubMed] [Google Scholar]
- 37. Demokan S, Demir D, Suoglu Y, Kiyak E, Akar U, et al. (2005) Polymorphisms of the XRCC1 DNA repair gene in head and neck cancer. Pathol Oncol Res 11: 22–25. [DOI] [PubMed] [Google Scholar]
- 38. Gajecka M, Rydzanicz M, Jaskula-Sztul R, Wierzbicka M, Szyfter W, et al. (2005) Reduced DNA repair capacity in laryngeal cancer subjects. A comparison of phenotypic and genotypic results. Adv Otorhinolaryngol 62: 25–37. [DOI] [PubMed] [Google Scholar]
- 39. Majumder M, Sikdar N, Paul RR, Roy B (2005) Increased risk of oral leukoplakia and cancer among mixed tobacco users carrying XRCC1 variant haplotypes and cancer among smokers carrying two risk genotypes: one on each of two loci, GSTM3 and XRCC1 (Codon 280). Cancer Epidemiol Biomarkers Prev 14: 2106–2112. [DOI] [PubMed] [Google Scholar]
- 40. Rydzanicz M, Wierzbicka M, Gajecka M, Szyfter W, Szyfter K (2005) The impact of genetic factors on the incidence of multiple primary tumors (MPT) of the head and neck. Cancer Lett 224: 263–278. [DOI] [PubMed] [Google Scholar]
- 41. Kietthubthew S, Sriplung H, Au WW, Ishida T (2006) Polymorphism in DNA repair genes and oral squamous cell carcinoma in Thailand. Int J Hyg Environ Health 209: 21–29. [DOI] [PubMed] [Google Scholar]
- 42. Li C, Hu Z, Lu J, Liu Z, Wang LE, et al. (2007) Genetic polymorphisms in DNA base-excision repair genes ADPRT, XRCC1, and APE1 and the risk of squamous cell carcinoma of the head and neck. Cancer 110: 867–875. [DOI] [PubMed] [Google Scholar]
- 43. Majumder M, Sikdar N, Ghosh S, Roy B (2007) Polymorphisms at XPD and XRCC1 DNA repair loci and increased risk of oral leukoplakia and cancer among NAT2 slow acetylators. Int J Cancer 120: 2148–2156. [DOI] [PubMed] [Google Scholar]
- 44. Yen CY, Liu SY, Chen CH, Tseng HF, Chuang LY, et al. (2008) Combinational polymorphisms of four DNA repair genes XRCC1, XRCC2, XRCC3, and XRCC4 and their association with oral cancer in Taiwan. J Oral Pathol Med 37: 271–277. [DOI] [PubMed] [Google Scholar]
- 45. Csejtei A, Tibold A, Koltai K, Varga Z, Szanyi I, et al. (2009) Association between XRCC1 polymorphisms and head and neck cancer in a Hungarian population. Anticancer Res 29: 4169–4173. [PubMed] [Google Scholar]
- 46. Kowalski M, Przybylowska K, Rusin P, Olszewski J, Morawiec-Sztandera A, et al. (2009) Genetic polymorphisms in DNA base excision repair gene XRCC1 and the risk of squamous cell carcinoma of the head and neck. J Exp Clin Cancer Res 28: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jelonek K, Gdowicz-Klosok A, Pietrowska M, Borkowska M, Korfanty J, et al. (2010) Association between single-nucleotide polymorphisms of selected genes involved in the response to DNA damage and risk of colon, head and neck, and breast cancers in a Polish population. J Appl Genet 51: 343–352. [DOI] [PubMed] [Google Scholar]
- 48. Gugatschka M, Dehchamani D, Wascher TC, Friedrich G, Renner W (2011) DNA repair gene ERCC2 polymorphisms and risk of squamous cell carcinoma of the head and neck. Exp Mol Pathol 91: 331–334. [DOI] [PubMed] [Google Scholar]
- 49. Krupa R, Kasznicki J, Gajecka M, Rydzanicz M, Kiwerska K, et al. (2011) Polymorphisms of the DNA repair genes XRCC1 and ERCC4 are not associated with smoking- and drinking-dependent larynx cancer in a Polish population. Exp Oncol 33: 55–56. [PubMed] [Google Scholar]
- 50. Yuan H, Li H, Ma H, Niu Y, Wu Y, et al. (2012) Genetic polymorphisms in key DNA repair genes and risk of head and neck cancer in a Chinese population. Exp Ther Med 3: 719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dos Reis MB, Losi-Guembarovski R, de Souza Fonseca Ribeiro EM, Cavalli IJ, Morita MC, et al. (2013) Allelic variants of XRCC1 and XRCC3 repair genes and susceptibility of oral cancer in Brazilian patients. J Oral Pathol Med 42: 180–185. [DOI] [PubMed] [Google Scholar]
- 52. Kostrzewska-Poczekaj M, Gawecki W, Illmer J, Rydzanicz M, Gajecka M, et al. (2013) Polymorphisms of DNA repair genes and risk of squamous cell carcinoma of the head and neck in young adults. Eur Arch Otorhinolaryngol 270: 271–276. [DOI] [PubMed] [Google Scholar]
- 53. Hu Z, Ma H, Chen F, Wei Q, Shen H (2005) XRCC1 polymorphisms and cancer risk: a meta-analysis of 38 case-control studies. Cancer Epidemiol Biomarkers Prev 14: 1810–1818. [DOI] [PubMed] [Google Scholar]
- 54. Hung RJ, Hall J, Brennan P, Boffetta P (2005) Genetic polymorphisms in the base excision repair pathway and cancer risk: a HuGE review. Am J Epidemiol 162: 925–942. [DOI] [PubMed] [Google Scholar]
- 55. Shall S, de Murcia G (2000) Poly(ADP-ribose) polymerase-1: what have we learned from the deficient mouse model? Mutat Res 460: 1–15. [DOI] [PubMed] [Google Scholar]
- 56. Lunn RM, Langlois RG, Hsieh LL, Thompson CL, Bell DA (1999) XRCC1 polymorphisms: effects on aflatoxin B1-DNA adducts and glycophorin A variant frequency. Cancer Res 59: 2557–2561. [PubMed] [Google Scholar]
- 57. Abdel-Rahman SZ, El-Zein RA (2000) The 399Gln polymorphism in the DNA repair gene XRCC1 modulates the genotoxic response induced in human lymphocytes by the tobacco-specific nitrosamine NNK. Cancer Lett 159: 63–71. [DOI] [PubMed] [Google Scholar]
- 58. Duell EJ, Wiencke JK, Cheng TJ, Varkonyi A, Zuo ZF, et al. (2000) Polymorphisms in the DNA repair genes XRCC1 and ERCC2 and biomarkers of DNA damage in human blood mononuclear cells. Carcinogenesis 21: 965–971. [DOI] [PubMed] [Google Scholar]
- 59. Lei YC, Hwang SJ, Chang CC, Kuo HW, Luo JC, et al. (2002) Effects on sister chromatid exchange frequency of polymorphisms in DNA repair gene XRCC1 in smokers. Mutat Res 519: 93–101. [DOI] [PubMed] [Google Scholar]
- 60. Xue H, Ni P, Lin B, Xu H, Huang G (2011) X-ray repair cross-complementing group 1 (XRCC1) genetic polymorphisms and gastric cancer risk: A HuGE review and meta-analysis. Am J Epidemiol 173: 363–375. [DOI] [PubMed] [Google Scholar]
- 61. Qian B, Zhang H, Zhang L, Zhou X, Yu H, et al. (2011) Association of genetic polymorphisms in DNA repair pathway genes with non-small cell lung cancer risk. Lung Cancer 73: 138–146. [DOI] [PubMed] [Google Scholar]
- 62. Fan J, Otterlei M, Wong HK, Tomkinson AE, Wilson DM 3rd (2004) XRCC1 co-localizes and physically interacts with PCNA. Nucleic Acids Res 32: 2193–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Caldecott KW (2003) XRCC1 and DNA strand break repair. DNA Repair (Amst) 2: 955–969. [DOI] [PubMed] [Google Scholar]
- 64. Yan L, Yanan D, Donglan S, Na W, Rongmiao Z, et al. (2009) Polymorphisms of XRCC1 gene and risk of gastric cardiac adenocarcinoma. Dis Esophagus 22: 396–401. [DOI] [PubMed] [Google Scholar]
- 65. Takanami T, Nakamura J, Kubota Y, Horiuchi S (2005) The Arg280His polymorphism in X-ray repair cross-complementing gene 1 impairs DNA repair ability. Mutat Res 582: 135–145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sensitivity analysis of XRCC1 Arg194Trp using the allelic model.
(DOC)
Begg's funnel plot of publication bias test for XRCC1 Arg399Gln using the dominant model.
(DOC)
PRISMA Checklist.
(DOC)
PRISMA Flowchart.
(DOC)