Abstract
Experimental studies of the interaction between host and symbiont in a maturing symbiotic organ have presented a challenge for most animal-bacterial associations. Advances in the rearing of the host squid Euprymna scolopes have enabled us to explore the relationship between a defect in symbiont light production and late-stage development (e.g., symbiont persistence and tissue morphogenesis) by experimental colonization with specific strains of the symbiont Vibrio fischeri. During the first four weeks post-inoculation of juvenile squid, the population of wild-type V. fischeri increased 100-fold; in contrast, a strain defective in light production (Δlux) colonized normally the first day, but exhibited an exponential decline to undetectable levels over subsequent weeks. Co-colonization of organs by both strains affected neither the trajectory of colonization by wild type, nor the decline of Δlux levels. Uninfected animals retained the ability to be colonized for at least two weeks post-hatch. However, once colonized by the wild-type strain for 5 days, a subsequent experimentally induced loss of the symbionts could not be followed by a successful recolonization, indicating the host’s entry into a refractory state. However, animals colonized by the Δlux before the loss of their symbionts were receptive to recolonization. Analyses of animals colonized with either a wild-type or a Δlux strain revealed slight, if any, differences in the developmental regression of the ciliated light-organ tissues that facilitate the colonization process. Thus, some other feature(s) of the Δlux strain’s defect also may be responsible for its inability to persist, and its failure to induce a refractory state in the host.
Keywords: Euprymna scolopes, Vibrio fischeri, luminescence, persistence, maintenance
Introduction
Symbiosis is now recognized as a principal feature of biological systems (Gilbert et al. 2013; McFall-Ngai et al. 2013). To understand the ‘rules’ governing the establishment and persistence of symbiotic associations in animals, biologists have developed a series of models systems that lend themselves to experimental manipulation under laboratory conditions (Ruby 2008; Kostic et al. 2013). The integration of information gained through the study of these systems is enabling the community of biologists in this field to construct a conceptual framework that addresses the questions: (i) what biochemical, molecular, and cellular features of symbiosis are conserved across the animal kingdom; and, (ii) how has evolutionary selection driven diversity in symbiotic relationships? In this contribution, we present the first experimental studies of maturation of the symbiotic state in the squid-vibrio model.
The binary symbiosis between the Hawaiian bobtail squid, Euprymna scolopes, and the marine luminous bacterium, Vibrio fischeri, has been exploited for much of the last 25 years as an experimental system for the study of the onset and early development of an animal-bacterial partnership (for review, see McFall-Ngai 2008). Similar to most symbiotic associations, the squid-vibrio system is horizontally transmitted, i.e., acquired each generation, as the newly hatched animal harvests free-living V. fischeri cells from the bacterioplankton. The embryo develops a nascent light organ with juvenile-specific ciliated surfaces where the symbiont aggregates in the hours following hatching of the juvenile into seawater. At 3–4 h after hatching, the aggregated cells enter pores on the surface of the organ and travel through ducts to one of six internal crypt spaces, where the symbionts grow. Beginning with the first dawn and thereafter throughout life, the host animal responds to the environmental light cue with an expulsion of the majority of symbionts into the surrounding seawater (Lee & Ruby 1994). Concomitant with the first expulsion of symbionts is an irreversible morphogenetic signal, specifically, the presentation of high-concentrations of cell envelope molecules (Koropatnick et al. 2004) in synergy with symbiont light production (Koropatnick et al. 2007), that triggers the loss of the superficial ciliated fields that potentiate the initial symbiont colonization. The symbionts also induce significant anatomical changes in the crypt epithelia with which they associate, including a swelling of these host cells and an increase of the density of the microvilli that interface directly with the symbionts.
Genetic tools have been developed in the bacterial partner of the squid-vibrio system (Ruby 2008). Microbiologists studying this system have identified three broad mutant classes whose defects affect early stages of the association: (i) initiation mutants, such as those defective in motility (Graf et al. 1994), which show no colonization of the organ; (ii) accommodation mutants, such as amino acid auxotrophs (Graf & Ruby 1998), which do not achieve normal colonization levels in the early symbiosis; and, (iii) persistence mutants, such as those defective in light production, which grow to normal levels initially, but whose populations in the organ subsequently decline (Ruby, 2008). Perhaps the most widely studied of the V. fischeri persistence mutants are those defective in one or more of the 6 lux structural genes responsible for luminescence (i.e., luxCDABEG), including ΔluxA::erm, which is deleted for a subunit of the luciferase protein (Visick et al. 2000). The ΔluxA::erm and ΔluxCDABEG (Δlux) mutations result in a complete loss of luminescence, and have been constructed in a number of V. fischeri strains (Bose et al. 2008). These mutations in the symbiont have specific effects on several aspects of early host development (for review see McFall-Ngai et al. 2012) including delays in regression of the light organ’s superficial ciliated epithelia that facilitate colonization, and in hemocyte trafficking into these epithelial fields (Koropatnick et al. 2007), as well as defects in crypt cell development, most notably a failure to induce host crypt-cell swelling (Visick et al. 2000).
Although research of the squid-vibrio system has principally focused on early development, i.e., over the first few days after hatching, studies of the symbiosis have suggested that long-term maintenance of the association requires a maturation process that extends for weeks beyond the initiation of the partnership (Montgomery & McFall-Ngai 1998). The gross morphology of the light organ progresses over this period from heart-shaped structure of the juvenile to the bi-lobed organization of the adult (Fig. 1). In addition, the tissues that modify bacterial light, including the reflector, lens, and diverticula of the ink sac, while rudimentary in the juvenile, mature to prominent features of the adult organ. This maturation of the host organ dramatically changes the relationship of the symbionts to the host tissue and to the external environment. Specifically, whereas the symbionts in the juvenile’s crypts are a few cell layers away from environment, the complex, protracted morphogenesis during organ maturation results in the symbionts in the crypts being remote from the organ surface. These structural changes are likely to influence host-symbiont interactions dramatically. Further, a recent project designed to characterize the host and symbiont transcriptome in the adult animal suggested pronounced metabolic shifts in the symbiont over the day-night cycle, which are associated with marked changes in crypt-cell microanatomy (Wier et al. 2010). Preliminary studies of the underlying mechanisms of this process suggest that they do not begin to occur until several weeks into the symbiotic association.
Fig. 1.
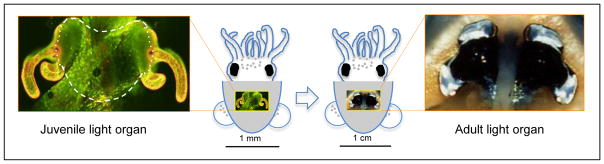
The light organ of E. scolopes, which is located in the center of the mantle cavity, undergoes morphogenesis from the hatchling to the mature condition. Left: The center of the newly hatched juvenile light organ is heart-shaped (subtended by the white dashed line in the confocal micrograph image). This retained portion of the organ will undergo maturation over the first weeks following inoculation with the specific symbiont, V. fischeri. The unique features of the juvenile organ include transparent, superficial fields of ciliated epithelia (golden, with fluorescent label; CellTracker Orange, Molecular Probes, Inc.) that potentiate colonization by the symbiont. These fields begin to regress within days of the initiation of the symbiosis. Right: The bilobed, mature light organ of the adult animal.
The suggestion of important maturation events in the squid-vibrio symbiosis has driven our interest in the development of protocols for the routine captive rearing of newly hatched host animals to enable studies of persistence in an animal-bacterial partnership. With modifications of E. scolopes culturing methods previously developed (Arnold et al. 1972; Hanlon et al. 1997; Claes & Dunlap 2000), we have successfully overcome the technical hurdles of raising the juveniles beyond the first week after hatching, and established a reliable system for long-term culture of the host animal. Here we present the studies of maturation events of this symbiosis, with particular emphasis on the role of symbiont persistence in driving the form and function of the fully developed organ.
Materials and methods
Bacterial strains and growth media
Vibrio fischeri strains TIM302 and TIM313 are derivatives of the wild-type symbiont strain ES114 (Boettcher & Ruby 1990), marked with chromosomal insertions for erythromycin resistance (ermr) (Table 1). Strain TIM302 also contains a chromosomal insertion of a gene encoding green fluorescent protein (GFP) (Miyashiro et al. 2010). Strains TIM386 and TIM387 are derivatives of EVS102 (Bose et al. 2008), which has a deletion of the luxCDABEG genes; they also contain chromosomal insertions for ermr. TIM387 encodes a chromosomal insertion of GFP. The ermr and GFP encoding sequences in plasmids pEV107 and pTM242 were integrated into the Tn7 site within EVS102 using helper plasmids pEVS104 and pUX-BF13 as described elsewhere (McCann et al. 2003) to generate TIM386 and TIM387, respectively.
Table 1.
Bacterial strains and plasmids used in this study
| Strain | Relevant genotype | Source |
|---|---|---|
| ES114 | Wild-type V. fischeri symbiont | (Ruby et al. 2005; Mandel et al. 2008) |
| EVS102 | ES114 ΔluxCDABEG | (Bose et al. 2008) |
| TIM302 | ES114 miniTn7:: erm PtetA-gfp+ | (Miyashiro et al. 2010) |
| TIM313 | ES114 miniTn7::erm | (Miyashiro et al. 2010) |
| TIM386 | EVS102 miniTn7::erm | This work |
| TIM387 | EVS102 miniTn7:: erm PtetA-gfp+ | This work |
| Plasmid
| ||
| pEVS104 | R6Kori RP4 oriT trb tra kan | (Stabb & Ruby 2002) |
| pEVS107 | R6Kori oriT mini-Tn7 mob erm kan | (McCann et al. 2003) |
| pTM242 | pEVS107 PtetA-gfp+ | (Miyashiro et al. 2010) |
| pUX-BF13 | R6Kori tns bla | (Bao et al. 1991) |
To prepare V. fischeri for colonization experiments, cells were grown in LBS medium (Stabb et al. 2001) containing (per liter) 950 ml deionized water, 50 ml 1M Tris-HCl buffer (pH 7.5), 10 g Difco Bacto-Tryptone, 5 g yeast extract, and 20 g NaCl. LBS agar medium was solidified with 15 g of agar per liter. V. fischeri strains for culture competitions were grown in SWTO medium containing (per liter) 700 ml of a marine salts solution (Instant Ocean® aquarium sea salt mixture, IO), 300 ml deionized water, 10 g NaCl, 5 g Bacto-Tryptone, and 3 g yeast extract (Bose et al. 2008). To select for antibiotic resistance strains, erythromycin (erm) was added to LBS agar at a concentration of 10 μg/ml.
Rearing E. scolopes juveniles to maturity
Fully mature Euprymna scolopes were collected in Maunalua Bay, Hawaii, and transported to the University of Wisconsin-Madison. These animals were maintained in artificial seawater (IO) as previously described (McFall-Ngai & Montgomery 1990; Montgomery & McFall-Ngai 1993). Egg clutches produced by the breeding colony were maintained individually in a 2.5-gal aquarium at 23 °C under heavy aeration and daily 50% water changes. Within 12 h of hatching, after ~23 d of embryogenesis, juvenile animals were placed in 100 ml of filter-sterilized IO containing (per ml) 3000 – 5000 CFU of the desired V. fischeri strain(s). Based on previous studies (Choe & Oshima 1963; Arnold et al. 1972; Hanlon et al. 1997), a reliable E. scolopes rearing method was developed (see Supporting information for details). The animals were exposed to symbiont cells for 14 – 16 h, and then transferred to 4-L rearing vessels at 25 to 60 squid/vessel (Table S1, Supporting information). Nonsymbiotic animals were placed in 100 ml of filtered IO without addition of V. fischeri, and then transferred into rearing vessels at the same time and density as the symbiont-colonized animals. All animals were maintained on 12 h/12 h light/dark cycle throughout the experiment. Although we were able to raise squid through their entire life cycle, the majority of experiments in this study ended at 4 weeks. This timing was chosen as the experimental end point because, by 4 weeks, the animals have transitioned into adult diel behavior, which is characterized by a nocturnal, active period and a diurnal, quiescent period (Hanlon et al. 1997; unpubl. obs.).
Colonization, co-colonization and persistence of wild-type and ΔluxCDABEG V. fischeri
For colonization experiments using single-strains, the squid were inoculated simultaneously but separately with 3000–5000 CFU of either wild type or ΔluxCDABEG (Δlux) per ml. The inoculum was plated on LBS agar before introduction of the squid. The number of V. fischeri symbionts in the light organs was determined at 1, 7, 14, 21, and 28 d after inoculation by plating squid homogenates (n = 10–30 squid per time point) on LBS agar (Stabb et al. 2001). E. scolopes were not fed the day of sampling and were rinsed for 1 h in filter-sterilized IO to reduce contamination. After rinsing, each squid was checked for luminescence using a TD-20/20 luminometer (Turner Design, Sunnyvale, CA, USA). The squid were then homogenized within 2 h prior to the scheduled lights-off period, and serial dilutions were plated onto LBS agar. The plates were incubated overnight at 28 °C, and colonies were counted within 24–48 h. As in the initial publication describing this operon deletion (Bose et al., 2008), we were unable to construct a suitable genetic-complementation vector containing the entire 6-gene locus. However, their work showed that the same type and level of defect was observed with independently constructed Δlux and luxA::erm mutants, in two different strain backgrounds. Thus, it seemed likely that the persistence phenotype of the Δlux mutant resulted from the shared defect in light emission, and not because of any second-site mutation that happened to appear in all strains.
For co-colonization experiments, the squid were inoculated at a 1:1 ratio of labeled wild type (TIM313) to Δlux (TIM387); the combined inoculum was 3000–5000 CFU/ml. Previous work has shown that the single-copy labels of ermr and GFP have no detectable effect on growth or colonization (Miyashiro et al., 2010). The inoculum was plated on LBS agar both just before introduction of the squid and after transferring the squid into the rearing tubs (12 h later) to assure that a 1:1 ratio of the two strains was maintained throughout the inoculation. The number of CFU per squid was determined 1, 3, 7, 11, and 15 d after inoculation. At each sampling, the animals were homogenized, and the diluted homogenate was plated on LBS agar (n = 20 squid per time point). After 48 h, the colonies were examined with a fluorescence microscope to identify Δlux colonies by their green fluorescence. Plates were made with a thin (~2 mm) layer of LBS agar to reduce the autofluorescence associated with non-fluorescent colonies. Both non-fluorescent and fluorescent colonies were counted to determine the CFU per light organ of each strain, and the ratio of the two strains (Δlux : wild type) was calculated as the competitive index (CI).
To increase the ability to detect co-colonizing Δlux cells as their percentage of the symbiotic population declines, an additional experiment was performed. The inoculation procedure was the same as above, only erm-sensitive ES114 was used in place of ermr TIM313. In this way, a small percentage of the Δlux TIM387 could be identified against a large background of wild-type ES114 in a symbiont population by plating the homogenate on LBS agar either with or without erm. Sampling on day 1 post-inoculation showed that the ratio of strains was 1:1 as expected; at day 15, undiluted homogenate was plated on erm-containing plates to identify the small percentage of erm-resistant TIM387 CFU present.
Secondary colonization of cured light organs
To determine whether an initial colonization would render the light organ resistant to a subsequent colonization, newly hatched animals were placed into one of three conditions: (i) not colonized (nonsymbiotic), (ii) colonized by wild type (ES114), or (iii) colonized by Δlux (EVS102). The initial symbiont was then removed from the light organ by treatment with the antibiotics gentamicin (20 μg/ml) and chloramphenicol (20 μg/ml) for 48 h (Nyholm et al. 2009). After curing, the animals were rinsed for 1 h in filter-sterilized IO, and wild-type colonized animals checked for the absence of luminescence as an indication of the loss (or great reduction) of symbionts. Because the Δlux-colonized squid were similarly treated, we assumed those symbionts had also been eliminated by the treatment.
Sets of wild-type colonized and nonsymbiotic squid were then (re)inoculated at either 1, 5, or 10 d post-hatching, and Δlux-colonized squid re-inoculated at 5 or 10 d post-hatching (n = 5–30 squid per time point) (Table 2) with 3000–5000 CFU of GFP-labeled wild-type V. fischeri (TIM302) per ml. Colonies of this GFP-labeled wild-type strain were easily distinguished from those of the initial non-fluorescent colonizers (ES114 and EVS102). After an overnight inoculation, the squid were returned to the rearing containers and, 24 and 48 h later, each squid was checked for luminescence. All the animals were then homogenized, and the diluted homogenate plated on LBS agar. The number of CFUs per squid for both the initial colonization and the secondary colonization were quantified. The effectiveness of the curing procedure was validated by the observation that the light organs of secondarily colonized animals only carried GFP-labeled symbionts. All colonization and curing procedures were performed at the same times, in parallel, with the same bacterial culture or antibiotic dilutions. This experiment was repeated twice using the time points described above, and a third time at 28 d post-hatching, with the same outcome.
Table 2.
Effect of initial colonization on the ability of E. scolopes to become secondarily colonized
| Initial treatment | Time of curing* | Time of secondary reinoculation# | Luminescence after secondary colonization† | CFU/squid (48 h after secondary colonization) | |
|---|---|---|---|---|---|
| 24 h | 48 h | ||||
| Nonsymbiotic | Day 1 | Day 3 | 5/5 | 5/5 | 105 |
| Day 5 | Day 7 | 5/5 | 5/5 | 106 | |
| Day 10 | Day 12 | 10/10 | 10/10 | 106 | |
|
| |||||
| Wild type | Day 1 | Day 3 | 0/5 | 5/5 | 105 |
| Day 5 | Day 7 | 0/10 | 0/10 | <30 | |
| Day 10 | Day 12 | 0/29 | 0/29 | <30 | |
|
| |||||
| ΔluxCDABEG | Day 5 | Day 7 | 2/5 | 4/5 | 105 |
| Day 10 | Day 12 | 11/20 | 19/20 | 106 | |
Animals in each treatment group, including the nonsymbiotic, were cured with antibiotics at the indicated times after an initial colonization.
Cured animals in each treatment group were secondarily exposed to wild-type V. fischeri.
Based on the production of luminescence, the number of animals (positives/total) recolonized at 24 or 48 h after reinoculation with wild type.
Development of the light-organ ciliated field
The morphology of the light-organ ciliated field was monitored over 4 weeks for animals under 3 conditions: (i) nonsymbiotic; (ii) colonized by wild type; or, (iii) colonized by Δlux. Animals were sampled at 1, 7, 14, 21, and 28 d. Samples were rinsed for 1 h in filtered IO, and then prepared for scanning electron microscopy (SEM) by fixing in 4% paraformaldehyde in 1X marine phosphate buffered saline (mPBS; 50 mM sodium phosphate, 0.45 M sodium chloride, pH 7.4). After 5 min in fix, the mantle was opened to allow proper fix penetration of the light organ. The squid were fixed for 14–16 h at room temperature, then washed twice for 10 min in mPBS, and dehydrated through an ethanol series (Montgomery & McFall-Ngai 1993). The samples were dried for SEM, using a Tousimis Samdri 780 critical point drier, and sputter coated with a SeeVac Auto conductavac IV. Samples were mounted on stubs and examined with a Hitachi S-570 LaB6 scanning electron microscope. In conjunction with the sampling of squid for SEM, three animals from each of the wild-type and Δlux treatment groups were sacrificed to determine the number of V. fischeri CFUs present in the light organ, and to ensure that the animals were colonized with the appropriate strains.
Results
While raising E. scolopes juveniles we observed that the general behavior of these squid gradually shifts during the first 4 weeks following hatching, as had been noted previously (Hanlon et al. 1997). Although no consistent day-night difference in behavior occurred in the first days to weeks, by 28 d post-hatch all animals had transitioned to the distinct pattern of diurnal quiescence and nocturnal activity that is characteristic of mature symbiotic E. scolopes. Thus, we chose to examine the relationship between the developing squid and its symbiotic bacteria during this important period of maturation. Specifically, we compared the colonization dynamics of wild-type luminescent symbionts with those of a non-luminescent mutant strain (Δlux).
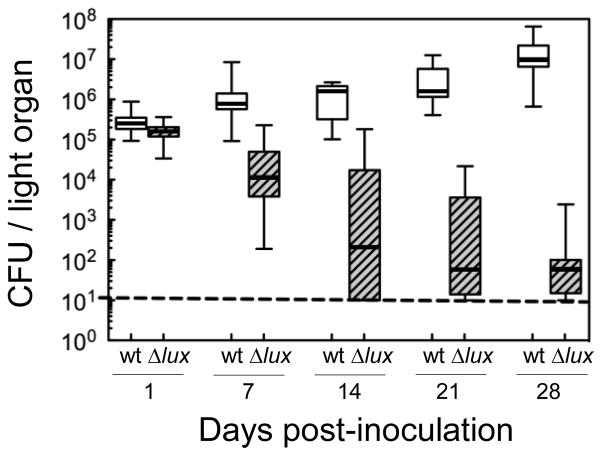
We first examined the growth trajectory of a symbiotic population of wild-type V. fischeri during the first 28 d following inoculation, relative to that of a Δlux population (Fig. 2). The wild-type strain initially colonized at ~105 CFU/squid. As the host grew, the light organs subsequently harbored ~106 CFU/squid by 7 d, and ~107 CFU/squid by 28 d. This growth of the symbiont population correlated with the increase in volume of the crypt space in which V. fischeri is maintained (Montgomery & McFall-Ngai, 1998). Previous studies of the symbiosis had demonstrated that while V. fischeri mutants defective in bioluminescence colonize normally over the first day following inoculation, they decline to about 25% of initial levels by 48–72 h (Visick et al. 2000; Bose et al. 2008). To determine whether a ΔluxCDABEG (Δlux) strain is maintained at this reduced level, or continues to decline in numbers as the organ develops, we monitored its population levels in the light organ through 28 d. In fact, the light-organ populations of the Δlux strain continued to decline exponentially; by 7 d they were at 10% of original colonization levels, and by 28 d the populations were < 0.1%, near the limit of detection (10 CFU/squid). The growth rates of wild-type and Δlux strains were indistinguishable (Fig. S2).
Fig. 2.
Colonization dynamics of the light organ of E. scolopes by a wild-type (wt; open) or ΔluxCDABEG (Δlux; hatched) strain of V. fischeri over the first 28 days (n = 10–30 squid per time point). The dashed line indicates the limit of detection (~10 CFU/light organ). The median and 25th to 75th percentile limits are indicated by the horizontal line and box, respectively, and the bars indicate the range of all values.
We then asked what effect co-colonization by the Δlux and wild-type strains would have on each other. Three general outcomes were possible: (i) the wild type could rapidly outcompete the Δlux strain, such that its loss would be accelerated; (ii) the light-emitting wild-type strain could functionally complement the Δlux such that this non-luminescent strain could now persist as a co-colonizer; or, (iii) the two strains would not affect one another’s growth patterns in the crypts. At 3 d following the co-inoculation, the Δlux population was reduced to approximately 25% that of wild type, with an average competitive index (CI) of 0.65 (Fig. 3), similar to previous reports for this point in the symbiosis (Visick et al. 2000; Bose et al. 2008). As the host matured, the Δlux populations continued to decline while wild type steadily increased (Fig. 3a). By 11 d post-inoculation, the Δlux strain, whose presence becomes increasingly difficult to discern during co-colonization, approached the limit of detection for these conditions (<500 CFU/squid); at 15 d very few Δlux CFU could be observed within the background of an average of 1.5 × 106 wild-type V. fischeri CFU/squid. More sensitive analyses using a selective medium (Materials and Methods) indicated that fewer than 10 Δlux CFU/squid. Taken together, these data show that there is apparently no significant effect of co-colonization on the symbiotic trajectory of either of the two strains.
Fig. 3.
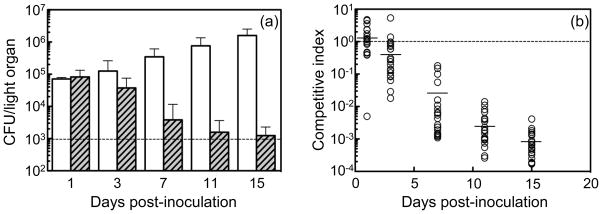
Co-colonization of E. scolopes by a 1:1 mixed inoculum of wild type and ΔluxCDABEG. (a) Mean CFU per light organ of wild-type (open) and ΔluxCDABEG (hatched) portions of the symbiotic population (n = 20 squid per time point). The dashed line is the limit of detection (~103 CFU per organ) and the standard error of the mean is displayed. (b) Competitive index (CI) of the ΔluxCDABEG strain relative to the wild type in the light organ populations (each circle represents the CI for one animal). The CI was calculated as the ratio of ΔluxCDABEG:wild type; the mean value at each time is indicated by the horizontal line, and the dotted line represents the initial inoculation ratio of 1:1.
We then studied the effects of initial colonization on the ability of previously colonized animals to be recolonized after losing their first symbionts. Nonsymbiotic animals could be colonized at any time during the first 28 d following hatching; however, did establishment of a previous colonization affect the host’s ability to initiate a subsequent one? In these experiments, we colonized juvenile squids with either the wild-type or the Δlux strain (Table 2). After 1, 3 or 5 d, the animals were cured with antibiotics, and re-exposed to an inoculum of wild-type V. fischeri. Juveniles colonized with wild type for 1 d and then cured could uniformly be recolonized, although their development of full symbiont populations (as indicated by the appearance of luminescence) was delayed (Table 2). Curing after 5 or 10 d of wild-type colonization resulted in squid that no longer could be recolonized under the inoculation conditions used. In contrast, 80–95% of the animals colonized by Δlux for 5 or 10 d could become recolonized. These outcomes indicate that, when colonized by wild-type symbionts, but not non-luminous ones, by 5 d the development of E. scolopes has progressed to a point at which it can no longer be secondarily colonized, even if the primary colonization is lost. The data presented in Table 2 are from a representative experiment that was repeated three separate times with the same outcome.
We next determined whether this difference in the ability to become recolonized correlates with differences in the morphological development of light organs of nonsymbiotic, wild-type colonized and Δlux-colonized animals. Colonization by symbionts typically triggers the loss of the juvenile-specific ciliated fields on the organ surface that potentiate colonization (Montgomery & McFall-Ngai 1994). One possible reason that animals initially colonized by the Δlux strain retain the capacity for secondary colonization by the wild-type strain is that Δlux is defective in inducing the regression of these fields. A previous study demonstrated that light organs colonized by the Δlux strain (Koropatnick et al., 2007) are delayed in morphogenesis in the first three days. In the present study, however, by 7 d post-inoculation, the light organs of animals colonized by either wild type or Δlux strains had undergone the typical regression of the ciliated field, i.e., they were indistinguishable (Fig. 4). This finding suggests that the extent of ciliated-field regression can’t explain the resistance of animals previously infected by wild-type V. fischeri for 5 d to a secondary colonization. Nonsymbiotic light organs exhibited little change over this time period (Fig. 4). Between 8 and 28 d, light organs colonized by the Δlux strain continued to retain a small region of the ciliated fields surrounding the pores (Fig. 4, white arrows), while those animals colonized by the wild-type strain progressively lost this feature, taking on the morphology of the fully mature organ. Nonsymbiotic animals exhibited only a very gradual change in the ciliated surface, likely due to their exposure to the small amounts of morphogens (lipopolysaccharide and peptidoglycan derivatives; Koropatnick et al. 2004) in the ambient seawater. By 28 d post-hatch, the light organs of Δlux-colonized squid had a level of ciliated-field regression more similar to that of nonsymbiotic animals.
Fig. 4.
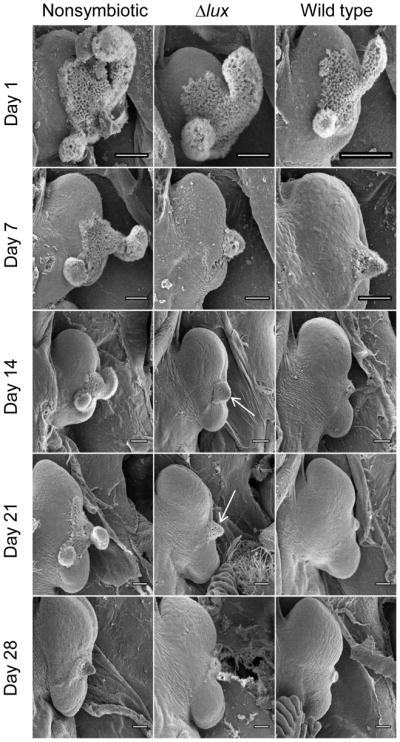
Scanning electron micrographs (SEMs) of the right half of E. scolopes light organs at different stages of maturation over the first 4 weeks post hatch. Left column: nonsymbiotic animals; middle column: Δlux colonized animals; right column: wild type colonized animals. The ciliated epithelial fields, seen on all organs at Day 1, regress at different rates. The white arrows indicate remnants of the ciliated field that are retained longer by Δlux-colonized light organs. Scale bars are 100 μm.
Discussion
The squid-vibrio symbiosis undergoes an important maturation process within the first four weeks after colonization (Montgomery & McFall-Ngai 1998). However, because newly hatched squid survive for only 4–5 days without access to food, direct, experiment-based analysis of later stages in this process requires raising juvenile E. scolopes into adults (Hanlon et al. 1997). In this study, we combined the ability to rear juvenile E. scolopes, with a capacity to colonize the newly hatched animals with a genetically modified strain of their symbiont, V. fischeri. In this way, the long-term impacts of a bacterial mutation could be assessed as the symbiosis matures.
During the first few weeks of the association, the nascent light organ progresses from a morphology that facilitates colonization (Nyholm & McFall-Ngai 2004) to one that both fosters persistence of the symbiont and mediates the nocturnal bioluminescent behavior of the host (Jones & Nishiguchi 2004). This maturation not only involves distinct morphological changes in the organ (Fig. 4; Hanlon et al. 1997; Montgomery & McFall-Ngai 1998), but also includes critical changes in the nature of the interaction between the partners (Ruby 2008; McFall-Ngai et al. 2012). By monitoring the impact of specific bacterial mutations on the maturing symbiosis, we hope to discover signaling and metabolic interactions between the host and its symbiont population as the partnership develops (Dunn 2012; McFall-Ngai et al. 2012). The importance of such developmental changes has become increasingly evident in recent years: for instance, while transcriptional analyses of the adult symbiosis have suggested that the host provides chitin oligosaccharides as a nutrient for their symbionts (Weir et al. 2010), juvenile squid do not (Miyashiro et al. 2011).
In the present study we chose to examine how the absence of a major product of the association, the symbiont’s bioluminescence, affects both the persistence of the bacterial partner and the host’s response to it. While it was previously reported that non-luminous mutants of V. fischeri initially colonize normally, but had declined to 1/4 of wild-type levels within 48 h (Visick et al. 2000; Bose et al. 2008), it was not known if or how the population level of such a ‘dark’ strain changed as the symbiosis continued to mature. Here we show that this decline continues at an exponential rate over the next 3–4 weeks, with a ΔluxCDABEG mutant eventually reaching undetectable levels, i.e., fewer than 10−5 the level of the wild-type population (Fig. 2). Such a remarkably effective apparent selection against a non-luminous symbiont by the host suggests an ability to perceive their light emission (Tong et al. 2009; McFall-Ngai et al. 2012) and/or the creation of physiological conditions that punish ‘cheaters’ (Ruby & McFall-Ngai 1999; Stabb 2005).
A similarly dramatic example of sanctioning has been reported in the legume-rhizobia symbioses (Kiers et al. 2003), and is believed to be the result of an evolutionary selection for mutualism called ‘partner choice’ (Foster & Wenseleers 2006). Beyond the extensive studies of this system, and the related plant-arbuscular mycorrhizal symbioses (Sanders 2003; Engelmoer et al. 2013), little experimental work on cheating by either partner has been reported for other host-microbe mutualisms (Sachs et al. 2011; Palmer et al. 2010), especially animal-bacteria ones. Nevertheless, the opportunity exists to discover how symbioses like those between nematodes (Chaston et al. 2013) or ants (Caldera & Currie 2012) and their microbiota have evolved to resist the appearance and persistence of cheaters.
The squid host’s ability to select against dark symbionts is even more remarkable because it occurs with equivalent efficiency in either single or mixed (i.e., Δlux + wild-type) colonizations (Fig. S1, Supporting information). That is, the increasingly diminished number of non-luminous cheaters fails to persist even amongst a vast preponderance of luminous cooperators; such high specificity is rarely observed in other symbioses (Oono et al. 2011). While the actual mechanism of any host-sanctioning in the light organ remains mostly speculative (Stabb 2005), our data help limit the possibilities. For instance, in our mixed-colonization experiments we expect that, on average, at least one of the six crypts in a light organ will be colonized by both a wild-type and a Δlux strain (Wollenberg & Ruby 2009). If the host sanctions cheaters by simply restricting the growth of symbionts within dark (i.e., colonized by Δlux alone) crypts (Lee & Ruby 1994), we would expect Δlux cells to persist only in the one co-colonized crypt. However, if this were the sole mechanism for sanctioning, the population of dark symbionts could theoretically remain at ~8% of the total. However, the specificity and effectiveness of the sanctioning (>99.9% of the Δlux cells in a co-colonization are lost; Fig. 3b) suggest the targeting of individual dark (cheater) bacteria, even within a mixed population.
Evolutionary theory suggests that it benefits the host to restrict the number of different strains that colonize a symbiotic organ (Wollenberg & Ruby 2009); thus, it is not surprising that once colonized by a wild-type strain, the light organ soon becomes refractory to secondary colonization (Table 2). However, If colonized by an uncooperative (i.e., non-luminous) symbiont, the host must not only be able to completely rid itself of the ‘cheater’, but also have a mechanism in place to allow recolonization. In fact, unless a functionally cooperative relationship is formed, the squid allows additional re-initiations of the symbiosis with different strains of V. fischeri to occur, perhaps indefinitely, until a stable association forms (Table 2). The mechanism underlying the decision of whether to ‘commit’ to a symbiont population is not known; however, it appears to function within the first few days after colonization, and is not dependent on delaying the regression of the light organ’s ciliated field (Fig. 4; Doino & McFall- Ngai 1995).
In summary, all horizontally transmitted symbioses face two fundamental challenges: effective initiation of the association by symbionts from an environmental reservoir, and successful maintenance of a functionally cooperative symbiont population. We are now in a position to bring to bear animal husbandry, imaging, metabolic, and genetic approaches to determine how both these critical steps are managed with such efficiency and fidelity in the squid-vibrio association.
Supplementary Material
Acknowledgments
We thank the staff of the Biological and Bio-materials Preparation, Imaging, and Characterization Laboratory of UW-Madison for assistance with drying, coating, and mounting of samples for scanning electron microscopy. This work was supported by NIH R01 grants AI50661 (to M.M.-N.) GM099507 (to E.G.R.), and OD11024 (to E.G.R. & M.M.-N.), and NSF grant IOS 0817232 (to M.M.-N. & E.G.R.). T.M. was supported during this project by NIH K99 grant GM097032.
Footnotes
Author contributions
E.J. Koch developed and fabricated the aquatic system for long-term maintenance of E. scolopes. E.J. Koch, M.J. McFall-Ngai, and E.G. Ruby designed the experiments and analyzed the data. E.J. Koch performed the experiments. T. Miyashiro constructed the bacterial strains. E.J. Koch, M.J. McFall-Ngai and E.G. Ruby wrote the manuscript.
Data Accessibility
All the data are experimental, and are found in the figures and tables in the main paper and supporting information files.
References
- Arnold JM, Singly CT, Williams-Arnold LD. Embryonic development and post-hatching survival of the sepiolid squid Euprymna scolopes under laboratory conditions. Veliger. 1972;14:361–364. [Google Scholar]
- Bao Y, Lies DP, Fu H, Roberts GP. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene. 1991;109:167–168. doi: 10.1016/0378-1119(91)90604-a. [DOI] [PubMed] [Google Scholar]
- Boettcher K, Ruby EG. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol. 1990;172:3701–3706. doi: 10.1128/jb.172.7.3701-3706.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose JL, Rosenberg CS, Stabb EV. Effects of luxCDABEG induction in Vibrio fischeri: enhancement of symbiotic colonization and conditional attenuation of growth in culture. Arch Microbiol. 2008;190:169–183. doi: 10.1007/s00203-008-0387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldera EJ, Currie CR. The population structure of antibiotic-producing bacterial symbionts of Apterostigma dentigerum ants: impacts of coevolution and multipartite symbiosis. Amer Nat. 2012;180:604–617. doi: 10.1086/667886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaston JM, Murfin KE, Heath-Heckman EA, Goodrich-Blair H. Previously unrecognized stages of species-specific colonization in the mutualism between Xenorhabdus bacteria and Steinernema nematodes. Cellul Microbiol. 2013 doi: 10.1111/cmi.12134. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Oshima Y. Rearing of cuttlefishes and squids. Nature. 1963;197:307. [Google Scholar]
- Claes MF, Dunlap PV. Aposymbiotic culture of the sepiolid squid Euprymna scolopes: role of the symbiotic bacterium Vibrio fischeri in host animal growth, development, and light organ morphogenesis. J Exper Zool. 2000;286:280–296. [PubMed] [Google Scholar]
- Doino JA, McFall-Ngai MJ. A transient exposure to symbiosis-competent bacteria Induces light organ morphogenesis in the host squid. Biol Bull. 1995;189:347–355. doi: 10.2307/1542152. [DOI] [PubMed] [Google Scholar]
- Dunn AK. Vibrio fischeri metabolism: symbiosis and beyond. Adv Microb Physiol. 2012;61:37–68. doi: 10.1016/B978-0-12-394423-8.00002-0. [DOI] [PubMed] [Google Scholar]
- Engelmoer DJP, Behm JE, Kiers ET. Intense competition between arbuscular mycorrhizal mutualists in an in vitro root microbiome negatively affects total fungal abundance. Mol Ecol. 2013;xx:xxx–xxx. doi: 10.1111/mec.12451. (THIS ISSUE) [DOI] [PubMed] [Google Scholar]
- Foster KR, Wenseleers T. A general model for the evolution of mutualisms. J Evolution Biol. 2006;19:1283–1293. doi: 10.1111/j.1420-9101.2005.01073.x. [DOI] [PubMed] [Google Scholar]
- Gilbert SF, Sapp J, Tauber AI. A symbiotic view of life: we have never been individuals. Quart Rev Biol. 2012;87:325–341. doi: 10.1086/668166. [DOI] [PubMed] [Google Scholar]
- Graf J, Ruby EG. Host-derived amino acids support the proliferation of symbiotic bacteria. Proc Natl Acad Sci USA. 1998;95:1818–1822. doi: 10.1073/pnas.95.4.1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf J, Dunlap PV, Ruby EG. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J Bacteriol. 1994;176:6986–6991. doi: 10.1128/jb.176.22.6986-6991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon RT, Claes MF, Ashcraft SE, Dunlap PV. Laboratory culture of the sepiolid squid Euprymna scolopes: A model system for bacteria-animal symbiosis. Biol Bull. 1997;192:364–374. doi: 10.2307/1542746. [DOI] [PubMed] [Google Scholar]
- Jones BW, Nishiguchi MK. Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda) Mar Biol. 2004;144:1151–1155. [Google Scholar]
- Kiers ET, Rousseau RA, West SA, Denison RF. Host sanctions and the legume-rhizobia mutualism. Nature. 2003;425:78–81. doi: 10.1038/nature01931. [DOI] [PubMed] [Google Scholar]
- Koropatnick TA, Engle JT, Apicella MJ, et al. Microbial factor-mediated development in a host-bacterial mutualism. Science. 2004;306:1186–1188. doi: 10.1126/science.1102218. [DOI] [PubMed] [Google Scholar]
- Koropatnick TA, Kimbell JR, McFall-Ngai MA. Responses of host hemocytes during the initiation of the squid-vibrio symbiosis. Biol Bull. 2007;212:29–39. doi: 10.2307/25066578. [DOI] [PubMed] [Google Scholar]
- Kostic AD, Howitt MR, Garrett WS. Exploring host-microbiota interactions in animal models and humans. Genes Develop. 2013;27:701–718. doi: 10.1101/gad.212522.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K-H, Ruby EG. Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl Environ Microbiol. 1994;60:1565–1571. doi: 10.1128/aem.60.5.1565-1571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MJ, Stabb EV, Ruby EG. Comparative genomics-based investigation of resequencing targets in Vibrio fischeri: focus on point miscalls and artefactual expansions. BMC Genomics. 2008;9:138. doi: 10.1186/1471-2164-9-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann J, Stabb EV, Millikan DS, Ruby EG. Population dynamics of Vibrio fischeri during infection of Euprymna scolopes. Appl Environ Microbiol. 2003;69:5928–5934. doi: 10.1128/AEM.69.10.5928-5934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M. Host-microbe symbiosis: the squid-Vibrio association--a naturally occurring, experimental model of animal/bacterial partnerships. Adv Exper Med Biol. 2008;635:102–112. doi: 10.1007/978-0-387-09550-9_9. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M, Heath-Heckman EA, Gillette AA, Peyer SM, Harvie EA. The secret languages of coevolved symbioses: insights from the Euprymna scolopes-Vibrio fischeri symbiosis. Sem Immunol. 2012;24:3–8. doi: 10.1016/j.smim.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai MJ, Montgomery MK. The anatomy and morphology of the adult bacterial light organ of Euprymna scolopes Berry (Cephalopoda: Sepiolidae) Biol Bull. 1990;179:332–339. doi: 10.2307/1542325. [DOI] [PubMed] [Google Scholar]
- Miyashiro T, Wollenberg MS, Cao X, Oehlert D, Ruby EG. A single qrr gene is necessary and sufficient for LuxO-mediated regulation in Vibrio fischeri. Mol Microbiol. 2010;77:1556–1567. doi: 10.1111/j.1365-2958.2010.07309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashiro T, Klein W, Oehlert D, Cao X, Schwartzman J, Ruby EG. The N-acetyl-D-glucosamine repressor NagC of V. fischeri facilitates colonization of Euprymna scolopes. Mol Microbiol. 2011;82:894–903. doi: 10.1111/j.1365-2958.2011.07858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery MK, McFall-Ngai MJ. Embryonic development of the light organ of the sepiolid squid Euprymna scolopes Berry. Biol Bull. 1993;184:296–308. doi: 10.2307/1542448. [DOI] [PubMed] [Google Scholar]
- Montgomery MK, McFall-Ngai MJ. Bacterial symbionts induce host organ morphogenesis during early postembryonic development of the squid Euprymna scolopes. Development. 1994;120:1719–1729. doi: 10.1242/dev.120.7.1719. [DOI] [PubMed] [Google Scholar]
- Montgomery MK, McFall-Ngai MJ. Late postembryonic development of the symbiotic light organ of Euprymna scolopes (Cephalopoda: Sepiolidae) Biol Bull. 1998;195:326–336. doi: 10.2307/1543144. [DOI] [PubMed] [Google Scholar]
- Nyholm SN, McFall-Ngai MJ. The winnowing: establishing the squid-Vibrio symbiosis. Nat Rev Microbiol. 2004;2:632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- Nyholm SN, Stewart JJ, Ruby EG, McFall-Ngai MJ. Recognition between symbiotic Vibrio fischeri and the haemocytes of Euprymna scolopes. Environ Microbiol. 2009;11:483–493. doi: 10.1111/j.1462-2920.2008.01788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oono R, Anderson CG, Denison RF. Failure to fix nitrogen by non-reproductive symbiotic rhizobia triggers host sanctions that reduce fitness of their reproductive clonemates. Proc Roy Soc B. 2011;278:2698–2703. doi: 10.1098/rspb.2010.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer TM, Doak DF, Stanton ML, et al. Synergy of multiple partners, including freeloaders, increases host fitness in a multispecies mutualism. Proc Natl Acad Sci USA. 2010;107:17234–17239. doi: 10.1073/pnas.1006872107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby EG. Symbiotic conversations are revealed under genetic interrogation. Nat Rev Microbiol. 2008;6:752–762. doi: 10.1038/nrmicro1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby EG, McFall-Ngai MJ. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends Microbiol. 1999;7:414–420. doi: 10.1016/s0966-842x(99)01588-7. [DOI] [PubMed] [Google Scholar]
- Ruby EG, Urbanowski M, Campbell J, et al. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc Natl Acad Sci USA. 2005;102:3004–3009. doi: 10.1073/pnas.0409900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs JL, Skophammer RG, Regus JU. Evolutionary transitions in bacterial symbiosis. Proc Natl Acad Sci USA. 2011;108:10800–10807. doi: 10.1073/pnas.1100304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders IR. Preference, specificity and cheating in the arbuscular mycorrhizal symbiosis. Trends Plant Sci. 2003;8:143–145. doi: 10.1016/S1360-1385(03)00012-8. [DOI] [PubMed] [Google Scholar]
- Stabb EV. Shedding light on the bioluminescence “paradox”. ASM News. 2005;7:223–229. [Google Scholar]
- Stabb EV, Reich KA, Ruby EG. Vibrio fischeri genes hvnA and hvnB encode secreted NAD+-glycohydrolases. J Bacteriol. 2001;183:309–317. doi: 10.1128/JB.183.1.309-317.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabb EV, Ruby EG. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Meth Enzymol. 2002;358:413–426. doi: 10.1016/s0076-6879(02)58106-4. [DOI] [PubMed] [Google Scholar]
- Tong D, Rosa NS, Oakley TH, et al. Evidence of light perception in a bioluminescent organ. Proc Natl Acad Sci USA. 2009;106:9836–9841. doi: 10.1073/pnas.0904571106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visick KL, Foster J, Doino J, McFall-Ngai MJ, Ruby EG. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J Bacteriol. 2000;182:4578–4586. doi: 10.1128/jb.182.16.4578-4586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wier AM, Nyholm SV, Mandel MJ, et al. Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc Natl Acad Sci USA. 2010;107:2259–2264. doi: 10.1073/pnas.0909712107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenberg MS, Ruby EG. Population structure of Vibrio fischeri within the light organs of Euprymna scolopes squid from two Oahu (Hawaii) populations. Appl Environ Microbiol. 2009;75:193–202. doi: 10.1128/AEM.01792-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.