Abstract
Processing bodies (P-bodies) are cytoplasmatic mRNP granules containing non-translating mRNAs and proteins from the mRNA decay and silencing machineries. The mechanism of P-body assembly has been typically addressed by depleting P-body components. Here we apply a complementary approach and establish an automated cell-based assay platform to screen for molecules affecting P-body assembly. From a unique library of compounds derived from myxobacteria, 30 specifically inhibited P-body assembly. Gephyronic acid A (GA), a eukaryotic protein synthesis inhibitor, showed the strongest effect. GA also inhibited, under stress conditions, phosphorylation of eIF2α and stress granule formation. Other hits uncovered interesting novel links between P-body assembly, lipid metabolism, and internal organelle physiology. The obtained results provide a chemical toolbox to manipulate P-body assembly and function.
Keywords: P-body assembly, eIF2α, gephyronic acid A, inhibitors, myxobacterial metabolites, processing bodies, stress granules
Introduction
Processing bodies (P-bodies) are discrete mRNP granules found in the cytoplasm of eukaryotic cells (for a review, see ref. 1 and references within). They are conserved from yeast to humans and contain translationally repressed mRNAs together with multiple proteins from the mRNA decay pathway. In metazoans, P-bodies also contain proteins from the miRNA silencing machinery.1 Two main pieces of evidence indicate that P-bodies are highly dynamic structures. First, their size and number are proportional to the pool of non-translating mRNAs. Thus, conditions that increase translation repression such as stress enhance the number and size of P-bodies.2 Second, P-body components rapidly cycle in and out of P-bodies indicating that there is a constant exchange with the cytoplasm where all P-body components are also diffusely present.2-4 Once in P-bodies, mRNAs can be degraded or stored for a later return into translation;1 however, how either fate is decided remains unknown. While the biological significance of P-bodies in the physiology of the cell remains an unresolved issue, the conserved ability of eukaryotic cells to assemble mRNPs into P-bodies strongly suggests that additional important functions remain to be discovered such as their recently described roles in long-term survival of yeast cells during stationary phase.5
The mechanistic details of P-body assembly are not well understood. The most accepted model proposes that it requires translationally repressed, ribosome-free mRNAs and some core P-body proteins that promote aggregation by specific protein–protein interaction domains.1 Consequently, conditions that inhibit the interaction of mRNAs with ribosomes increase P-body formation while P-bodies rapidly disappear when mRNAs are trapped in complexes with polysomes or when core P-body proteins are depleted or post-translationally modified.1 In human cells, the decapping activator DDX6 (also referred as Rck/p54) plays a key role in P-body assembly since its depletion abrogates P-body formation, even in the presence of stress conditions.6 Interestingly, DDX6 also has fundamental roles under pathological conditions. It is overexpressed in several cancers7 and utilized by several members of the positive-strand RNA virus group, including the hepatitis C virus (HCV),8 West Nile Virus, and Dengue Virus to promote their expansion. Although the relationship between cancer and P-bodies remains to be explored, a growing body of evidence supports a complex interplay between P-bodies and viruses. Besides DDX6, multiple P-body components have been shown to promote or inhibit viral infections.9 In turn, viral infections modulate P-body formation. While some viruses inhibit P-body assembly, others alter their composition at early times after infection.10-12 Thus, the elucidation of the detailed mechanisms that lead to P-body assembly under normal or pathological conditions is fundamental to understanding the molecular events controlling key cellular and viral mRNA transitions.
Traditionally, studies on P-body assembly and structure have involved the use of yeast deletion mutant libraries or, for mammalian cells, depletion of one or more P-body components by siRNA technologies. While these tools are of great help in understanding mRNA regulation, a complementary approach that identifies and uses small chemicals might unveil novel and unpredicted P-body assembly features since depletion of a protein might not have a similar effect as to inactivate it with a drug.13 Here, we established an automated cell-based screening platform to identify small molecules affecting P-body assembly. This platform was used to screen a unique library of natural products derived from myxobacteria.14 These soil bacteria are one of the top producers of microbial secondary metabolites. Importantly, the myxobacteria metabolome targets proteins that are not usually targeted by metabolites from other microorganisms.15 Examples are novel inhibitors of eukaryotic protein synthesis and microtubule assembly. Screening campaigns have already revealed that a large proportion of the myxobacteria secondary metabolites have activities against non-infectious and infectious human diseases. For example, argyrin, a cyclic peptide derived from the myxobacteria Archangium gephyra, has been shown to act as a potent anti-tumor drug through inhibiting the proteasome.16 In addition, a recent screen identified multiple myxobacterial molecules that target HCV infection at different steps of its lifecycle.17
Here we described a screening assay for inhibitors of P-body assembly. As a proof of concept, a myxobacterial library containing 154 structurally defined myxobacteria metabolites was tested. Almost 20% of the compounds showed significant activities against P-body formation. Most interesting compounds include modifiers of translation, cytoskeleton, lipid metabolism, and organelle physiology. Together, our study uncovered novel features of P-body assembly and provides a set of interesting natural products for manipulating and studying these puzzling intracellular foci.
Results
To study the effect of small compounds on P-body assembly, we established an automated cell-based screening assay that monitors by indirect immunofluorescence the localization of the core P-body component DDX6.1 Under normal cellular conditions, DDX6 locates both diffusely in the cytoplasm and in discrete cytoplasmic foci that correspond to P-bodies. Loss of distinct foci signals correspond to P-body disruption.6 Molecules that interfere with DDX6 localization in P-bodies might be affecting different processes, from changes in mRNA status to directly affecting DDX6 or other core P-body components. This broad range of effects might be very valuable to uncover unpredicted cellular functions involved in P-body assembly. In addition, overlapping the obtained results with previous screens using the same set of molecules might help to identify the molecular mechanisms involved in their effect on P-bodies.
The screen for P-body disruption was performed with the human lung carcinoma cell line A-549.18 These cells have been used in screening campaigns to analyze the effect of natural products on cellular morphology and to elucidate their mode of action and targets. The screening protocol was defined as follows. Test compounds and vehicle controls (MeOH or DMSO) were added in duplicate to 384-well plates containing A-549 cells with the Evolution P3 Pipetting Platform (PerkinElmer). Twenty-four hours after addition of compounds, cells were fixed and stained with a specific antibody against DDX6 and with DAPI to count nuclei. Relative nuclei numbers were used as a measure of compound-related toxicity. Immunofluorescent foci and nuclei were analyzed using an automated microscopy, which was set to take four images per well, thus generating around 1500 images per 384-well plate (see Materials and Methods).
Of the 154 molecules tested, 102 (≤ 66% of the total library) did not show any significant effect on P-body assembly, while 22 (≤ 14%) reduced cell viability by ≥ 60% compared with the DMSO and/or MeOH controls. These substances were no longer considered for further analysis. From the remaining 30 compounds, 21 (≤ 14% of the library) reduced P-body assembly by at least 50% with low toxic effects (less than 30% reduction in cell viability). The other nine (≤ 6% of the library) reduced P-body assembly by more than 70% with less than 10% reduction in cell viability (Table 1). Thus, almost 20% of the compounds showed significant activities against P-body formation. However, no compound was found to significantly increase P-body numbers. Although most chemical classes and biological activities of the myxobacterial secondary metabolome are known, the specific mode of action of many compounds has yet to be described.14,19 Figure 1 shows the distribution of chemical types and activities for all the 30 molecules found here to significantly affect P-body assembly. The most represented class of metabolites (14 hits) corresponds to macrolactones, non-ribosomal polypeptides known as polyketides (Fig. 1A and Table 1). Macrolactones are also the most abundant secondary metabolites produced by myxobacteria.19 The availability of different macrolactone structures is controlled by alternative gene expression that allows an effective and selective antibiotic synthesis. Other hit compounds belong to the group of peptides and lactones (three hits each). These types of compounds are also commonly produced by other microorganisms.
Table 1. Summary of myxobacterial compounds reducing P-body formation by 50% with less than 30% reduction in cell viability. Compounds are listed by inhibitory strength from high to low.
| Name | % Rel. P-body inhibition/cell | % Rel. cell viability | Structural class | Activity |
|---|---|---|---|---|
| Gephyronic Acid | 91.3 ± 3.7 | 73.2 ± 3.1 | Aliphatic acid | Translation inhibitor20 |
| Cerebrosid 751 | 87.6 ± 4.5 | 99.9 ± 6.6 | glycosphingolipid | n.d. |
| Aurachin D | 87.5 ± 1.2 | 88.0 ± 6.5 | Quinolone | Anti-fungal19 |
| Methyl-Chivosazol | 84.4 ± 6.5 | 73.5 ± 1.3 | Macrolactone | Actin polimerization inhibitor19 |
| Tubulysin B | 80.3 ± 3.8 | 89.5 ± 5.2 | Peptide | Tubulin polymerization inhibitor19 |
| Kulkenon | 79.8 ± 6.0 | 93.0 ± 9.3 | Macrolactone | n.d. |
| Spirangien B | 78.1 ± 7.4 | 85.7 ± 2.1 | Spiroketal | IL-8 inhibitor45 |
| Thuggacin B | 77.4 ± 2.6 | 95.7 ± 1.7 | Macrolactone | Anti-bacterial19 |
| Myxalamid B | 76.0 ± 4.4 | 80.1 ± 3.7 | Polyene | Anti-fungal19 |
| Cruentaren A | 73.6 ± 7.8 | 93.0 ± 2.5 | Benzolactone | F-ATPase inhibitor46 |
| Soraphen F | 73.1 ± 5.8 | 107.8 ± 3.0 | Macrolactone | Acetil-CoA carboxylase inhibitor26 |
| Disorazol B1 | 69.4 ± 5.4 | 104.6 ± 8.0 | Macrolactone | Tubulin polymerization inhibitor19,47 |
| Vioprolid D | 69.3 ± 2.9 | 68.0 ± 0.3 | Cyclic peptide | Anti-fungal19 |
| Crocapeptin A | 63.8 ± 5.1 | 72.2 ± 0.1 | Peptolide | n.d. |
| Soraphen B1 | 63.0 ± 7.8 | 98.8 ± 0.2 | Macrolactone | Acetil-CoA carboxylase inhibitor26 |
| Soraphen E1 | 62.2 ± 9.3 | 93.1 ± 5.5 | Macrolactone | Acetil-CoA carboxylase inhibitor26 |
| Soraphen B2 α | 60.8 ± 5.5 | 74.6 ± 3.3 | Macrolactone | Acetil-CoA carboxylase inhibitor26 |
| Jerangolid A | 59.9 ± 8.8 | 107.8 ± 7.4 | Lactone | Anti-fungal19 |
| Leupyrrin A1 | 59.6 ± 7.1 | 84.6 ± 4.5 | Macrolactone | Translation inhibitor19 |
| Archazolid A | 58.4 ± 9.4 | 80.3 ± 2.4 | Macrolactone | V-ATPase inhibitor48 |
| Crocacin C | 58.3 ± 5.9 | 74.0 ± 2.8 | Peptide | Anti-fungal19 |
| Pellasoren | 58.0 ± 13.1 | 84.4 ± 4.9 | Lactone | Anti-cancer. Cytotoxic49 |
| Carolacton | 57.3 ± 10.7 | 91.2 ± 3.4 | Macrolactone | Anti-bacterial biofilm50 |
| Maltepolid B | 56.5 ± 7.6 | 91.8 ± 3.7 | Macrolactone | n.d. |
| Pyrrolnitrin | 56.5 ± 6.9 | 95.4 ± 6.6 | Phenylpyrole | Anti-fungal, Anti-bacterial19 |
| Ambruticin VS4 | 56.2 ± 9.5 | 102.3 ± 2.7 | Lactone | Anti-fungal19 |
| Trichangion | 55.3 ± 7.0 | 86.2 ± 4.4 | Phenothiazine | n.d. |
| Tubulysin I | 54.9 ± 12.8 | 92.0 ± 2.2 | Peptide | Tubulin polymerization inhibitor19 |
| Soraphen A4 α | 52.8 ± 9.0 | 103.2 ± 9.4 | Macrolactone | Acetil-CoA carboxylase inhibitor26 |
| Maltepolid C | 50.5 ± 6.8 | 93.6 ± 2.5 | Macrolactone | n.d. |
n.d., not described; ± , SEM.
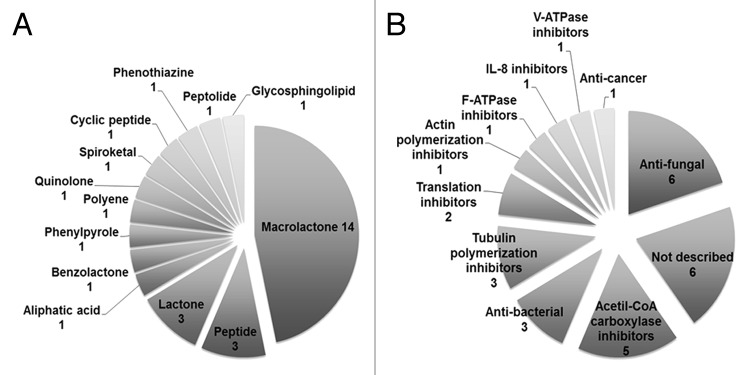
Figure 1. Distribution of (A) chemical classes and (B) known activities for all 30 compounds found to effectively reduce P-body formation with no more than 30% reduction in cell viability.
The myxobacterial secondary metabolites in our screen display a broad range of biological activities.19 Nine of the P-body hits have been reported to have antifungal and/or antibacterial activities (Fig. 1B and Table 1). The predominance of antimicrobials and antifungals as hit compounds is not surprising given the fact that these constitute almost half of the fraction of known biological activities of the myxobacteria secondary metabolome.14 Interestingly, we found five derivatives of soraphen, an acetyl-CoA carboxylase inhibitor first described as antifungal, in our hits (Fig. 1B and Table 1). This group of macrolactones carries an uncommon phenyl ring making it a unique class of polyketides.19 Other P-body hits were known inhibitors of the cytoskeleton (four hits), translation inhibitors (two hits), and compounds inhibiting ATPases (two hits)19 (Fig. 1B and Table 1). Finally, six of the hit molecules: cerebrosid 751, crocapeptin A, trichangion, kulkenon, and the maltepolides B and C (Fig. 1B and Table 1), have no specific targets or mode of action described so far, thus making them an intriguing group of compounds for additional studies.
Gephyronic acid A (GA), a polyketide that inhibits eukaryotic protein synthesis,20 showed the strongest activity against P-body formation (around 90% inhibition) with acceptable cell viability (≥ 70%) in the primary screen (Table 1). The mechanism by which GA inhibits mRNA translation is still under investigation. Translation blockade can occur either at the initiation, elongation, or termination phase, with different effects on P-bodies.1 It has been shown that inhibiting the ribosomes–mRNA interaction with puromycin enhances P-body assembly, while blocking translation elongation with cycloheximide, and thus trapping the ribosomes on mRNAs, prevents P-body assembly.1 In order to have an initial hint of the mode of action of GA and to validate its effect in other cell line, we compared the effect of GA to that of puromycin and cycloheximide in HeLa cells (Fig. 2A), a cell line that has been extensively used in the P-body research field. Cells were treated with drug or vehicle controls and immunostained for DDX6. As expected, puromycin increased the number of P-bodies. Similar to the effect of cycloheximide, GA treatment decreased P-body abundance (Fig. 2A). These results suggest that GA might affect P-body formation by stalling ribosomes on the mRNA. Alternatively, they might also reflect effects at early steps of translation initiation such as binding of ribosomal subunits or initiation factors, since it is not yet well established what translation events need to be stalled for altering P-body assembly.
Figure 2. The translation inhibitor gephyronic acid A strongly reduces P-body formation in HeLa cells. (A) Cells were treated with either 1 µM of gephyronic acid A (GA) for 3.5 h, 25 µg/ml of cycloheximide for 15 min, 100 µg/ml of puromycin for 1 h, or mock-treated. After the indicated time, cells were immunostained with an antibody for DDX6 (green) and the nuclei was visualized with DAPI (blue). The number of DDX6-containing P-bodies from at least 100 cells per condition was quantified and related to those detected in mock-treated cells. Representative images for each condition are shown. Numbers below the panel show the median and the standard error of the mean of the P-body number per cell relative to the untreated control (set to 100%). (B) HeLa cells were incubated for 1 h (upper graph) or 3.5 h (lower graph) with 0, 50, 100, 500, or 1000 nM of GA. Cells were immunostained to quantify DDX6- and Dcp1-containing P-bodies in at least 100 cells. The relative abundance of DDX6- and Dcp1-containing P-bodies vs. log GA concentration is shown related to mock-treated cells. Right graphics indicate cell viability for the different concentrations of GA and times of incubation using an ATP assay that measures intracellular ATP levels. Error bars indicate standard error of the mean. (C) For both time-points, expression levels of DDX6 and Dcp1 were tested by western blot under the higher concentrations of GA. β-actin was used as a control for protein loading.
To further characterize the effect of GA on P-bodies, we performed short-term dose-response experiments in HeLa cells (Fig. 2B). Cells were treated for 1 h or 3.5 h with increasing concentrations of GA and immunostained for DDX6 and Dcp1, another core P-body component. Cell viability was followed in parallel by analyzing intracellular ATP levels. Treatment with GA for 1 h resulted in a moderate reduction of both DDX6- and Dcp1-containing P-bodies at concentrations of 500 nM and 1 µM (Fig. 2B, top). Extending treatment to 3.5 h resulted in a steeper decrease of DDX6- containing P-bodies and a dramatic reduction of Dcp1-containing ones, even at the lowest concentration relative to mock-treated controls (Fig. 2B, bottom). The difference in the reduction levels between Dcp1- and DDX6-containing P-bodies is not unexpected since FRAP studies demonstrate that P-body components exit P-bodies with different kinetics.3 In any case, cell viability was decreased. Importantly, the effect of GA on P-body formation was not due to a reduction of DDX6 or Dcp1 protein expression levels (Fig. 2C). Of note is that Dcp1 migrates as two bands being the upper one predominant under 1000 nM GA treatment. Interestingly, this upper band corresponds to a phosphorylated Dcp1 form that correlates with a decreased localization in P-bodies.21,22 Together, these data indicate that the blockade of mRNA translation by GA directly affects the assembly of P-bodies.
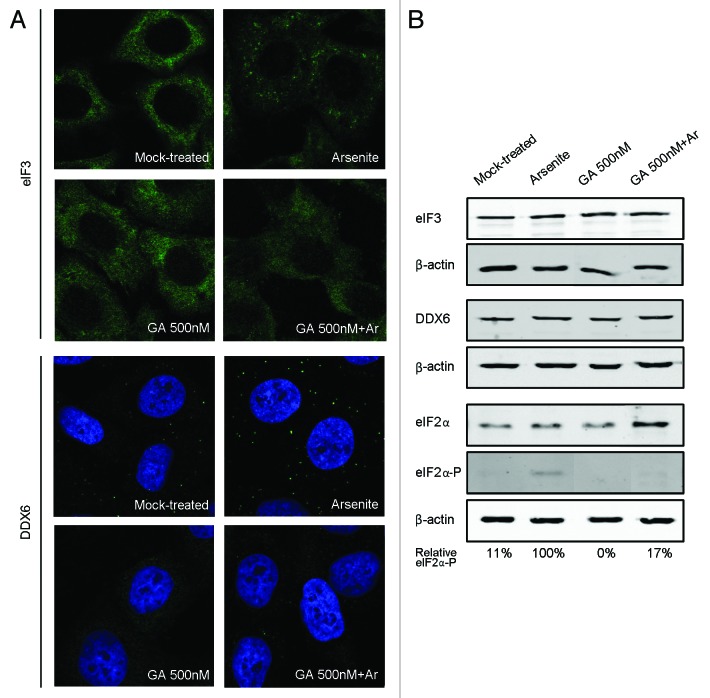
Stress granules (SGs) are another kind of dynamic cytosolic mRNP granules closely related to P-bodies.1,23 Like P-bodies, they contain non-translating mRNAs; however, SGs also contain translation initiation factors and the 40S ribosomal subunit, suggesting that they are aggregates of mRNPs stalled in the process of translation initiation. In addition, SGs are solely detected under stress conditions. The observation that SGs localize in close proximity to P-bodies, together with the fact that both granules share some components, demonstrate an intimate link between them. To investigate whether GA had also an effect on SGs assembly, we monitored by immunofluorescence the localization of the eukaryotic translation initiation factor eIF3, a well-established SGs marker (Fig. 3A, upper panels). As in the mock control, no eIF3-containing SGs were detected when GA was added up to concentrations of 500 nM. As expected, arsenite treatment induced the formation of SGs in these cells. However, no SG formation was observed when cells were previously treated with GA and then with arsenite. This effect was not related to changes in eIF3 expression levels (Fig. 3B). The phosphorylation of eIF2α by cellular kinases in response to stress is one of the most characterized pathways of SGs induction.24 To assess whether GA could interfere with this pathway we monitored eIF2α and eIF2α phosphorylation levels under the conditions mentioned above (Fig. 3B). As expected, arsenite treatment induced the phosphorylation of eIF2α while in cells treated only with GA eIF2α remained non-phosphorylated. Importantly, in those cells treated with both compounds, phosphorylation of eIF2α was dramatically inhibited.
Figure 3. GA prevents stress granules formation and inhibits P-body assembly when stress conditions are applied. (A) HeLa cells were either mock-treated, treated with 500 nM of gephyronic acid A (GA) for 3.5 h, treated with 0.1 mM of arsenite (Ar) for 30 min, or treated with GA for 3.5 h adding plus Ar during the last 30 min. After the indicated times, cells were immunostained with an antibody for the stress granule component eIF3 (upper panel) or the P-body component DDX6 (lower panel). (B) Protein levels of eIF3, DDX6, eIF2α, and eIF2α-phosphorylated were tested by western blot using β-actin as a control for protein loading. Numbers below the panel indicate the percentage of eIF2α-phosphorylated expression levels relative to those detected in arsenite-treated cells.
It is well established that P-body size and number increase in response to stress conditions.1,2 To test whether GA treatment also affects P-body formation under stress, cells were previously treated with GA and then with arsenite (Fig. 3A, lower panels). As previously described, the number and size of DDX6-containing P-bodies increased in arsenite-treated cells when compare with mock-treated cells. However, when cells were previously treated with GA, P-body assembly was inhibited, even in the presence of arsenite, without affecting DDX6 expression levels (Fig. 3B). Together, these results indicate that GA prevents SG formation and inhibits P-body assembly when stress conditions are applied.
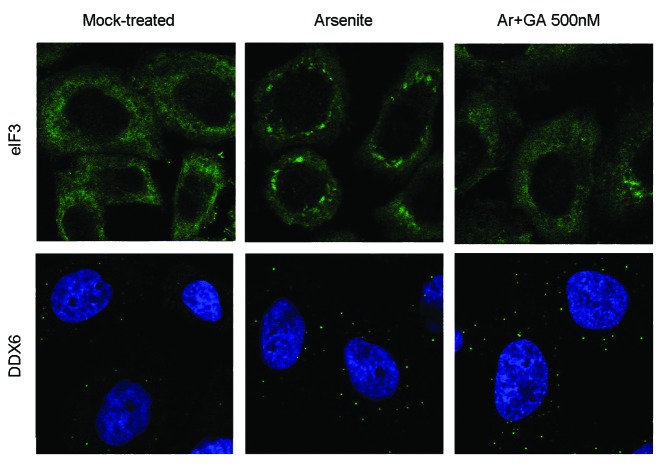
To determine whether GA might also disassemble SGs and P-bodies once they are already formed in response to stress conditions, cells were mock-treated, treated with arsenite for 1.5 h, or treated with arsenite for 1.5 h adding GA for the last hour of incubation. Immunofluorescence detection of eIF3 showed that the addition of GA in cells under arsenite treatment promoted SG disassembly (Fig. 4, upper panel). Under these conditions, only 23% of cells displayed SGs while in solely arsenite-treated cells SGs were present in more than 90% of cells. In contrast, treatment with GA had no effect on the number or size of P-bodies (Fig. 4, lower panel). The different effect of GA on these two closely related granules might reflect important differences in their disassembly process.
Figure 4. In stressed cells, GA promotes disassembly of SGs but not of P-bodies. HeLa cells were either mock-treated, treated with 0.1 mM of arsenite (Ar) for 1.5 h, or treated with Ar for 1.5 h adding GA 500 nM during the last hour. After the indicated times, cells were immunostained with an antibody against the stress granule component eIF3 (upper panel) or the P-body marker DDX6 (lower panel).
Discussion
P-bodies are cytoplasmic aggregates of non-translating mRNPs whose protein components play key roles in the modulation of post-transcriptional gene expression. The detailed mode of P-body assembly and function remain unclear. Previous work addressed these fundamental questions by selectively depleting P-body components. Here, we present an alternative approach and establish a robust cell-based platform to screen for molecules affecting P-body assembly. The platform was used to test the effect of a unique library of natural products derived from myxobacteria that display diverse activities,19 20% of which showed significant effects on P-body formation and established interesting links with unanticipated cellular processes.
Five derivatives of soraphen were found to inhibit P-body assembly. Early studies showed that the effect of soraphen could be reversed by adding fatty acids to the culture medium of cells.19 Later findings led to the identification of the acetyl-CoA carboxylase as the target of soraphen.25,26 This enzyme catalyzes the synthesis of cholesterol, fatty acids, and complex lipids,27 suggesting a link between P-body assembly and lipid metabolism. Interestingly, it has been shown that certain features of the mRNA metabolism are regulated in response to lipid signaling,28 and that under heat-stress the formation of P-bodies requires sphingolipid synthesis.29
The finding that the tubulin polymerization inhibitors tubulysin and disorazol, and the actin polymerization inhibitor methyl-chivosazol, inhibited P-body formation was somewhat surprising, since it has been suggested that disturbance of the cellular microtubule network leads to an increase in P-body foci number and size rather than to a decrease.3,30 However, it has been also described that retrograde dynein and anteretrograde kinesin motors exert opposing effects on the assembly of P-bodies and SGs, indicating that a balance between these opposing movements govern foci formation and disassembly.31 Thus, together these observations, support a complex functional interplay between P-bodies and the cytoskeleton, which mechanisms might be difficult to elucidate due to the putative pleiotropic effects that may arise by the manipulation of the cytoskeleton. The effect of cruentaren A, an F-ATPase inhibitor, and archazolid A, an inhibitor of vacuolar-type ATPase (V-ATPase), was interesting. Both ATPases share a similar structure and mechanisms of action, however they display distinct functions.32 The F-ATPase localizes in mitochondria and uses a proton gradient to drive ATP synthesis. Interestingly, recent studies have revealed physical and functional relationships between P-bodies, mitochondria, and RNAi.33,34 In contrast to the F-ATPase, the V-ATPase localizes in a multitude of different membranes translocating protons from the cytoplasm into intracellular compartments and across the plasma membrane. It plays crucial roles in the function and trafficking of internal organelles such as vacuoles, endosomes, or lysosomes. Inhibitors of this multi-subunit key player in cell physiology have different effects that are under study. Concretely, archazolid treatment leads to the formation of vacuoles in the endoplasmic reticulum and the acidification of lysosomes.35 More recent work linked archalozid treatment with cellular stress responses, including autophagy.36 Together, these two latter hits show an interaction between P-body assembly and internal organelle physiology. Indeed, exciting new findings from a genetic screen in yeast to identify genes affecting P-body and SG formation detected five subunits of the V-ATPase complex and also demonstrated that P-body and SGs are targeted to vacuoles by autophagy.37
From all identified hits, GA showed the most potent effect against P-body assembly. GA is a secondary metabolite from the myxobacterium Archangium gephyra with a characterized chemical structure.20 It inhibits the proliferation of mammalian cells and fungi by acting as a translation inhibitor; however, prokaryotes are not affected. Our results now show that at concentrations in the nanomolar range GA inhibits eIF2α-phosphorylation as well as SGs formation under stress conditions. Since eIF2α-phosphorylation is one of the most important pathways for SG induction in response to stress conditions, these results open the possibility that the effect of GA on both P-bodies and SGs assembly might be occurring through a direct or indirect effect on the translation initiation factor eIF2α. This would trap the mRNA into dysfunctional preinitiation complexes that might affect both P-body and SG formation. In fact, other conditions affecting formation of proper initiation complexes such as depletion of eIF3 or reduction in 60S subunit joining38,39 have also been shown to inhibit SG formation. Intriguingly, when GA was added after inducing stress, SGs were disassembled but not P-bodies. This indicates that the window within which translation needs to be repressed in order to alter P-body or SG assembly still needs to be precisely defined.
Although P-body components are known to play a pivotal role in eukaryotic mRNA metabolism, the function of P-body granules remains unclear. The diverse mode of action and targets of the myxobacterial products found here to influence P-body assembly, including cytoskeleton components, lipid metabolism, ATPases, and protein synthesis, could be further used to investigate and map unknown functional aspects of mammalian P-bodies. In addition, these compounds might also be relevant to other related cytoplasmic mRNP granules, including, for example, those found in germ cells and neuronal cells that play important roles in the localization and control of mRNAs in embryos.40-43 Further characterization of the identified hits may unravel novel functional aspects of the mammalian mRNP machinery.
Materials and Methods
Cell lines and antibodies
The A-549 (DSMZ-ACC107) is a human lung carcinoma cell line.18 A-549 and HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS) at 37 °C and 5% CO2. The antibodies used in this work included the following: rabbit anti-DDX6 (MBL Co. LTD, PD009), mouse anti-Dcp1a (Abnova, H00055802-M06), mouse anti-β-actin (Sigma-Aldrich, A5441), rabbit anti-eIF2α (Cell Signaling, 9722), rabbit anti-eIF2α-P (Cell Signaling, 9721), Alexa 548 anti-rabbit (Invitrogen, A11036), Alexa 647 anti-mouse (Invitrogen, A21236), IRDYE680 anti-mouse (Li-Cor, 926-68070), and IRDYE800 anti-rabbit (Li-Cor, 926-32211).
Compound library
The library of 154 myxobacteria secondary metabolites used for the screening belongs to a collection of natural compounds isolated at the Helmholtz Centre for Infection Research.44 Compounds of > 95% purity as measured by LC-MS were provided in 96-well screening plates in a concentration of 1 mM in DMSO or MeOH.
Screening approach and data analysis
For primary screening, A-549 cells were seeded in 384-well plates at a density of 5 × 103 cells per well in 30 µL of culture medium and incubated overnight at 37 °C and 5% CO2. After incubation, 60–70 nL of the test compounds and the DMSO and/or MeOH controls from the 96-well screening plates were added to the cells in the 384-well plates with the Evolution P3 Pipetting Platform (PerkinElmer). Final concentration of compounds was 2 µM with around 0.2% of DMSO or MeOH in all cases. Twenty-four hours after addition of compounds, cells were washed twice with PBS and fixed for 30 min with a solution of 4% PFA in PBS. Cells were then permeabilized with a solution of 0.1% Triton-X100 in PBS for 5 min and blocked with a solution of 10% FCS in PBS for 30 min at room temperature (RT). Primary rabbit antibody against DDX6 (MBL Co. LTD, PD009) was added to the cells in a ratio of 1:2000 in PBS with 10% FCS and incubated for 1 h at RT. After washing twice with PBS, a secondary anti-rabbit Alexa 488 antibody (Invitrogen, A11008) was added to the cells and incubated for 45 min at RT. After another washing step with PBS, 1 µg/mL of 4′, 6-diamidino-2-phenylindole (DAPI) (Invitrogen, D1306) was added and incubated for 10 min. Twenty microliters of PBS per well was added to the cells. Plates were imaged for immunofluorescence signals with the ImageXpress Micro Widefield High Content Screening System (Molecular Devices, LLC) as follows: four images per well were acquired with a distance of 600 µm in the X-axes and 800 µm in the Y-axes between images. The automated microscope adaptive background correction was set so that P-body foci were considered positive between a width of 2 and 6 pixels and with an intensity of 500 gray levels above well background. Nuclear staining was set at 16–47 pixels wide and 200 gray levels above well background. When indicated, cell viability was measured by the Cell Titer Glo assay (Promega, G7570) or by normalizing nuclei counts of treated cells to the nuclei count of the MeOH or DMSO controls. Screening data from around 3000 images was analyzed with the MetaXpress® Software (Molecular Devices, LLC). The parameters used for analysis were: foci count (P-bodies) and nuclei count (cell count) per image per well. To correct for errors in site imaging values from the four images of each well were marked as outliers by applying a Grubbs Test provided by the GraphPad Prism software (P > 0.05, critical Z value 1.48). Outliers were discarded from the analysis. Final values were averaged and plotted as the % ratio between P-body foci and cell count relative to the DMSO or MeOH controls. Errors are given as ± SEM.
Indirect immunofluorescence and confocal imaging
Immunofluorescence analysis was performed as previously described.12 Nuclei were labeled with DAPI (Invitrogen, D1306). Cells were washed with PBS and mounted in Mowiol (Sigma-Aldrich, 324590). Images were acquired on a Leica SP2 confocal microscope (Leica). At least 100 cells were analyzed under each condition. In the particular case of P-bodies, the number of these granules was quantified with the ImageJ 1.45J software (National Institutes of Health, NIH) by applying median and background filters, and setting a threshold mask with “RenjyEntropy Thresholding.” The particles counted were selected by pixel size between 1–100 and circularity of 0.85. Under this set up conditions every 8 pixels analyzed corresponded to 1 µm. The final values for P-body number per cell were obtained from relating the number of P-bodies of each image to the number of nuclei present.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the Spanish Ministerio de Ciencia e Innovación (grants BFU2010-20803 and SAF2020-21336) and Deutsche Forschungsgemeinschaft SA 356/7-1.
Glossary
Abbreviations:
- P-bodies
processing bodies
- SGs
stress granules
- GA
gephyronic acid A
- mRNP
messenger ribonucleoprotein particles
- FRAP
fluorescence recovery after photobleaching
- eIF2α
eukaryotic initiation factor 2α
- eIF3
eukaryotic initiation factor 3
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/26851
References
- 1.Decker CJ, Parker R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol. 2012;4:a012286. doi: 10.1101/cshperspect.a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–84. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aizer A, Brody Y, Ler LW, Sonenberg N, Singer RH, Shav-Tal Y. The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Mol Biol Cell. 2008;19:4154–66. doi: 10.1091/mbc.E08-05-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Lührmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–27. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramachandran V, Shah KH, Herman PK. The cAMP-dependent protein kinase signaling pathway is a key regulator of P body foci formation. Mol Cell. 2011;43:973–81. doi: 10.1016/j.molcel.2011.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Serman A, Le Roy F, Aigueperse C, Kress M, Dautry F, Weil D. GW body disassembly triggered by siRNAs independently of their silencing activity. Nucleic Acids Res. 2007;35:4715–27. doi: 10.1093/nar/gkm491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuller-Pace FV. DEAD box RNA helicase functions in cancer. RNA Biol. 2013;10:121–32. doi: 10.4161/rna.23312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheller N, Mina LB, Galão RP, Chari A, Giménez-Barcons M, Noueiry A, Fischer U, Meyerhans A, Díez J. Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates. Proc Natl Acad Sci U S A. 2009;106:13517–22. doi: 10.1073/pnas.0906413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beckham CJ, Parker R. P bodies, stress granules, and viral life cycles. Cell Host Microbe. 2008;3:206–12. doi: 10.1016/j.chom.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dougherty JD, White JP, Lloyd RE. Poliovirus-mediated disruption of cytoplasmic processing bodies. J Virol. 2011;85:64–75. doi: 10.1128/JVI.01657-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emara MM, Brinton MA. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc Natl Acad Sci U S A. 2007;104:9041–6. doi: 10.1073/pnas.0703348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pérez-Vilaró G, Scheller N, Saludes V, Díez J. Hepatitis C virus infection alters P-body composition but is independent of P-body granules. J Virol. 2012;86:8740–9. doi: 10.1128/JVI.07167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaelin WG., Jr. Molecular biology. Use and abuse of RNAi to study mammalian gene function. Science. 2012;337:421–2. doi: 10.1126/science.1225787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diez J, Martinez JP, Mestres J, Sasse F, Frank R, Meyerhans A. Myxobacteria: natural pharmaceutical factories. Microb Cell Fact. 2012;11:52. doi: 10.1186/1475-2859-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weissman KJ, Müller R. A brief tour of myxobacterial secondary metabolism. Bioorg Med Chem. 2009;17:2121–36. doi: 10.1016/j.bmc.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 16.Nickeleit I, Zender S, Sasse F, Geffers R, Brandes G, Sörensen I, Steinmetz H, Kubicka S, Carlomagno T, Menche D, et al. Argyrin a reveals a critical role for the tumor suppressor protein p27(kip1) in mediating antitumor activities in response to proteasome inhibition. Cancer Cell. 2008;14:23–35. doi: 10.1016/j.ccr.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 17.Gentzsch J, Hinkelmann B, Kaderali L, Irschik H, Jansen R, Sasse F, Frank R, Pietschmann T. Hepatitis C virus complete life cycle screen for identification of small molecules with pro- or antiviral activity. Antiviral Res. 2011;89:136–48. doi: 10.1016/j.antiviral.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 18.Giard DJ, Aaronson SA, Todaro GJ, Arnstein P, Kersey JH, Dosik H, Parks WP. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J Natl Cancer Inst. 1973;51:1417–23. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- 19.Weissman KJ, Müller R. Myxobacterial secondary metabolites: bioactivities and modes-of-action. Nat Prod Rep. 2010;27:1276–95. doi: 10.1039/c001260m. [DOI] [PubMed] [Google Scholar]
- 20.Sasse F, Steinmetz H, Höfle G, Reichenbach H. Gephyronic acid, a novel inhibitor of eukaryotic protein synthesis from Archangium gephyra (myxobacteria). Production, isolation, physico-chemical and biological properties, and mechanism of action. J Antibiot (Tokyo) 1995;48:21–5. doi: 10.7164/antibiotics.48.21. [DOI] [PubMed] [Google Scholar]
- 21.Aizer A, Kafri P, Kalo A, Shav-Tal Y. The P body protein Dcp1a is hyper-phosphorylated during mitosis. PLoS One. 2013;8:e49783. doi: 10.1371/journal.pone.0049783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rzeczkowski K, Beuerlein K, Müller H, Dittrich-Breiholz O, Schneider H, Kettner-Buhrow D, Holtmann H, Kracht M. c-Jun N-terminal kinase phosphorylates DCP1a to control formation of P bodies. J Cell Biol. 2011;194:581–96. doi: 10.1083/jcb.201006089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giménez-Barcons M, Díez J. Yeast processing bodies and stress granules: self-assembly ribonucleoprotein particles. Microb Cell Fact. 2011;10:73. doi: 10.1186/1475-2859-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–42. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tong L. Acetyl-coenzyme A carboxylase: crucial metabolic enzyme and attractive target for drug discovery. Cell Mol Life Sci. 2005;62:1784–803. doi: 10.1007/s00018-005-5121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen Y, Volrath SL, Weatherly SC, Elich TD, Tong L. A mechanism for the potent inhibition of eukaryotic acetyl-coenzyme A carboxylase by soraphen A, a macrocyclic polyketide natural product. Mol Cell. 2004;16:881–91. doi: 10.1016/j.molcel.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 27.Tong L, Harwood HJ., Jr. Acetyl-coenzyme A carboxylases: versatile targets for drug discovery. J Cell Biochem. 2006;99:1476–88. doi: 10.1002/jcb.21077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo G, Costanzo M, Boone C, Dickson RC. Nutrients and the Pkh1/2 and Pkc1 protein kinases control mRNA decay and P-body assembly in yeast. J Biol Chem. 2011;286:8759–70. doi: 10.1074/jbc.M110.196030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cowart LA, Gandy JL, Tholanikunnel B, Hannun YA. Sphingolipids mediate formation of mRNA processing bodies during the heat-stress response of Saccharomyces cerevisiae. Biochem J. 2010;431:31–8. doi: 10.1042/BJ20100307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sweet TJ, Boyer B, Hu W, Baker KE, Coller J. Microtubule disruption stimulates P-body formation. RNA. 2007;13:493–502. doi: 10.1261/rna.355807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loschi M, Leishman CC, Berardone N, Boccaccio GL. Dynein and kinesin regulate stress-granule and P-body dynamics. J Cell Sci. 2009;122:3973–82. doi: 10.1242/jcs.051383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saroussi S, Nelson N. The little we know on the structure and machinery of V-ATPase. J Exp Biol. 2009;212:1604–10. doi: 10.1242/jeb.025866. [DOI] [PubMed] [Google Scholar]
- 33.Huang L, Mollet S, Souquere S, Le Roy F, Ernoult-Lange M, Pierron G, Dautry F, Weil D. Mitochondria associate with P-bodies and modulate microRNA-mediated RNA interference. J Biol Chem. 2011;286:24219–30. doi: 10.1074/jbc.M111.240259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ernoult-Lange M, Bénard M, Kress M, Weil D. P-bodies and mitochondria: which place in RNA interference? Biochimie. 2012;94:1572–7. doi: 10.1016/j.biochi.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 35.Sasse F, Steinmetz H, Höfle G, Reichenbach H. Archazolids, new cytotoxic macrolactones from Archangium gephyra (Myxobacteria). Production, isolation, physico-chemical and biological properties. J Antibiot (Tokyo) 2003;56:520–5. doi: 10.7164/antibiotics.56.520. [DOI] [PubMed] [Google Scholar]
- 36.von Schwarzenberg K, Wiedmann RM, Oak P, Schulz S, Zischka H, Wanner G, Efferth T, Trauner D, Vollmar AM. Mode of cell death induction by pharmacological vacuolar H+-ATPase (V-ATPase) inhibition. J Biol Chem. 2013;288:1385–96. doi: 10.1074/jbc.M112.412007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buchan JR, Kolaitis RM, Taylor JP, Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153:1461–74. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol. 2008;10:1224–31. doi: 10.1038/ncb1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mokas S, Mills JR, Garreau C, Fournier MJ, Robert F, Arya P, Kaufman RJ, Pelletier J, Mazroui R. Uncoupling stress granule assembly and translation initiation inhibition. Mol Biol Cell. 2009;20:2673–83. doi: 10.1091/mbc.E08-10-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–6. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- 41.Arkov AL, Ramos A. Building RNA-protein granules: insight from the germline. Trends Cell Biol. 2010;20:482–90. doi: 10.1016/j.tcb.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weil TT, Parton RM, Herpers B, Soetaert J, Veenendaal T, Xanthakis D, Dobbie IM, Halstead JM, Hayashi R, Rabouille C, et al. Drosophila patterning is established by differential association of mRNAs with P bodies. Nat Cell Biol. 2012;14:1305–13. doi: 10.1038/ncb2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeitelhofer M, Karra D, Vessey JP, Jaskic E, Macchi P, Thomas S, Riefler J, Kiebler M, Dahm R. High-efficiency transfection of short hairpin RNAs-encoding plasmids into primary hippocampal neurons. J Neurosci Res. 2009;87:289–300. doi: 10.1002/jnr.21840. [DOI] [PubMed] [Google Scholar]
- 44.Reichenbach H. Myxobacteria, producers of novel bioactive substances. J Ind Microbiol Biotechnol. 2001;27:149–56. doi: 10.1038/sj.jim.7000025. [DOI] [PubMed] [Google Scholar]
- 45.Reboll MR, Ritter B, Sasse F, Niggemann J, Frank R, Nourbakhsh M. The myxobacterial compounds spirangien a and spirangien M522 are potent inhibitors of IL-8 expression. Chembiochem. 2012;13:409–15. doi: 10.1002/cbic.201100635. [DOI] [PubMed] [Google Scholar]
- 46.Kunze B, Steinmetz H, Höfle G, Huss M, Wieczorek H, Reichenbach H. Cruentaren, a new antifungal salicylate-type macrolide from Byssovorax cruenta (myxobacteria) with inhibitory effect on mitochondrial ATPase activity. Fermentation and biological properties. J Antibiot (Tokyo) 2006;59:664–8. doi: 10.1038/ja.2006.89. [DOI] [PubMed] [Google Scholar]
- 47.Hopkins CD, Wipf P. Isolation, biology and chemistry of the disorazoles: new anti-cancer macrodiolides. Nat Prod Rep. 2009;26:585–601. doi: 10.1039/b813799b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huss M, Sasse F, Kunze B, Jansen R, Steinmetz H, Ingenhorst G, Zeeck A, Wieczorek H. Archazolid and apicularen: novel specific V-ATPase inhibitors. BMC Biochem. 2005;6:13. doi: 10.1186/1471-2091-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jahns C, Hoffmann T, Müller S, Gerth K, Washausen P, Höfle G, Reichenbach H, Kalesse M, Müller R. Pellasoren: structure elucidation, biosynthesis, and total synthesis of a cytotoxic secondary metabolite from Sorangium cellulosum. Angew Chem Int Ed Engl. 2012;51:5239–43. doi: 10.1002/anie.201200327. [DOI] [PubMed] [Google Scholar]
- 50.Jansen R, Irschik H, Huch V, Schummer D, Steinmetz H, Bock M, Schmidt T, Kirschning A, Müller R . Carolacton – A Macrolide Ketocarbonic Acid that Reduces Biofilm Formation by the Caries- and Endocarditis-Associated Bacterium Streptococcus mutans. European Journal of Organic Chemistry. 2010;2010:1284–9. [Google Scholar]