Abstract
Bacteriophage λ makes two proteins with overlapping amino acid sequences that are essential for tail assembly. These two proteins, gpG and gpGT, are related by a programmed translational frameshift that is conserved among diverse phages and functions in λ to ensure that gpG and the frameshift product gpGT are made in a molar ratio of approximately 30:1. Although both proteins are required and must be present in the correct ratio for assembly of functional tails, neither is present in mature tails. During λ tail assembly major tail protein gpV polymerizes to form a long tube whose length is controlled by tape measure protein gpH. We show that the ‘G’ domains of gpG and gpGT bind to all or parts of tail length tape measure protein gpH, that the ‘T’ domain of gpGT binds to major tail shaft subunit gpV, and present a model for how gpG and gpGT chaperone gpH and direct the polymerization of gpV to form a tail of the correct length.
Keywords: regulation of macromolecular assembly, protein assembly chaperones, translational recoding, translational frameshift regulation, bacteriophage tail assembly
Introduction
The long tails of the dsDNA bacteriophages, including T4 and λ, are some of the most recognized icons of molecular biology and provide classical examples of molecular machines, each assembled from its parts with high precision and in a manner that makes a structure that is both stable and capable of being activated to carry out a complex dynamic function. Past studies of tail assembly have concentrated on bacteriophages T4 1; 2; 3; 4; 5; 6 and λ 7; 8; 9; 10; 11, and have included increasingly higher resolution views of the intricate structure of the T4 tail 12; 13; 14; 15. However, despite this wealth of information on both systems, there remain fundamental unanswered questions about the mechanism of assembly, about the tails’ biological functions, and about how assembly mechanism and ultimate biological function are related.
Bacteriophage λ has a simple, non-contractile tail encoded by a contiguous block of eleven genes located in the late operon, just downstream from the head genes. The mechanism by which the λ tail proteins are assembled into functional tails was first investigated by Katsura and colleagues in the 1970’s, primarily using in vitro complementation between mutant extracts 7; 8; 9; 16; 17. These experiments established an order of action for most of the tail genes and identified a series of assembly intermediates. In the assembly pathway that was inferred from these data, a group of eight tail proteins interact to produce an assembly intermediate termed the initiator, corresponding to the conical tip and short protruding terminal fiber of the mature tail. In the next step, the major tail shaft subunit gp V (gene product of gene V), polymerizes onto the initiator to an exact length. Finally, the last two tail proteins stabilize the tail and prepare it for productive addition to heads. Because of technical limitations on the experiments that were possible at the time, these studies, though very illuminating, could not specify the protein composition of the intermediates identified, they were somewhat equivocal about the middle parts of the pathway that we address in this paper, and they did not address one of the central issues of tail assembly, namely how the precise length of the tail is determined.
Subsequent studies with λ tails showed that the length of the tail shaft—that is, the length of the tubular polymer of protein gpV—is determined by the size of a “length template” or “tape measure” protein, gpH 11; 16 The current models propose that the tape measure protein is located in the lumen of the tail shaft, where 3–6 copies of gpH apparently each span the length of the tail shaft in the form of α-helical coiled-coils. In addition to its role in tail length determination, the tape measure has a role in infection; it is ejected from the tail prior to the DNA during DNA injection18, and it has an essential role in getting the phage DNA into the cell 19. A tape measure protein has also been demonstrated in phage T4 tail assembly 20, and genes encoding tape measure proteins can be confidently identified in the genomes of virtually all long-tailed phages 21; 22. Despite the apparently universal use of tape measure proteins in these phages, the mechanism by which length determination is accomplished has remained elusive.
We have previously investigated a pair of proteins encoded by a pair of overlapping open reading frames (ORFs), G and T, located in the middle of the tail gene cluster of λ. Protein gpG, encoded by the upstream ORF G, is produced in a large amount during phage growth, comparable to the amount of the major tail subunit gpV. Protein gpGT is produced when 3–4% of the ribosomes translating the G ORF undergo a −1 translational frameshift near the end of G, into the overlapping T ORF 23. Both gpG and gpGT have essential roles in tail assembly 23; 24, but neither is present in the mature tail 23, arguing that they play the roles of assembly co-factors or assembly chaperones. Efficient tail assembly requires that gpG and gpGT be present in the appropriate molar ratio and also that the physical connection between the G and T domains of gpGT be intact24. Here we present studies aimed at elucidating the mechanism by which gpG and gpGT promote assembly of functional tails. We show that both proteins bind to the tail length tape measure gpH and hold it in a functionally competent state. The T domain of gpGT binds to the major tail subunit gpV, and we present evidence that this interaction converts gpV from a soluble to an assembly-competent conformation. These observations lead to a model for tail assembly in which gpG and gpGT act as assembly chaperones to promote the assembly of gpV and gpH into a functional tail shaft, with a polymer of gpV forming a cylindrical tube surrounding the extended gpH in the mature structure. This model is consistent with all available data and suggests a specific mechanism for tail length determination.
Results
The T domain of gpGT interacts with major tail protein gpV
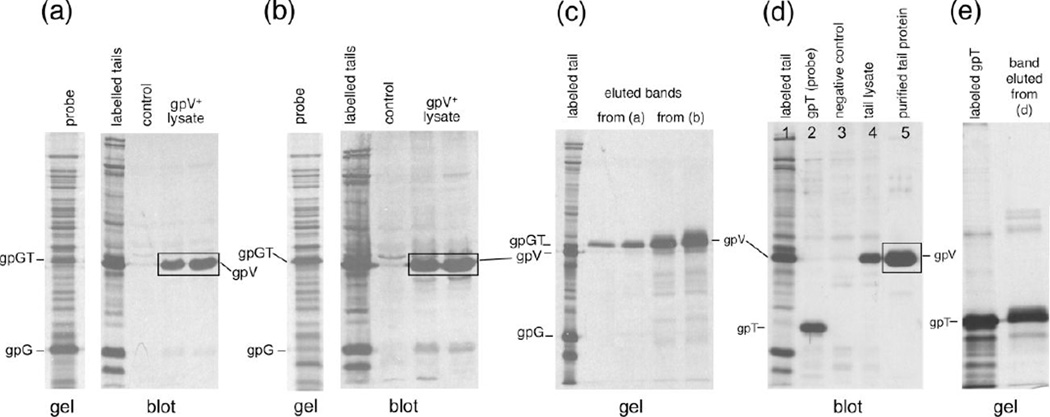
The first experiments to show that the gpGT protein interacts directly with the major tail protein subunit gpV came from the far-western blot 25 experiments shown in Figure 2. In this technique candidate binding partner proteins are subjected to SDS-PAGE, transferred to a membrane and probed with an extract containing radio-labeled protein(s) of interest. If one of the proteins in the radio-labeled extract can bind to a protein on the membrane, the corresponding band in the membrane will be labeled and can be detected by autoradiography. In our case an extract that contains gpV, gpG and gpGT was separated by SDS-PAGE and transferred to a PVDF membrane. The probes we used were radio-labeled extracts that contained either (a) wild type gpG and gpGT (produced from plasmid pT7-5-GT) or (b) gpGT alone (produced from plasmid pT7-5-GT-mut, which contains a single base pair insertion at the G–T junction, fusing the G and T orf’s and allowing gpGT to be expressed without a frameshift24). Figure 2 panels (a) and (b) show the results of probing with the pT7-5-GT and pT7-5-GT-mut extracts, respectively. It is clear that some protein in both probes interacts with the gpV. To identify the protein that interacts with gpV, we cut out the band from the membranes, recovered the radioactive proteins, and analyzed them on another SDS polyacrylamide gel (Figure 2 (c)). In both cases the protein interacting with gpV runs at the gpGT position, with little or none of the more abundantly expressed gpG present. The fact that the extract over-expressing gpGT gives a stronger binding band reinforces the conclusion that the protein that binds to gpV is indeed gpGT.
Figure 2. The T portion of gpGT interacts with gpV.
A crude extract of induced cells harboring plasmid pT7-5-VT (which expresses wild type gpV, gpG and gpGT) was separated by 12.5% SDS-PAGE. The proteins were transferred to PVDF membrane, blocked, probed with a radio-labeled cell extract, washed, dried and exposed to X-ray film. In (a) a radio-labeled extract of cells expressing pT7-5-GT was used as probe. pT7-5-GT expresses wild type gpG and gpGT as shown in the left panel. The right panel shows the autoradiograph of the probed membrane. The first lane contains radio-labeled tail proteins which were transferred to the membrane. The control lane contained an extract prepared from cells containing the vector. The lanes labeled gpV lysate contained 5 µl and 10 µl of a cell extract prepared from cells expressing pT7-5-VT. In (b) The set up was the same as in (a) except the probe was a radio-labeled extract of cells expressing pT7-5-GT-mut, which expresses gpGT without a frameshift. Radio-labeled protein in both probes binds to gpV.
(c) The radiolabeled proteins that bind to gpV in (a) and (b) (boxed in the figure) were identified by cutting out the bands, eluting the bound proteins, and running these on a new SDS gel along with standards. An autoradiograph of the gel is displayed. The proteins eluted from (a) from (b) are labeled in the figure and were found to run the position of gpGT.
(d) Far western blot using a radio-labeled pET21a–T extract as probe. pET21a–T which expresses just "gpT" was made by fusing the T open reading frame to the N-terminal T7 tag in pET21a. Lane 1, radio-labeled whole tail proteins transferred to the membrane. Lane 2, the radio-labeled pET21a–T probe. Lane 3, the extract prepared from cells containing the vector, a negative control. Lane 4, unlabeled extract prepared from cells expressing the whole tail proteins. Lane 5, purified tails. (e) The protein that interacts with the major tail protein in (d) was recovered by cutting the (boxed) band out and boiling in SDS-sample buffer. The recovered protein was separated by 12.5% SDS-PAGE. The protein recovered from the bands ran at the same position as “gpT”.
Since gpGT contains almost the entire amino acid sequence of gpG, the result suggests that it is the T domain of gpGT that interacts with gpV. To test this, we used a construct (pET21a-T) that expresses the T domain alone (see Materials & Methods). The T frame is fused to a leader peptide (T7 tag) in this vector to allow it to be expressed independently of gpG; we call the resulting protein "gpT". A far-western blot was performed using as a probe a radio-labeled extract expressing gpT (Figure 2 (d)). Similarly to the results obtained when using gpGT containing extracts (above), gpV is labeled in the far-western blot (Figure 2(d) and the radio-labeled protein extracted from the blot is gpT (Figure 2(e)), arguing directly that it is the T domain of gpGT that interacts with gpV.
gpGT interacts with gpV under non-denaturing conditions
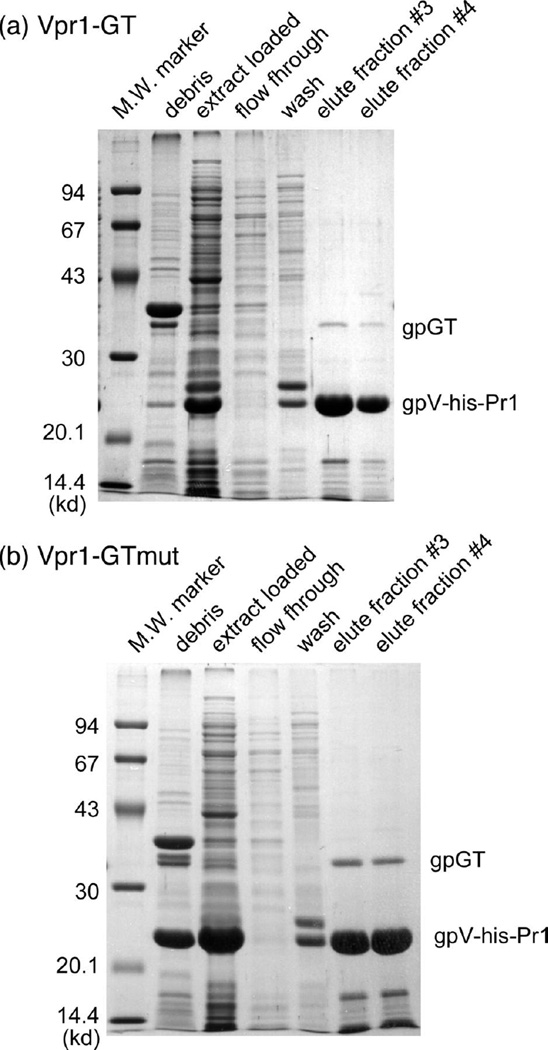
The far-western blots clearly show that gpGT interacts with gpV. However, gpV was denatured during SDS-PAGE and it is not clear to what extent it renatured on the PVDF membrane. We therefore asked whether gpGT can also be seen to interact with gpV under non-denaturing conditions. The strategy was to tag gpV with an N -terminal 6-His tag and express the His6-gpV together with gpG and gpGT using 2 plasmids, one containing the GT-mut variant that produces GT without a frameshift. The resulting extracts were subjected to native nickel affinity chromatography, which selectively retains the 6-His tag. If gpV interacts with gpGT, we would expect the two proteins to co-elute from the column. A C-terminally truncated mutant of gpV (gpV-pr1) was used because the full length His-tagged gpV runs at exactly the same position on an SDS gel as gpGT. The truncated gpV-pr1 is biologically active 26 and the 6-His tagged version is also active as tested by complementation assay (data not shown). Figure 3 shows an SDS gel analysis of the fractions eluted from the two nickel affinity columns. gpGT co-elutes with the 6-His tagged gpV (fractions #3 and #4); this demonstrates that gpGT interacts with gpV under native conditions as well as after denaturation and renaturation of gpV in the far-western blots. The fact that this experiment was done with the C-terminally truncated version of gpV (gpV-pr1) also argues that it is not the C-terminal domain of gpV that interacts with gpGT, as is expected if the gpGT–gpV interaction is an essential event of tail assembly. Katsura 26 showed the C-terminal region of gpV is a morphologically distinct domain that extends out radially from the outside of the tail tube. (We have sequenced the pr1 mutant from a clone derived from the original isolate supplied by I. Kasura and found that the deletion removes residues 156–246 of gpV, which are replaced by the sequence TR.)
Figure 3. gpGT interacts with a gpV variant under non-denaturing conditions.
His-tagged gpV-pr1 was expressed together with gpG and gpGT and purified on a native nickel affinity column. (a) Plasmid pTQ30-Vpr1-GT which has the N-terminally 6-His-tagged Vpr1 and wild type G and GT under the control of a T7 promoter was expressed in BL21(DE3) cells and the his-tag-containing complexes produced were purified using native nickel affinity chromatography. Fractions from the purification were run on SDS gels as indicated. (b) The same as (a) except the plasmid used was pTQ30-Vpr1-GT-mut, which contains the GT fusion mutation that allows gpGT to be expressed without a frameshift.
Overexpressing gpGT causes gpV to polymerize
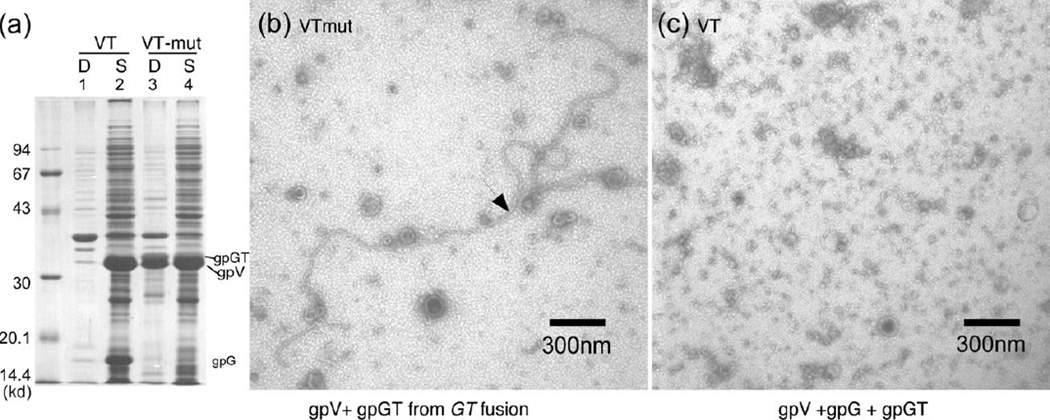
The results reported above indicate that the T domain of gpGT interacts with gpV. What role might this interaction play in tail assembly? Figure 4 shows that overexpression of gpGT in the presence of gpV causes gpV to move from the supernatant to the pellet fraction of the extract. Lane 1 and lane 2 in panel (a) show the pellet and supernatant when gpV was expressed together with wild type gpG and gpGT (plasmid pT7-5-VT). Most of the gpV is in the supernatant. Lane 3 and lane 4 show the pellet and supernatant of cells expressing pT7-5-VT-mut where gpGT is over produced by about 30-fold. The gpV is now partitioned between supernatant and pellet. Panels (b) and (c) show negatively stained electron micrographs of the material displayed lane 2 and lane 4, respectively. The long strings with tail like striations (“polytail”) were seen when gpGT was overexpressed (panel (b)) but absent when a normal amount of gpGT was expressed (panel (c)). The effect is not due to absence of gpG in pT7-5-GT-mut because gpV when expressed alone stays soluble in dimer-monomer equilibrium9. Combining this with the result that gpGT interacts with gpV, we conclude that overexpression of gpGT causes gpV to polymerize.
Figure 4. Overexpressing gpGT causes gpV to polymerize.
(a) Plasmid pT7-5-VT, which contains the wild type genes V, G, and T , and plasmid pT7-5-VT-mut, which is the same as pT7-5-VT except for the GTmut fusion mutation, were expressed in BL21(DE3) cells. The cells were lysed by sonication. The extracts were centrifuged for 25 minutes in a microcentrifuge at maximum speed. The debris was washed 3 times with 20 mM Tris HCl pH 8.0, 100 mM NaCl, 1 mM EDTA and 2% Triton X-100. The proteins were separated on a 12.5% SDS-polyacrylamide gel and visualized using Coomassie blue stain. “D” stands for debris and "S" stands for supernatant. (b) The supernatant from pT7-5-VT-mut was negatively stained and visualized in an with 1% uranyl acetate and viewed in a Zeiss EM901 election microscope at 30,000 x magnification. (c) The supernatant from pT7-5-VT was similarly stained and visualized as in (b).
gpH function requires co-expression with gpG and gpGT
We now describe a set of in vitro complementation experiments which show that the function of gpG is linked to the function of the tail length tape measure protein gpH. Plasmid pETail5, described elsewhere24, contains all the tail genes in wild type form except that a single base pair insertion in the region of the G–T frameshift fuses the G and T ORFs into a single gene, with the result that gpGT is made in large amounts and production of gpG is essentially eliminated; this plasmid does not produce active tails. Because gpG is largely lacking in the pETail5 extracts, we tried to complement this extract by providing gpG and gpGT expressed from another plasmid using the in vitro complementation methodologies developed by Katsura 27. Cells expressing pETail5 and cells expressing pT7-5-GT (produces wild type gpG, and gpGT) were mixed together, concentrated, and then frozen and thawed several times to lyse the cells. Assembly of active tails in this mixture was assayed by testing its ability to form infective phages when supplemented with free phage heads28, provided separately, as summarized in Table 1. No activity above background was detected (3×104 pfu/ ml). However, when the second extract was produced from plasmid pT7-5-GH, which expresses gpG, gpGT and gpH, a four hundred fold increase in activity above the background was detected (1.2×107 pfu/ ml). The fact that gpH must be provided to the pETail5 extract suggests that either the gpH produced from pETail5 is not active or gpH is not produced because of over-expression of gpGT. An SDS gel of radio-labeled proteins from pETail5 showed in a separate study24 that gpH is produced and we show below that in fact more gpH is produced in this construct than from the plasmid with wild type sequence. This argues that gpH produced from pETail5, though wild type in sequence, is not active. Taking these results together, we conclude that for an extract to have complementation activity it must have wild type gpG, gpGT and gpH all expressed together.
Table 1. Tape measure protein gpH requires gpG and gpGT for activity.
Efficient production of tails by plasmid pETail5, which has a deficiency in gpG can only occur if gpH is supplied along with gpG and gpGT and not by supplying gpG without gpH. Extracts were mixed, incubated, and assayed for phage yield after free heads were added as described under Materials and Methods.
| Feature | Components and results | ||||
|---|---|---|---|---|---|
| plasmid extract 1 |
none | pT7-5-GT | pT7-5-GH | pT7-5-GT | pT7-5-GH |
| proteins in extract 1 |
- | gpG, gpGT | gpG, gpGT, gpH |
gpG, gpGT | gpG, gpGT, gpH |
| plasmid extract 2 |
pETail5 | none | none | pETail5 | pETail5 |
| phage yield/ ml |
3×104 | < 2×102 | < 2×102 | 3×104 | 1.2×107 |
gpH interacts physically with gpG and gpGT
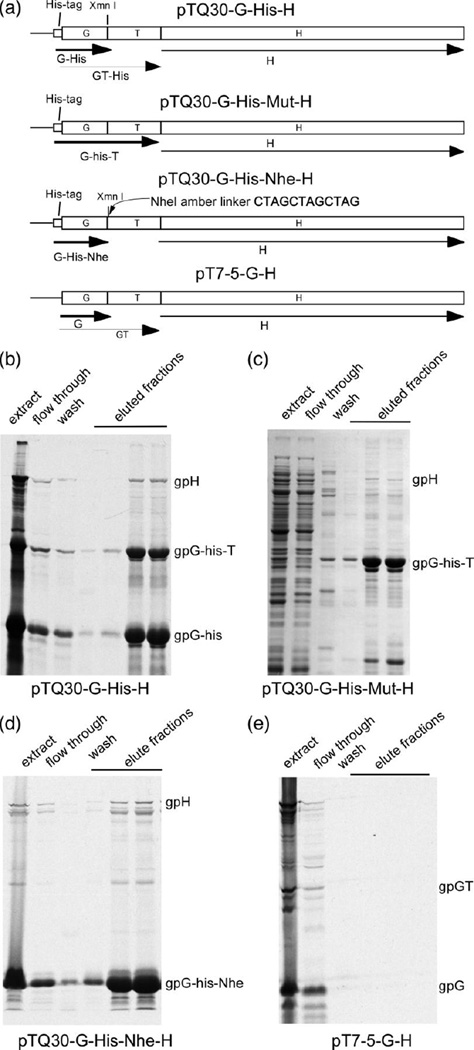
The fact that gpH's activity requires co-expression with gpG and gpGT suggested that there might be physical interactions among gpG, gpGT and gpH. We tested this possibility by native nickel affinity chromatography. The N-terminus of protein G was fused to a 6-His tag in vector pTQ30 (a T7 promoter-controlled expression vector that allows N-terminal His tagging, see Materials and Methods). The resulting 6-His tagged protein, his-gpG, is biologically active as tested by its ability to complement pETail5 when co-expressed with gpH (data not shown). Figure 5 shows the results from native nickel chromatography. Panel (a) shows the constructs used in the experiment. Panel (b) shows that gpH co-purifies with his-gpG and with his-gpGT. Panel (c) used a construct that over-expresses his-gpGT and shows that gpH also interacts with his-gpGT alone. To test whether gpG alone interacts with gpH, a NheI oligonucleotide linker containing an amber codon in all three reading frames is inserted at the XmnI site right after the slippery sequence (Figure 5(a)). The inserted linker will prevent the production of gpGT because a ribosome in any frame will be stopped by an amber codon in the linker. Figure 5(d) shows the nickel affinity chromatography result from the NheI linker insertion construct, and we see that gpH is also co-purified with his-gpG*-Nhe (the His-tagged gpG from the NheI linker insertion mutant, which has a slightly different and shorter C-terminus than the wild type version). In a negative control (Figure 5(e)), gpG without the 6-His tag does not pull down gpH, showing that gpH alone will not bind to the column. In summary, these experiments show that both gpG and gpGT interact with gpH with sufficient affinity to co-purify using a nickel affinity column.
Figure 5. gpG interacts with the tape measure protein gpH.
(a) Plasmid constructs used in this experiment. pTQ30-G-His-H contains λ DNA from G to H with a the addition of 6-His tag to the N-terminal of G. pTQ30-G-His-Mut-H is the same as pTQ30-G-His-H except for the GT fusion mutation which allows gpGT to be expressed without frameshifting. pTQ30-G-His-Nhe-H is the same as pTQ30-G-His-H, but has an Nhe amber linker, which has stop codons in all three reading frames, inserted into the XmnI site right after the slippery sequence. pT7-5-GH contains the wild type λ DNA from G to H in which the N-terminal of G is not His tagged. (b) Native nickel affinity chromatography of a radio-labeled extract prepared from cells expressing plasmid pTQ30-G-His-H. The proteins were separated on a 12.5% SDS polyacrylamide gel and visualized by autoradiography. (c) Native nickel affinity chromatography of an extract prepared from cells expressing plasmid pTQ30-G-His-mut-H. The proteins were separated on a 12.5% SDS polyacrylamide gel. The gel was Coomassie blue stained. (d) Native nickel affinity chromatography of a radio-labeled extract expressing plasmid pTQ30-G-His-H. The proteins were separated on a 12.5% SDS polyacrylamide gel and visualized by autoradiography. (e) Native nickel affinity chromatography of a radio-labeled extract expressing plasmid pT7-5-GH. The proteins were separated on a 12.5% SDS polyacrylamide gel and visualized by autoradiography. Note: his-gpG is His tagged gpG; his-gpGT is His tagged gpGT, his-gpG*-Nhe is His tagged gpG (slightly altered at the C-terminus) expressed from pTQ30-G-his-Nhe-H.
gpG interacts with more than one part of gpH
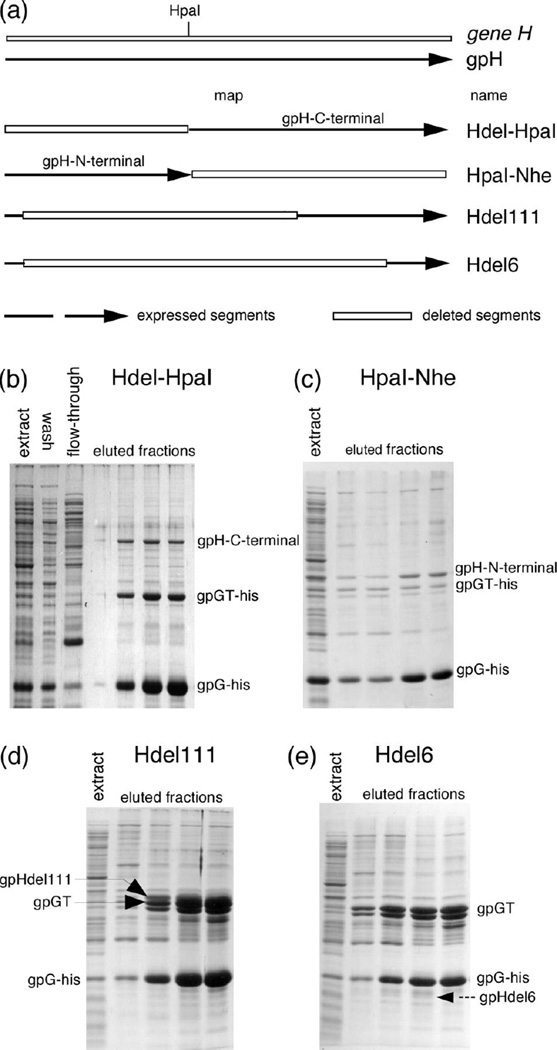
We attempted to map the region of gpH that interacts with gpG. A series of H truncations and in-frame deletions was created as shown in Figure 6 panel (a). Note that the two constructs, H-del-HpaI and H-HpaI-Nhe, have no region of gene H in common. All the H-deletions were transferred into a plasmid which produces gpG His-tagged at its N-terminus, and nickel affinity chromatography was performed to test for interactions. Figure 6 panels (b, c, d) show that his-gpG interacts with all but one of the truncated variants of gpH, indicating that his-gpG interacts with more than one part of gpH. Figure 6 panel (e) shows that his-gpG failed to pull down a detectable amount of gpHdel6 in which about 80 percent of the gpH is truncated. gpHdel6 still has a significant part of the gpH C-terminus left, which suggests gpG interacts primarily with the more N-terminal parts of gpH. This result agrees with Katsura's suggestion that the C-terminal end of gpH binds to the initiator 17, a suggestion made because a large (70,000 kDa) gpH amber fragment does not form the initiator and because a short lacZ insertion into the middle of gene H could make a small number of short tails without the presence of the N-terminus of gpH.
Figure 6. gpG interacts with more than one part of gpH.
N-terminally His-tagged gpG was co-expressed with truncated variants of gpH. Complexes in the resulting extracts were purified by native nickel affinity chromatography. The eluted proteins were separated on SDS polyacrylamide gel and visualized by Coomassie blue staining. The indicated positions of different H truncations were determined by radio-labeling experiments (data not shown). (a) Gene H and the regions of gpH expressed for these experiments. The boxed regions indicate deletions. Hdel-HpaI: (plasmid pTQ30-G-his-Hdel-HpaI) the DNA from the start codon of H to the HpaI site is deleted. HpaI-Nhe: (plasmid pTQ30-G-his-H-HpaI-Nhe) NheI amber linker which has amber stop codons in all three open reading frames was inserted at the HpaI site, which makes gpH synthesis stop right at the HpaI site. Hdel111: (plasmid pTQ30-G-His-Hdel111) residues 37 to 561 of gpH are deleted. Hdel6: (plasmid pTQ30-G-His-Hdel6) amino acids from 34 to 734 were deleted. (b) Hdel-HpaI nickel affinity chromatography. The label "gpH-C-terminal" indicates the truncated gpH produced from the constru ct. (c) HpaI-Nhe nickel affinity chromatography. The label "gpH-N-terminal" indicates the truncated gpH produced from the construct. (d) Hdel111 nickel affinity chromatography. The label "gpHdel111" indicates the truncated gpH produced from the construct. (e) Hdel6 nickel affinity chromatography. The dashed arrow labeled "gpHdel6" indicates the position where the truncated gpH produced from this construct ran in control experiments.
Discussion
The frameshift strategy is important
The effect of the λ tail frameshift is two fold. On the one hand it creates two proteins gpG and gpGT, with gpGT containing almost the entire sequence of gpG. On the other hand the ratio of the two proteins is determined by the efficiency of frameshifting. Through complementation experiments reported in a separate publication24, we have shown that gpGT alone will not complement λ expressing neither gpG nor gpGT but will complement λ expressing gpG but not gpGT. This shows that gpGT alone is not able to promote tail assembly but gpG is also required. We also showed that the T domain of gpGT is not functional which argues that it is important for gpGT to share the same N-terminal region with gpG. Next through another set of complementation experiments in which we modulated the expression of gpGT through an arabinose controlled promoter, we showed that there is an optimum gpGT expression level that complements best24. Neither too high a level nor too low a level of gpGT works. We take this result to imply that the molar ratio of gpG and gpGT is important.
In summary, these results indicate that both aspects of the frameshift are important for tail assembly. This is consistent with the observation that the frameshift is highly conserved among dsDNA tailed phages. On the other hand, the compelling evidence for the existence of an analogous frameshift in most dsDNA tailed phages, as we have reported previously22, indicates that most long-tailed may use a strategy similar to λ's to assemble their tails to the correct length.
Chaperone function of gpG/gpGT
Our results show that gpH's activity requires co-expression with gpG and gpGT. This explains the observation that the amber mutation in G region did not complement an amber mutation in gene H in vitro 8; 10; 29. Because the requirement for co-expression, neither amber mutant can provide active gpH. Furthermore, we showed that gpG and the G part of gpGT physically interact with gpH. This interaction does not seem to be limited to one region of gpH, because we showed that gpG can interact with two segments of gpH with no common amino acid sequence (Figure 6). There is no obvious sequence similarity between the two segments except that both segments are predicted to contain mostly coiled-coil secondary structure. It is not known whether gpG recognizes the coiled-coil structure or some other unknown motif.
The requirement for co-expression of gpG and gpGT with gpH, and the demonstration of a physical interaction between gpG and gpH, argue that the nascent gpH polypeptide might require the binding of gpG and gpGT to fold correctly. This is consistent with the observation that gpG is expressed in a large amount similar to the level of the major tail protein gpV. Since gpG interacts with more than one part of gpH, the amount of gpG expressed is probably enough to coat the entire gpH. The large amount of gpG might ensure gpH is properly chaperoned over its entire length. Since gpGT also uses its G part to interact with gpH, it is likely that some part of gpH is also bound with gpGT molecules. The observation that tail assembly can be disrupted by altering the ratio of gpG and gpGT production argues the amount of gpGT incorporated probably is determined by the ratio of gpG vs. gpGT, rather than by some sort of mechanism that assembles the 'correct' ratio of gpGT to gpG regardless of the ratio in the unassembled pool. It also argues that having the correct ratio of gpGT to gpG in complex with gpH is important for successful assembly.
The requirement for a smaller protein to help the folding of a larger protein is not unique in the G/ H case. In phage T4 there are several examples, including products of gene 57, which are required for the formation of two segments of T4's tail fibers (gp34 and gp37) and short tail fibers (gp 12)30; 31. Also, the T4 protein gp38 is specifically required for the successful production of the distal tail fiber segment (gp37)30; 31; 32. The λ side tail fiber protein probably has a similar requirement. The λ side tail fiber protein is composed of two protein (Stf) and Tfa. Stf is a long protein while Tfa is a small protein which is expressed in large amount even though only ~3 copies are incorporated into each fiber. λ Tfa can functionally substitute T4's Tfa homolog (gp38), which is not even incorporated into the final structure.
Interaction between gpGT and gpV
The way gpGT is expressed allows it to possess an N-terminal half that is almost identical to gpG and a unique C-terminal T part. we have shown that the G part can interact with gpH. we also showed that the T part of gpGT interacts with major tail protein gpV and overexpression of gpGT causes the major tail protein to polymerize. However, the T part of gpGT cannot be the only trigger for gpV polymerization. In a U amber phage, the formation of two aberrant long polymers of gpV called polytails or polytubes requires both gpL and gpM29. Polytubes are observed in a U amber and T amber double mutant, suggesting that gpGT is not essential for gpV polymerization in the absence of gpU, though the polytails produced in the double (V,T) mutant are significantly shorter than the polytubes seen with other double mutants that were tested 29. However, gpGT is required for gpV polymerization in the presence of gpU, since no tail like structure was observed in phage with amber mutation in the T region. Since the G part of gpGT interacts with gpH, gpGT might function to recruit gpV to polymerize around the tape measure protein during normal assembly.
Sequence comparison among other phages supports the interaction measured in λ
This study shows that the G part of gpGT interacts with the tape measure protein gpH and the T part interacts with major tail protein gpV. It is interesting to note that for the Pseudomonas F pyocin, which is essentially a phage tail, the T part is similar to phage D3 T and HK022 T while the G part has no detectable similarity to D3 G or HK022 G (Figure 7). According to our results, we would expect that the major tail protein of D3 would be similar to the F pyocin major tail protein while the tape measure protein might not be too similar. This is indeed what we found (Figure 7), we saw high similarities between the major tail proteins of F pyocin, D3 and HK022 and little similarity between the N-terminal regions of the corresponding tape measure proteins, where the gpG is supposed to interact. We did find a region of similarity between the C-termini of the HK022 and F pyocin tape measure proteins, but this is a part of the protein which we think is not important for gpG interaction, since we have shown for phage λ that gpG appears to interact with multiple sites within the N-terminal 60–80% of gpH, but not within the C-terminal 20%.
Figure 7. BLASTX search results using F pyocin "V G-T-H" region.
(a) The graphic output from the BLASTX search using F pyocin genes (bp 19,842 – 22,861 from GenBank GI:5725144). The hits with low scores (black) were removed for clarity. (b) The BLAST output of alignment of F pyocin "T" region with D3 "T". (c). The BLAST output of alignment of F pyocin "T" region with HK022 "T". (d) The BLAST output of alignment of F pyocin "V" region with D3 "V". (e) The BLAST output of alignment of F pyocin "V" region with HK022 "V".
Other length regulated structures
There are other length regulated structures of note that have been studied in bacteria, including the injectisome needles of Type III secretion systems and the flagellar hook, both of which have molecular ruler molecules which appear to operate in a manner analogous to the tape measure protein of bacteriophage tails 33; 34; 35. The assembly mechanisms of these structures may not require the types of chaperone we describe here. The more recently characterized Type VI secretion systems resemble contractile phage tails and have a clear evolutionary connection to bacteriophages36; 37. However, whether the Type VI secretion apparatus utilize phage-like tape measure proteins or chaperones for their assembly remains to be discovered.
Model and conclusions
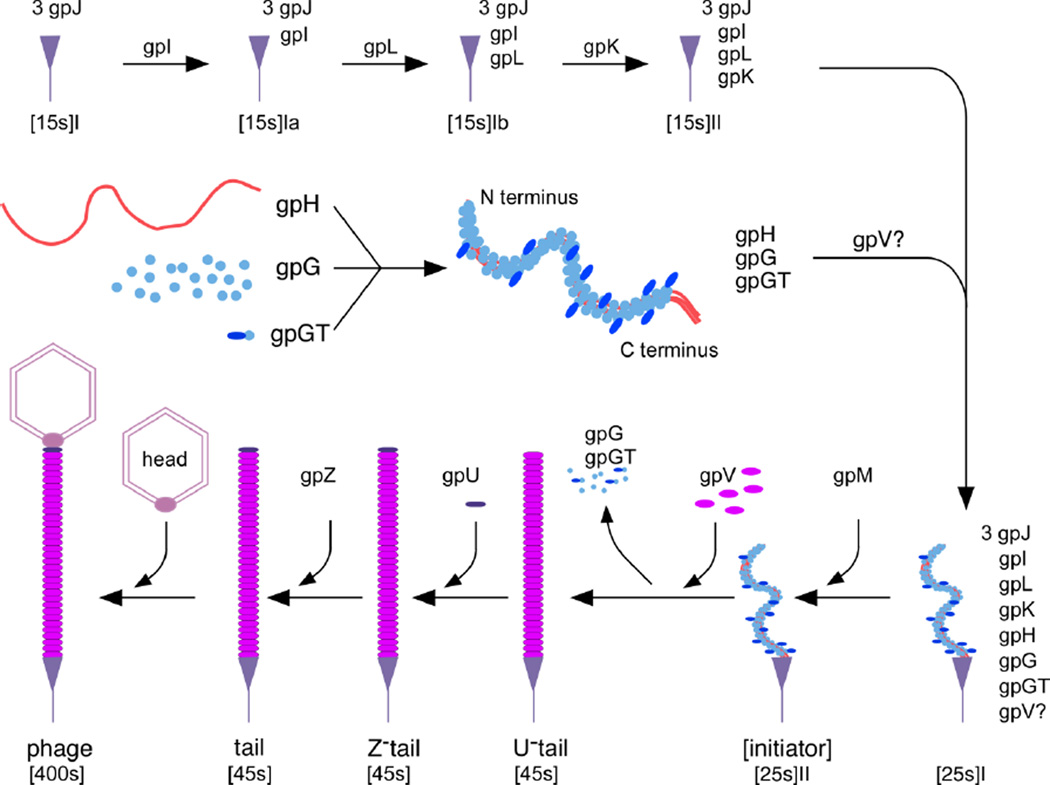
The identifiable strong conservation of gene order appears to be limited to the gpV-gpG/ gpGT-gpH region across a diverse set of phages22. This suggests that these four proteins' function might be closely linked together and that they may evolve as a unit. This is consistent with our results which show the four proteins' functions are related, with gpGT providing a bridge between major tail protein gpV and tape measure protein gpH. Our results also provide a rational explanation for the broad conservation of frameshifting among dsDNA tailed phages. That is, it appears that both the production of two proteins sharing the same N-terminal sequence and keeping the two proteins in a fixed ratio are very important. We incorporated our results in the assembly pathway shown in Figure 1. A major difference from the assembly pathways proposed earlier is that we think gpG, gpGT gpH and possibly gpV form a subassembly in the assembly pathway. A small amount of gpGT is incorporated into the complex between gpG and gpH. The C-terminal of gpH probably is used to bind to the initiator 17 and gpGT together with other factors in the initiator (e.g., gpL, gpM), induces gpV to polymerize around the tape measure protein. When the tail reaches the length specified by the tape measure protein, it will stop and be capped by gpU. The detailed picture of how gpG, gpGT , gpV and gpH interact with each other to form the complex structure is not known. How gpG and gpGT are excluded in the final structure and how gpH is cleaved also remain unanswered. Those questions cannot be answered until we have a more detailed structural and biochemical characterization of the tail and tail assembly intermediates.
Figure 1. A working model for λ tail assembly that includes roles for gpG and gpGT as tape measure protein chaperones.
The model is based on Katsura's8, but is modified to accommodate our new observations. A major difference is that gpG, gpGT and gpH are shown to form a subassembly before joining the initiator. Even though we have shown that gpGT interacts with gpV, it is not clear whether gpV is actually involved in initiator formation or only adds later. Our results do not show exactly how gpG and gpGT interact with gpH, but we show 3 copies of gpH coated with both gpG and gpGT as one possible way in which these proteins interact. The locations of some of the proteins in the structures are not known, including some tail tip proteins and gpZ.
The experiments reported here and supported by earlier work give, we believe, a good first order view of how lambda tails are assembled, how the assembly process is mediated by the assembly chaperones gpG and gpG–T and how that process leads to a tail tube whose length is determined by the length of gpH, the tail tape measure protein. The assembled tail tube, made of ~192 copies of the major tail subunit gpV, contains a few copies of gpH, with their C- termini oriented toward the tip of the tail, and it is capped on one end by the structure known alternately as the tail tip or the initiator and on the other end by the head.
As with any other biological structure produced by assembly of its parts, the question arises how and to what extent do the process of assembly and the structure of the assembled product contribute to subsequent biological functions of the structure. In the case of lambda tails such subsequent functions would include attaching the phage to the host cell and mediating the passage of the phage DNA across the cell envelope and into the cell cytoplasm. These processes cannot depend directly on the gpG and gpG–T chaperones because those proteins are not present in the mature tail. Beyond the fact that the gpJ protein, which is located at the tip of the tail, is responsible for binding to the lambda receptor, the outer membrane protein LamB, little is reliably known about these processes in lambda. However, there are a few experimental facts that provide useful constraints to our understanding, including results that implicate gpH, the tape measure protein, as a central player.
Biochemical experiments with membrane vesicles decorated with LamB receptors show that under conditions in which the phage DNA is injected into the vesicles, gpH is also injected18. We expect the gpH precedes the DNA into the cell, both because of the position of the gpH in the tail tube relative to the DNA and because of genetic experiments which argue that gpH interacts with an inner membrane sugar transport protein, PtsM in some way that is essential for successful DNA injection19. These facts as well as direct examination of the gpH amino acid sequence indicate that gpH is quite hydrophobic. We speculate that this may explain why gpH, to be functional, has to be co-expressed with gpG and gpG–T, which might shield gpH from the polar solvent until it can be surrounded with the gpV tail tube.
A prominent feature of lambda tail maturation is that gpH gets smaller in apparent molecular weight by about 10 kD38. Amino terminal sequencing rules out the possibility of an N-terminal truncation as a cause of the decrease in molecular weight39. Other possibilities include a C-terminal truncation or removal of an internal segment of the gpH sequence. We do not yet know the biological function of this maturation of gpH, but we speculate that either the processed gpH or the piece released during maturation of gpH may support the tail functions. It is intriguing in this context that several phages (not including lambda) have a recognizable sequence for a lysozyme or other muralytic enzyme located within the C-terminal half of their tape measure protein sequences40; 41. Finally we note that recent experiments show that lambda tails contain an Fe4S4 iron-sulfur cluster42. The cluster is coordinated by conserved cysteines in gpL, one of the components of the initiator or tail tip. Genetic and biochemical experiments suggest that the iron-sulfur cluster may have essential roles in both tail assembly and DNA injection.
Materials and Methods
DNA methods
Standard protocols 43 were used for DNA manipulations and most reagents and enzymes were used according to manufacturers instructions. PCR for plasmid construction was performed as follow, 0.5 µM of each primer, 0.2 mM of each dNTP, 1×PFU buffer, 5 µl of template (for plasmid template, miniprep plasmid DNA was diluted 100 fold and 5 µl was used; for phage template, CsCl purified phage particles (about 1012 titer) were diluted 100 fold and 5 µl were used), and 2 units of Pfu polymerase in a 50 µl reaction. After an initial denaturation at 94°C for 1 min, 20 cycles of 94°C for 30 sec, 50°C for 30 sec and 72°C for 2.5 minutes were run on a 9600 or 9700 GeneAmp PCR machine (Perkin-Elmer). Linker insertion was done as described previously (Lathe et al., 1984). Deletions were made using a two-step PCR protocol using two divergent overlapping primers and two flanking primers 44. The two fragments produced by the first PCR reactions were gel purified and reamplified using the flanking primers for 20 cycles. The final product was subcloned back into the plasmid and sequenced to verify the deletion. Description of individual plasmid constructions and any primers used are found in Tables 2.
Table 2.
Plasmids and primers used in this study.
| Name | Description, construction details |
|---|---|
| pT7-5 | Expression plasmid pT7-5 contains an ampicillin resistance gene and the T7 -10 promoter adjacent to a multiple cloning site (GenBank AY230150) |
| pET21+ | Expression plasmid pET21+ contains an ampicillin gene, a LacI gene and T7 -10 promoter adjacent to a Lac operator and a multiple cloning site (Novagen). |
| pET21a | Expression plasmid pET21a is the same as pET21+ except a T7 capsid protein leader peptide is included which allow ORFs to be fused to the tag (Novagen). |
| pTQ30 | Expression plasmid with His tag. EcoRI, XbaI fragment of pTQE30 (QIAGEN) was cloned into EcoRI, XbaI site of pT7-5. The resulting construct can be used to construct N-terminal 6xHis tagged protein under the control of T7 promoter. |
| pBluescript(sk-) | A general purpose cloning vector (Stratagene). |
| pBS-GT | PstI-MscI fragment of pBRUM-1 (Uma Chandran, PhD thesis, 1990, University of Pittsburgh, containing λ genes U through M) containing the G–T region was cloned into PstI and EcoRV sites of pBluescript(sk-). |
| pBS-GT-mut | PCR product using GT1/ GT2 as primer and pBRUM-1 as template was cut with PstI-StuI and inserted back into pBS-GT PstI and XmnI-partial digest vector. The resulting construct added one more G right after the slippery sequence which allow gpGT to be synthesized without a frameshift mechanism. |
| pBS-λ-tail I | 16KB BamHI-EcoRI fragment of λ DNA containing the entire 11 tail genes was cloned into pBluescript(sk-) BamHI-EcoRI sites. |
| pBS-λ tail3 | The large fragment of pBS-λ-tail-I BswiI-HindII digestion was filled in and recirculized. The resulting plasmid has less extra DNA upstream of gene Z. |
| pBS-λ-tail5 | pBS-λ-tail3 was cut with BamHI, filled in and then cut with NsiI to obtain the vector. The vector was ligated to 2.6 kb BsaAI-NsiI fragment of pBS-λ-tail I. The resulting plasmid resulted in further reduce of extra DNA upstream of gene Z. |
| pETail | The SacI XhoI fragment of pBStail 5 was inserted between the SacI- XhoI sites of pET21+. |
| pETail4 | BstbI-Xho I digestion of PCR fragment produced by using primers J1, J2 and pETail4 as template was inserted back into the BstBI-XhoI sites of pETail I. The resulting construct gets rid of lom and left about 20 bp downstream of gene J. |
| pETail5 | pETail4 ApaI -Nsi I fragment which contains the slippery sequence was replace with the ApaI-NsiI fragment from pBS-GT-mut. The resulting plasmid contains all the tail genes and GT-fusion mutation. |
| pT7-5-Tail4 | pET-Tail4 cut with SacI, XhoI ligate to pT7-5 SacI SalI digest |
| pT7-5-Tail5 | pET-Tail5 cut with SacI, XhoI ligate to pT7-5 SacI SalI digest |
| pT7-5-VT | MscI fragment of pBRUM containing-λ genes U through M (Uma Chandran, PhD thesis, 1990, University of Pittsburgh) was cloned into pT7-5 SmaI site to create pT7-5-VT. |
| pT7-5-VT-mut | ApaI NsiI frag containing the mutant GT was cloned into PT7-5-VT ApaI NsiI site |
| pT7-5-GH | StuI-XbaI fragment of pT7-5-H was inserted into StuI-XbaI sites of pT7-5-GM. |
| pT7-5-GT | The 1 kb XbaI HincII fragment containing G/ GT of pT7-5-VT was inserted into XbaI-SmaI sites of pT7-5. The resulting plasmid will express gpG and gpGT. |
| pT7-5-GT-mut | The 1 kb XbaI HincII fragment of pT7-5-VT-mut was inserted into XbaI-SmaI sites of pT7-5. The resulting plasmid will express gpGT only. |
| pTQ30-G-His-H | PCR product using primers GT2/ G-His on template pT7-5-GT was cut with SacI and Apa I to obtain the insert. pTQ30-GH was digested with the SacI and Apa I to obtain the vector |
| pTQ30-G-His-T | EcoRI-NsiI fragment of pTQ30-G-His-H was inserted into EcoRI- NsiI sites of pT7-5-GT. |
| pTQ30-G-His- mut-H |
ApaI-NsiI fragment of pT7-5-GT-mut is inserted into ApaI NsiI sites of pTQ30-G-His-H |
| pT7-5-G-Ham- HpaI |
Nhe amber linker was inserted in HpaI site of pT7-5-GH |
| pT7-5-tail4- Hdel111 |
Four primers (H-del1,H-del2)/ (H-del3, H-del4) were used to generated the fragment with the deletion following the protocols described in the method section using pET21a-TH and λ-M-tail as template respectively. The resulting PCR product was digested with NsiI and EcoNI and inserted into NsiI-EcoNI sites of pT7-5- tail4. |
| pT7-5-tail4- Hdel6 |
Four primers (H-del1,H-del5)/ (H-del6, H-del4) were used to generated the fragment with the deletion follow the protocols in the method section using pET21a–TH and λ-M-tail as template respectively. The resulting PCR product was digested with NsiI and EcoNI and inserted into NsiI-EcoNI sites of pT7-5-tail4. |
| pTQ30-G-His- H-HpaI-Nhe |
NsiI-StuI fragment of pT7-5-G-Ham-HpaI was inserted into NsiI- StuI sites of pTQ30-G-His-H |
| pTQ30-G-His- Hdel111 |
NsiI-StuI fragment of pT7-5-tail4-Hdel111 was inserted into NsiI- StuI sites of pTQ30-G-His-H |
| pTQ30-G-his- Hdel-HpaI |
PCR fragment using primers H-del1/ H-del8 and pT7-5-GH as template was digested with NsiI and ligated to NsiI-HpaI site of pTQ30-G-his-H. PFU was used in PCR reaction so one end is blunt and can be used directly for blunt end ligation. |
| pTQ30-G-His- Hdel6 |
NsiI-XmnI fragment of pT7-5-tail4-Hdel6 was inserted into NsiI- HindIII sites (HindIII site was filled in with T4 polymerase) of pTQ30-G-His-H. |
| pTQ30-G-His- Nhe-H |
ApaI-NsiI fragment of pT7-5-GT-Nhe is inserted into ApaI NsiI sites of pTQ30-G-His-H |
| pET21a–T | pET21a was digested with EcoRI, blunted with mung bean nuclease and cut with Hind III to obtain the vector. pBS-GT was digested with XmnI and HindIII to obtain the insert. The resulting plasmid contains only the T part of GT fused to the T7 tag from the vector. |
| pTQ30-Vhis- pr1-tail |
Primers V-his and GT2 were used to generated a fragment from template phage λVpr1S7CI857. The PCR fragment was cut with BamHI and ApaI and cloned into BamHI and ApaI site of pTQ30-G- his-tail |
| pTQ30-G-his- Tail |
NsiI HindIII fragment of pT7-5-tail4 was cloned into NsiI HindIII sites of pTQ30-G-His-H |
| pTQ30-Vhis- pr1-GT |
ApaI, HindIII fragment of pT7-5-VT was cloned into pTQ30-Vhis- pr1-tail ApaI, HindIII site. |
| pTQ30-Vhis- pr1-GT-mut |
ApaI, HindIII fragment of pT7-5-VT -mut was cloned into pTQ30- Vhis-pr1-tail ApaI, HindIII site. |
| pT7-5-VT-mut | ApaI NsiI frag contained the mutant GT was cloned into PT7-5-VR- right ApaI NsiI site |
| pT7-5-GT-Nhe | An Nhe amber linker was inserted at XmnI site right after the slippery sequence. |
| GT1 | gccctgcagcgcattgagcatctc |
| GT2 | gaaggcctttcccgcagaaacagg |
| G-HIS | AGGAGCTCatgttcctgaaaaccg |
| J1 | gaggagttttcgaaag |
| J2 | ggctcgagacgaacctctgtaac |
| H-del1 | tcagcgatccggatatgc |
| H-del2 | aaatcctccggtaccagaaaaatgacgcc |
| H-del3 | catttttctggtaccggaggatttacggg |
| H-del4 | cgttctccaccgacctct |
| H-del5 | gaatttatccgcatcactttccgtaccag |
| H-del6 | acggaaagtgatgcggataaattcgcacag |
| H-del8 | cataccggactcctcctg |
| V-his | GAGGATCCcctgtaccaaatcctaca |
| NheI amber linker |
CTAGCTAGCTAG |
Protein expression
T7 promoter controlled plasmid were expressed in BL21(DE3) or BL21(DE3)pLysS cells (Studier, 1991; Studier et al., 1990) as described in Duda et al (1995). Overnight cultures grown in LB/ Amp (50 µg/ ml Amp) (BL21(DE3)) or LB/ Amp/ Cam (50 µg/ ml Amp, 25 µg/ ml Cam) (BL21(DE3)pLysS) were diluted 1:2000 and grown at 37°C with vigorous shaking When the A550 reached 0.4, cultures were induced with IPTG (final concentration of 0.4 mM for pT7-5 based vectors and 1 mM for pET21 based vectors (Novagen)). The induced cells were grown for 3.5 hours, chilled and harvested by centrifugation. Cells for nickel-affinity chromatography analysis were resuspended in binding buffer (see below), lysed by sonication and centrifuged to remove cell debris.
Tail purification
Plasmid pETail4 or pT7-5-tail4 containing the whole tail genes was expressed in BL21(DE3) pLysS cells as described above. The cells were harvested and lysed in 50 mM TrisHCl pH 8.0 plus 5 mM Na2 ethylene diamine tetraacetic acid. The lysate was clarified by centrifugation, followed by 7.5% (w/ v) PEG precipitation. The pellet was resuspended in TKG buffer (20 mM Tris-HCl, pH 7.5, 100 mM potassium glutamate), loaded onto a 10~30% glycerol gradient and centrifuged in a SW28 rotor (Beckman) at 27 krpm for 5 hrs. The tail band was extracted with a needle and syringe and pelleted by ultracentrifugation in a Type 45 Ti rotor (Beckman) at 40 krpm for 4.5 hrs. The tail pellet was resuspended in a small volume of TKG buffer (1 ml for a 1 liter culture).
Radio-labeling
Radio-labeling was performed as described45. Briefly, BL21(DE3) containing appropriate plasmid was grown at 37°C in M9 based RG-glucose medium45 to OD550=0.4. IPTG was added at a final concentration of 0.4 mM to induce for 15 minutes after which rifampicin was added at a final concentration of 0.2 mg/ ml, to shut down expression of host genes for 15 min. 35S-methionine was added to 10 µCi/ ml and the culture was incubated for another 15 min. The cells were then harvested by centrifugation for further analysis.
SDS polyacrylamide gel electrophoresis
SDS gel electrophoresis was performed as described in (Laemmli, 1970). Protein samples were mixed with 4 × SDS sample buffer, boiled for 3 minutes, loaded onto a gel and run at 150 volts.
Far western Blots
Proteins in an SDS gel were transferred to a PVDF membrane (Millipore) at 100 v for 20 min. The membrane was blocked using 5% non-fat milk in TTBS buffer (0.2% Tween 20, 0.9% NaCl, 100 mM TrisHCl pH 7.5 and then probed with radio-labeled extract (~1 µCi/ 6 cm2 in TTBS) in a heat sealed bag for 2 hours. The membrane was washed 3x in TTBS for 10 min., dried and exposed to Bio-Max film (Kodak). The membrane areas corresponding to bound radioactive proteins were cut out, boiled in SDS sample buffer for 5 min, and the eluted radioactive protein was analyzed on a new gel.
Nickel affinity chromatography
Nickel affinity chromatography was performed as described in Novagen protocols using buffers that each contained 0.5 M NaCl, 20 mM Tris-HCl pH 7.9 as their base. Briefly, a 0.5 – 1 ml column of Chelating Sepharose FF (GE Healthcare) was charged with 4 volume of 400 mM NiSO4 and equilibrated with 3 volumes of binding buffer (with 5 mM imidazole). The sample was loaded and washed with 10 volumes of binding buffer and 8 volumes of washing buffer (with 150 mM imidazole). Bound protein was eluted with 3 volumes of elution buffer (with 500 mM imidazole).
λ Head purification
A 1 L culture of λ Vam750Sam7cI857 was grown in M9 medium (13.3% Na2HPO4•7H2O, 3% KH2PO4, 1% NH4Cl, 0.5% NaCl and incubated at 30°C with vigorous shaking to a concentration of 3×108 / ml. The cells then were heat induced at 43°C for 15 min in a water bath, grown for 3 hrs at 37°C, chilled, harvested by centrifugation and resuspended in a small volume λ-dil buffer (10 mM Tris-HCl, pH 7.4, 10 mM MgCl2). Chloroform was added to saturation to induce lysis. After lysis cell debris removed by centrifugation and 5 ml of heads were loaded onto 34 ml 10%–30% glycerol gradients in λ-dil and centrifuged in a SW28 rotor (Beckman) at 27 krpm for 80 minutes. The head bands were drawn using a 20 G needle and aliquots were frozen at −70°C for future use.
In vitro tail complementation
was done as described in Katsura et al. (1976) with modifications. Proteins were expressed in BL21(DE3) pLysS cells as described above. Cells were resuspended 1/ 200 volume of Buffer A (0.1 M Tris-HCl (pH 8.8) 20 mM MgSO4, 40 µg/ ml DNase I (Sigma D-5025)). 10 µl of each type of cells were mixed, frozen and thawed for 3 times. The mixture was incubated at room temperature for 30 minutes. 10 µl of the mixture was used to assay for activity by head-tail joining assay. Head tail joining was performed as described in (Weigle, 1966). 10 µl each of tail and head samples were mixed with 80 µl of λ-dil, incubated at room temperature for 30 minutes and titered for phage production.
Highlights.
Chaperones gpG and gpGT bind directly to bacteriophage λ ‘s tape measure protein gpH
λ tail tape measure protein gpH requires co-expression of chaperones for function
Overexpression of gpGT causes λ tail tube protein gpV to polymerize into long tubes
The T part of gpGT binds to gpV, suggesting it recruits gpV for assembly around gpH
Acknowledgements
This work was supported by NIH grant R01 GM47795 to RWH and RLD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Berget PB, King J. T4 tail morphogenesis. In: Mathews CK, Kutter EM, Mosig G, Berget PB, editors. Bacteriophage T4. Washington, D.C: American Society for Microbiology; 1983. pp. 246–258. [Google Scholar]
- 2.Coombs DH, Arisaka F. T4 tail structure and function. In: Karam JD, Drake JW, Kreuzer KN, editors. Molecular Biology of Bacteriophage T4. Washington, DC: American Society for Microbiology; 1994. pp. 259–281. [Google Scholar]
- 3.Edgar RS, Wood WB. Morphogenesis of bacteriophage T4 in extracts of mutant-infected cells. Proc Natl Acad Sci U S A. 1966;55:498–505. doi: 10.1073/pnas.55.3.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edgar RS, Lielausis I. Some steps in the assembly of bacteriophage T4. J Mol Biol. 1968;32:263–276. doi: 10.1016/0022-2836(68)90008-9. [DOI] [PubMed] [Google Scholar]
- 5.Kikuchi Y, King J. Genetic control of bacteriophage T4 baseplate morphogenesis. III. Formation of the central plug and overall assembly pathway. J Mol Biol. 1975;99:695–716. doi: 10.1016/s0022-2836(75)80180-x. [DOI] [PubMed] [Google Scholar]
- 6.King J. Bacteriophage T4 tail assembly: four steps in core formation. J Mol Biol. 1971;58:693–709. doi: 10.1016/0022-2836(71)90034-9. [DOI] [PubMed] [Google Scholar]
- 7.Katsura I, Kuhl PW. A regulator protein for the length determination of bacteriophage lambda tail. J Supramol Struct. 1974;2:239–253. doi: 10.1002/jss.400020217. [DOI] [PubMed] [Google Scholar]
- 8.Katsura I, Kuhl PW. Morphogenesis of the tail of bacteriophage lambda. III. Morphogenetic pathway. J Mol Biol. 1975;91:257–273. doi: 10.1016/0022-2836(75)90379-4. [DOI] [PubMed] [Google Scholar]
- 9.Katsura I, Tsugita A. Purification and characterization of the major protein and the terminator protein of the bacteriophage lambda tail. Virology. 1977;76:129–145. doi: 10.1016/0042-6822(77)90290-2. [DOI] [PubMed] [Google Scholar]
- 10.Katsura I. Tail assembly and injection. In: Hendrix R, Roberts J, Stahl F, Weisberg RA, editors. Lambda II. Cold Spring Harbor, N Y: Cold Spring Harbor Laboratory; 1983. pp. 331–346. [Google Scholar]
- 11.Katsura I, Hendrix RW. Length determination in bacteriophage lambda tails. Cell. 1984;39:691–698. doi: 10.1016/0092-8674(84)90476-8. [DOI] [PubMed] [Google Scholar]
- 12.Kostyuchenko VA, Chipman PR, Leiman PG, Arisaka F, Mesyanzhinov VV, Rossmann MG. The tail structure of bacteriophage T4 and its mechanism of contraction. Nat Struct Mol Biol. 2005;12:810–813. doi: 10.1038/nsmb975. [DOI] [PubMed] [Google Scholar]
- 13.Kostyuchenko VA, Leiman PG, Chipman PR, Kanamaru S, van Raaij MJ, Arisaka F, Mesyanzhinov VV, Rossmann MG. Three-dimensional structure of bacteriophage T4 baseplate. Nat Struct Biol. 2003;10:688–693. doi: 10.1038/nsb970. [DOI] [PubMed] [Google Scholar]
- 14.Leiman PG, Chipman PR, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell. 2004;118:419–429. doi: 10.1016/j.cell.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 15.Rossmann MG, Mesyanzhinov VV, Arisaka F, Leiman PG. The bacteriophage T4 DNA injection machine. Curr Opin Struct Biol. 2004;14:171–180. doi: 10.1016/j.sbi.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 16.Katsura I. Determination of bacteriophage lambda tail length by a protein ruler. Nature. 1987;327:73–75. doi: 10.1038/327073a0. [DOI] [PubMed] [Google Scholar]
- 17.Katsura I. Mechanism of length determination in bacteriophage lambda tails. Adv Biophys. 1990;26:1–18. doi: 10.1016/0065-227x(90)90004-d. [DOI] [PubMed] [Google Scholar]
- 18.Roessner CA, Ihler GM. Proteinase sensitivity of bacteriophage lambda tail proteins gpJ and gpH in complexes with the lambda receptor. J Bacteriol. 1984;157:165–170. doi: 10.1128/jb.157.1.165-170.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scandella D, Arber W. Phage lambda DNA injection into Escherichia coli pel- mutants is restored by mutations in phage genes V or H. Virology. 1976;69:206–215. doi: 10.1016/0042-6822(76)90207-5. [DOI] [PubMed] [Google Scholar]
- 20.Abuladze NK, Gingery M, Tsai J, Eiserling FA. Tail length determination in bacteriophage T4. Virology. 1994;199:301–310. doi: 10.1006/viro.1994.1128. [DOI] [PubMed] [Google Scholar]
- 21.Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA, Jacobs-Sera D, Falbo J, Gross J, Pannunzio NR, Brucker W, Kumar V, Kandasamy J, Keenan L, Bardarov S, Kriakov J, Lawrence JG, Jacobs WR, Jr, Hendrix RW, Hatfull GF. Origins of highly mosaic mycobacteriophage genomes. Cell. 2003;113:171–182. doi: 10.1016/s0092-8674(03)00233-2. [DOI] [PubMed] [Google Scholar]
- 22.Xu J, Hendrix RW, Duda RL. Conserved translational frameshift in dsDNA bacteriophage tail assembly genes. Mol Cell. 2004;16:11–21. doi: 10.1016/j.molcel.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 23.Levin ME, Hendrix RW, Casjens SR. A programmed translational frameshift is required for the synthesis of a bacteriophage lambda tail assembly protein. J Mol Biol. 1993;234:124–139. doi: 10.1006/jmbi.1993.1568. [DOI] [PubMed] [Google Scholar]
- 24.Xu J, Hendrix RW, Duda RL. A balanced ratio of proteins from gene G and frameshift-extended gene GT is required for phage lambda tail assembly. J Mol Biol. 2013 doi: 10.1016/j.jmb.2013.07.002. (accepted, under revision) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 26.Katsura I. Structure and function of the major tail protein of bacteriophage lambda. Mutants having small major tail protein molecules in their virion. J Mol Biol. 1981;146:493–512. doi: 10.1016/0022-2836(81)90044-9. [DOI] [PubMed] [Google Scholar]
- 27.Katsura I, Kuhl PW. Morphogenesis of the tail of bacteriophage lambda. II. In vitro formation and properties of phage particles with extra long tails. Virology. 1975;63:238–251. doi: 10.1016/0042-6822(75)90388-8. [DOI] [PubMed] [Google Scholar]
- 28.Weigle J. Assembly of phage lambda in vitro. Proc Natl Acad Sci U S A. 1966;55:1462–1466. doi: 10.1073/pnas.55.6.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katsura I. Morphogenesis of bacteriophage lambda tail. Polymorphism in the assembly of the major tail protein. J Mol Biol. 1976;107:307–326. doi: 10.1016/s0022-2836(76)80007-1. [DOI] [PubMed] [Google Scholar]
- 30.King J, Laemmli UK. Polypeptides of the tail fibres of bacteriophage T4. J Mol Biol. 1971;62:465–477. doi: 10.1016/0022-2836(71)90148-3. [DOI] [PubMed] [Google Scholar]
- 31.Ward S, Dickson RC. Assembly of bacteriophage T4 tail fibers. 3. Genetic control of the major tail fiber polypeptides. J Mol Biol. 1971;62:479–492. doi: 10.1016/0022-2836(71)90149-5. [DOI] [PubMed] [Google Scholar]
- 32.Bishop RJ, Wood WB. Genetic analysis of T4 tail fiber assembly. I. A gene 37 mutation that allows bypass of gene 38 function. Virology. 1976;72:244–254. doi: 10.1016/0042-6822(76)90327-5. [DOI] [PubMed] [Google Scholar]
- 33.Cornelis GR, Agrain C, Sorg I. Length control of extended protein structures in bacteria and bacteriophages. Curr Opin Microbiol. 2006;9:201–206. doi: 10.1016/j.mib.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 34.Journet L, Agrain C, Broz P, Cornelis GR. The needle length of bacterial injectisomes is determined by a molecular ruler. Science. 2003;302:1757–1760. doi: 10.1126/science.1091422. [DOI] [PubMed] [Google Scholar]
- 35.Shibata S, Takahashi N, Chevance FF, Karlinsey JE, Hughes KT, Aizawa S. FliK regulates flagellar hook length as an internal ruler. Mol Microbiol. 2007;64:1404–1415. doi: 10.1111/j.1365-2958.2007.05750.x. [DOI] [PubMed] [Google Scholar]
- 36.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, Pukatzki S, Burley SK, Almo SC, Mekalanos JJ. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci U S A. 2009;106:4154–4159. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hendrix RW, Casjens SR. Protein cleavage in bacteriophage lambda tail assembly. Virology. 1974;61:156–159. doi: 10.1016/0042-6822(74)90250-5. [DOI] [PubMed] [Google Scholar]
- 39.Walker JE, Auffret AD, Carne A, Gurnett A, Hanisch P, Hill D, Saraste M. Solid-phase sequence analysis of polypeptides eluted from polyacrylamide gels. An aid to interpretation of DNA sequences exemplified by the Escherichia coli unc operon and bacteriophage lambda. Eur J Biochem. 1982;123:253–260. doi: 10.1111/j.1432-1033.1982.tb19761.x. [DOI] [PubMed] [Google Scholar]
- 40.Boulanger P, Jacquot P, Plancon L, Chami M, Engel A, Parquet C, Herbeuval C, Letellier L. Phage T5 straight tail fiber is a multifunctional protein acting as a tape measure and carrying fusogenic and muralytic activities. J Biol Chem. 2008;283:13556–13564. doi: 10.1074/jbc.M800052200. [DOI] [PubMed] [Google Scholar]
- 41.Piuri M, Hatfull GF. A peptidoglycan hydrolase motif within the mycobacteriophage TM4 tape measure protein promotes efficient infection of stationary phase cells. Mol Microbiol. 2006;62:1569–1585. doi: 10.1111/j.1365-2958.2006.05473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tam W, Pell LG, Bone D, Tsai A, Dai X, Edwards AE, Hendrix RW, Maxwell KL, Davidson AR. Tail tip proteins related to bacteriophage <lambda> gpL coordinate an iron-sulphur cluster. J Mol Biol. 2013 doi: 10.1016/j.jmb.2013.03.032. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2 edit. Cold Spring Harbor, NY: CSHL Press; 1989. [Google Scholar]
- 44.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 45.Duda RL, Martincic K, Hendrix RW. Genetic basis of bacteriophage HK97 prohead assembly. J Mol Biol. 1995;247:636–647. doi: 10.1006/jmbi.1994.0169. [DOI] [PubMed] [Google Scholar]