Abstract
Mitochondrial ribosomes of Trypanosoma brucei are composed of 9S and 12S rRNAs, which are encoded by the kinetoplast genome, and more than 150 proteins encoded in the nucleus and imported from the cytoplasm. However, a single ribosomal protein RPS12 is encoded by the kinetoplast DNA (kDNA) in all trypanosomatid species examined. As typical for these organisms, the gene itself is cryptic and its transcript undergoes an extensive U-insertion/deletion editing. An evolutionary trend to reduce or eliminate RNA editing could be traced with other cryptogenes, but the invariably pan-edited RPS12 cryptogene is apparently spared. Here we inquired whether editing of RPS12 mRNA is essential for mitochondrial translation. By RNAi-mediated knockdowns of RNA editing complexes and inducible knock-in of a key editing enzyme in procyclic parasites, we could reversibly downregulate production of edited RPS12 mRNA and, by inference, synthesis of this protein. While inhibition of editing decreased edited mRNA levels, the translation of edited (Cyb) and unedited (COI) mRNAs was blocked. Furthermore, the population of SSU-related 45S complexes declined upon inactivation of editing and so did the amount of mRNA-bound ribosomes. In bloodstream parasites, which lack active electron transport chain but still require translation of ATP synthase subunit 6 mRNA (A6), both edited RPS12 and A6 mRNAs were detected in translation complexes. Collectively, our results indicate that a single ribosomal protein gene retained by the kinetoplast mitochondrion serves as a possible functional link between editing and translation processes and provide the rationale for the evolutionary conservation of RPS12 pan-editing.
Keywords: mitochondria, RNA editing, polyadenylation, translation, kDNA, TUTase
Introduction
The monophyletic origin of mitochondria from within α-proteobacteria notwithstanding, mitochondrial genomes differ dramatically among eukaryotic groups in terms of size, copies per cell, structural organization, and coding capacity.1 One of the most extreme cases is represented by trypanosomatids. These parasitic protozoa belong to the class Kinetoplastea typified by the presence of a “kinetoplast,” a nucleoprotein structure located in the mitochondrion adjacent to the flagellar basal body. The kinetoplast DNA (kDNA) is composed of catenated maxicircles and minicircles encoding ribosomal and mRNAs, and guide RNAs (gRNAs), respectively. Polycistronic precursors are transcribed from maxicircles and then processed into rRNA and pre-mRNAs. In Trypanosoma brucei and related organisms, 12 pre-mRNAs (“pre-edited mRNAs”) undergo U-insertion/deletion RNA editing to acquire open reading frames suitable for productive translation (reviewed in refs. 2–4). The editing required varies from a few insertions at the 5′ end (“5′ edited mRNAs”) to massive incorporation of hundreds of U-residues (“pan-editing”). The remaining six transcripts contain encoded translatable reading frames and are referred to as “unedited” mRNAs. In line with the general pattern observed in mitochondrial systems, T. brucei imports most elements of mitochondrial transcription, RNA processing and translation processes from the cytoplasm except for the 9S small subunit (SSU) and 12S large subunit (LSU) rRNAs and a single ribosomal protein S12 (RPS12). No tRNAs are encoded in the kDNA and these molecules are imported as well.5
The RPS12 mRNA represents a typical case of pan-editing with 132 uridylates added and 28 uridylates deleted by this process in T. brucei. The cleavage, U-insertion/deletion and ligation editing reactions are catalyzed by the RNA editing core complex (RECC),6,7while the cascade of editing reactions is directed by gRNAs.8,9 These 50–60 nucleotide (nt) molecules are 3′ uridylated by RNA editing TUTase 1 (RET1)10,11 and stabilized via association with the gRNA binding complex (GRBC).12 RNA editing also generates mRNAs encoding subunits of respiratory complexes, such as COII and COIII of cytochro me c oxidase (COI is encoded by the unedited mRNA) and subunit Cyb of the cytochrome bc1, which are critical for the actively respiring insect (procyclic) developmental stage of the parasite.13 Interestingly, RNA editing also continues in the oxidative phosphorylation-deficient mitochondrion of bloodstream form (BF) parasites and is essential for their viability.14 In the bloodstream form, ATP is produced via substrate level phosphorylation and is hydrolyzed by the F1F0 ATP synthase to generate mitochondrial trans-membrane potential, thus maintaining the organelle.15,16 Therefore, editing and translation of the mitochondrial mRNA encoding subunit 6 (A6) of the F1F0 ATPase complex is expected to occur, and that, in turn, calls for the functional RPS12 mRNA. In bacterial ribosomes, the S12 protein plays an indispensable role in formation of the decoding center and is anticipated to perform a similar function in mitochondrial ribosomes.17
Recently, we demonstrated that editing is necessary, but not sufficient for mitochondrial protein synthesis: a discrete post-editing 3′ processing event is required to enable mRNA binding to the ribosome. Specifically, KPAP1 poly(A) polymerase and RET1 TUTase add 200–300 nt-long A/U-tails to mRNAs in a reaction that also requires two additional RNA binding factors.18 As expected, long A/U-tails were detected in most mitochondrial transcripts in the procyclic form while in bloodstream parasites only A6 and RPS12 mRNAs possessed such structures.18,19 Furthermore, RPS12 mRNA is productively edited in all species examined20-22 and knockdown of RET1 TUTase blocked translation of both edited and unedited mRNAs.23 These observations have indicated that RPS12, a single mitochondrially encoded ribosomal protein, is essential throughout the lifecycle of trypanosomes. The evolutionary trend to replace pan-edited cryptogenes with their less-edited counterparts24 seems to exclude the RPS12 cryptogene25,26 raising the prospect that functionality of RPS12 mRNA editing extends beyond the mere production of RPS12 protein. We hypothesized that the pan-editing status of the RPS12 mRNA serves as a functional link between editing and global mitochondrial translation, such that environmental or developmental impact on the editing system would modulate translation of edited and unedited mRNAs. The fact that mitochondrial translation creates a component of its own machinery suggests a positive feedback mechanism (production of RPS12 further facilitates its own production). Therefore, a variation of the RPS12 mRNA editing and the coupled process of 3′ adenylation/uridylation might impact the entire system of mitochondrial translation. We further hypothesize that this mechanism of regulation is not attainable unless the mRNA is pan-edited.
In order to ascertain the existence of editing—translation connection we analyzed the effects of eliminating various RNA editing complexes and an essential editing enzyme on edited and unedited mRNAs and rRNA levels, integrity of mitochondrial ribosomes, and translation of edited and unedited mRNAs. Here, we demonstrate that inhibition of editing eliminated mRNA-associated ribosomal fraction, and selectively affected the pool of free small subunits, while exerting minor effect on the overall rRNA abundance. Notably, translation of both unedited cytochrome c oxidase subunit I mRNA (COI) and edited apocytochrome b mRNA (Cyb) was abolished in every RNA editing-deficient genetic background tested. We also found that RPS12 and A6 mRNA are associated with ribosomes in procyclic and bloodstream developmental stages indicating that both products are produced throughout the lifecycle of trypanosomes. Conversely, mRNAs encoding proteins unnecessary in bloodstream form are either undectable (edited Cyb) or abundantly present, but nearly excluded from ribosome-bound fraction (unedited COI).
Results
RNAi knockdowns of different editing complexes induce similar changes in steady-state mRNA levels
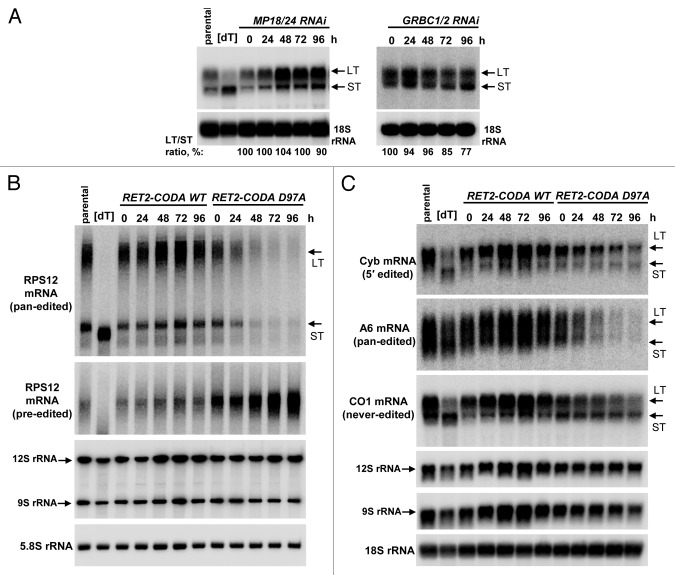
Approximately 25 proteins have been reported as essential for U-insertion/deletion editing and cell viability (reviewed in refs. 2, 3, and 27). Editing enzymes, RNA binding, and structural proteins are distributed among several complexes of varying stability that are engaged in higher-order interactions with polyadenylation and translational machineries. To address a possible functional relationship between RNA editing and translation processes, as presented by RPS12 mRNA being the product of the former and the source of a critical protein component for the latter, we chose to inactivate core components of two best-characterized editing complexes. Inducible RNAi cassette simultaneously targeting structural proteins MP18 (KREPA628,29) and MP24 (KREPA430,31) was introduced into procyclic T. brucei strain 29–1332 to inactivate the RNA editing core complex (RECC33), which performs elementary reactions of mRNA cleavage, U-insertion and deletion, and ligation. Likewise, subunits of the guide RNA binding complex (GRBC1 and GRBC212,34) responsible for gRNA binding and stabilization were depleted by RNAi. In addition to dual knockdowns intended to destabilize entire complexes, we employed the iCODA RNAi-based knock-in system to replace the endogenous RET2 protein with ectopically expressed catalytically active (WT) and inactive (D97A) variants of the same.35-37 RET2 TUTase is the only enzyme of its kind present in the core editing complex and is essential for parasite’s viability.11 To compare the effects of RECC and GRBC downregulation, and a single amino acid substitution D97A in RET2, cells were collected after RNAi or RNAi/RET2 co-expression was induced by tetracycline for 72 h. Quantitative real-time PCR analysis of rRNAs and unedited mRNAs showed minor changes while the steady-state levels of most edited mRNAs declined to various degrees (Fig. 1). Repression of RET2 TUTase by RNAi produced virtually indistinguishable results from RET2-CODA D97A while knock-in with active RET2 restored edited mRNA abundance (ref. 36 and data not shown). Importantly for the purposes of this study, edited RPS12 mRNA was uniformly downregulated in all genetic backgrounds lacking RNA editing. The mRNAs whose translation products can be monitored, edited Cyb, and unedited COI38,39 declined or remained unaffected (slightly upregulated), respectively.
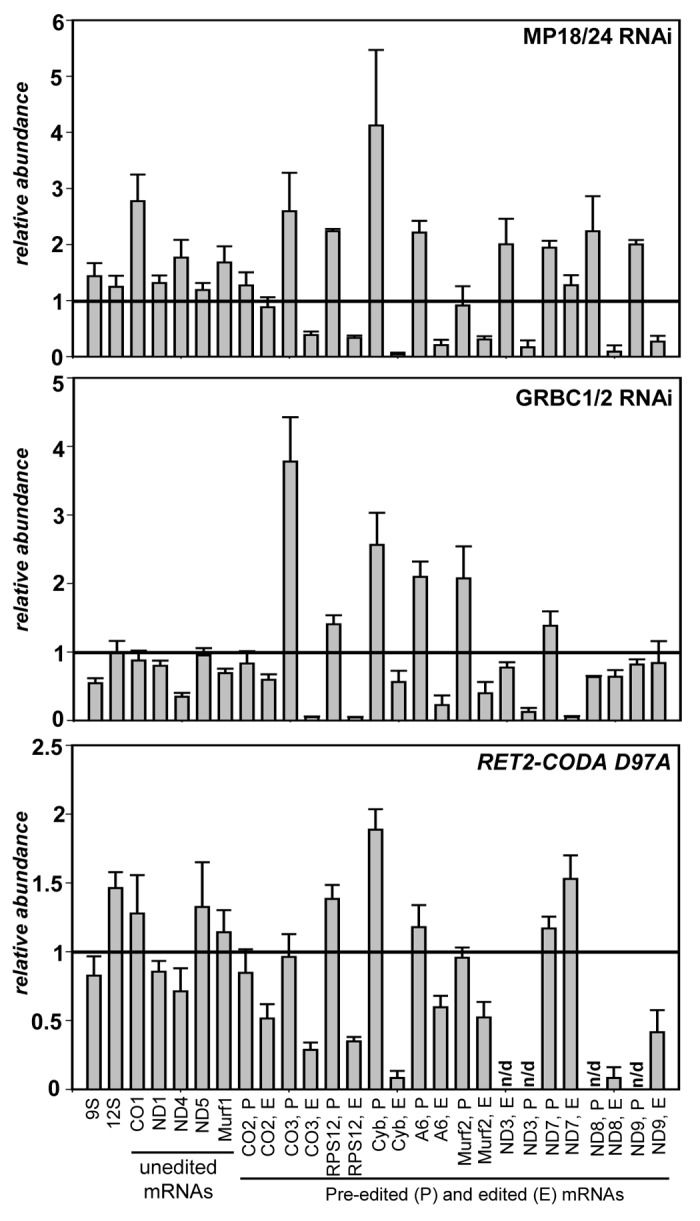
Figure 1. Inhibition of RNA editing by dual RNAi knockdowns of critical subunits in the RNA editing core (MP18 and MP24) and guide RNA binding (GRBC1 and GRBC2) complexes, and RNAi-based knock-in of catalytically inactive RET2 TUTase. Change in relative abundance for mitochondrial mRNAs and rRNAs was determined by real-time RT-PCR analysis; thick line at “1” reflects no change in relative abundance upon RNAi induction; bars above or below represent an increase or decrease, respectively. RNA levels were normalized to β-tubulin mRNA. Error bars represent the standard deviation from at least three replicates. P, pre-edited mRNA; E, edited mRNA; n/d, not determined.
Loss of RNA editing complexes causes disparate effects on mRNA 3′ adenylation/uridylation status
Although U-insertion/deletion editing is required for building open reading frames in mRNAs, it is not sufficient for their translation. In addition to editing, mRNA must undergo 3′ modification resulting in addition of 200–300 nt-long A/U-tail.18 Such tails are also present in unedited mRNAs. Because of the functional coupling between the completion of RNA editing and A/U-tail addition40 and an established interaction between polyadenylation and guide RNA binding complexes12,18 we next analyzed polyadenylation states of unedited COI mRNA in dual MP18/24 and GRBC1/2 knockdowns (Fig. 2A). In agreement with real-time PCR data, COI mRNA was upregulated 2-fold in MP18/24 dual RNAi, but the ratio between long- and short-tailed forms remained unaltered. In GRBC1/2-knockdown cells, however, the relative abundance of the long-tailed mRNA that is presumed to be the translation competent form, declined by approximately 25% over 96 h of RNAi induction.
Figure 2. MRNA editing and polyadenylation states in genetic backgrounds lacking RNA editing. (A) Expression of dual RNAi cassettes targeting subunits of the RNA editing core complex (MP18 and MP24, left panels) or guide RNA biding complex (GRBC1 and GRBC2, right panels) was induced by addition of tetracycline and cells were collected for RNA purification at 24 h intervals. Total RNA was separated on 1.7% agarose /formaldehyde gel and subjected to hybridization with DNA probe for unedited COI mRNA. [dT], RNA was treated with RNase H in the presence of 18-mer [dT] to remove poly(A) tails. ST and LT, mRNA forms terminating with short (A) or long (A/U) tails, respectively. (B) Pre-edited and edited forms of RPS12 mRNA were analyzed in RET2-CODA WT and RET2-CODA D97A genetic backgrounds. RNAi targeting the endogenous RET2 mRNA and expression of RNAi-resistant variants of the same were induced with tetracycline for indicated periods of time. Total RNA was separated on 5% polyacrylamide/ 8M urea gels and transferred onto membrane. Cytosolic 5.8S rRNAs served as loading control (C). Same RNA samples as in (B) were separated on 1.7% agarose/formaldehyde and probed for moderately edited Cyb, pan-edited A6 and unedited COI mRNAs. In addition, mitochondrial rRNAs (9S and 12S) were visualized on the same membrane.
To establish an experimental system and timeframe that allows for inhibition of editing accompanied by minimal secondary effects on unedited mRNAs, we analyzed unedited COI and edited Cyb and RPS12 mRNA in RET2-CODA WT and RET2-CODA D97A cell lines. The ATP synthase subunit 6 mRNA served as control for pan-edited mRNA. High-resolution northern blotting demonstrated a virtually complete restoration and elimination of both long- and short-tailed forms of fully edited RPS12 mRNA in RET2-CODA WT and RET2-CODA D97A genetic backgrounds, respectively (Fig. 2B). As expected, pre-edited RNA remained unaffected in RET2 RNAi rescue experiments (RET2 CODA), but accumulated when no longer consumed by editing (RET2-CODA D97A). In close correlation with real-time PCR analysis, Cyb and A6 mRNAs declined dramatically while both LT and ST forms of COI mRNA stayed virtually constant up to 72 h of RNAi induction (Fig. 2C). Because RET2-CODA D97A cell line displayed strong growth inhibition phenotype after 96 h we chose to perform the following experiments by inducing expression for 72 h.
Inactivation of RNA editing by a single amino acid substitution eliminates mRNA-bound ribosomal fraction
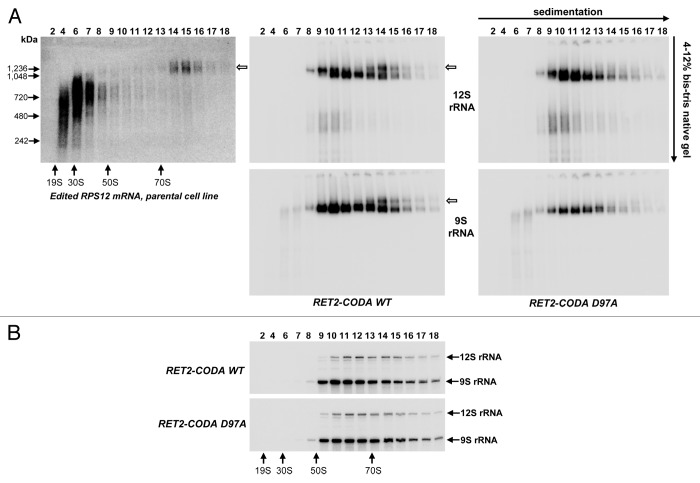
We previously showed that 9S and 12S rRNA sediment in a broad range (50S to more than 90S) whereas long-tailed edited RPS12 mRNA co-sediments with ribosomal particles in the 70S-90S zone of glycerol density gradients. Pre-edited and short-tailed edited mRNA co-sediment predominantly with RNA editing complexes in 20S-40S region.18 To further distinguish mRNA-bound ribosomes from assembled monosomes, we fractionated total cell lysate from parental 29–13 cell line on 10–30% glycerol gradient and then separated gradient fractions on 4–12% bis-Tris NuPAGE native gel, and analyzed edited RPS12 mRNA and rRNAs by northern blotting (Fig. 3A). Edited RPS12 mRNA formed heterogeneous complexes, most likely with editing machinery, ranging in apparent molecular mass from 500–900 kDa, but a minor fraction was clearly visualized as discrete particle of ~1.2 MDa sedimenting at ~80S (Fig. 3A, left panel, open arrow). In parallel experiments, sedimentation and native gel migration patterns were established for 12S and 9S rRNAs in cell lysates obtained from RET2 RNAi rescue (RET2-CODA WT, Fig. 3A, central panel). In fractions 14–16 we observed a separation of rRNA-containing particles into two bands. The slower-migrating band of these two overlapped precisely with position occupied by edited RPS12 mRNA (Fig. 3A, left and central panels, open arrows). Co-sedimentation and co-migration of rRNAs with edited mRNA as ~1.2 MDa particle provided a strong indication that the upper band corresponds to an mRNA-bound monosome which we termed the 80S complex. We concluded that a combination of glycerol gradient and native gel can be used to monitor ribosome-mRNA binding and next analyzed the effect of editing blockade on this interaction. Remarkably, introduction of a RET2-CODA D97A led to elimination of the putative mRNA-ribosome complex and a moderate decline of 9S rRNA-containing complexes (Fig. 3A, right panel). Under denaturing conditions, however, the relative ratio of 9S and 12S did not appreciably change throughout the glycerol gradient (Fig. 3B).
Figure 3. Loss of the 80S mRNA-associated ribosomal population upon inactivation of RNA editing. (A) Native gel analysis of ribosomal complexes. Total cell lysates obtained from parental and RET2 knock-in cell lines were fractioned on glycerol gradients and each fraction was further separated on 3–12% Bis-Tris NuPAGE native gel and transferred onto nylon membrane. NativeMARK Unstained Protein Standards (Invitrogen) were separated alongside. Hybridizations were performed with the same probes as in Figure 2 to detect fully-edited RPS12 mRNA, 9S, and 12S rRNAs. (B) Sedimentation of rRNAs. RNA was extracted from indicated fractions, separated on 5% polyacrylamide/8M urea gel and simultaneously probed for 9S and 12S rRNAs.
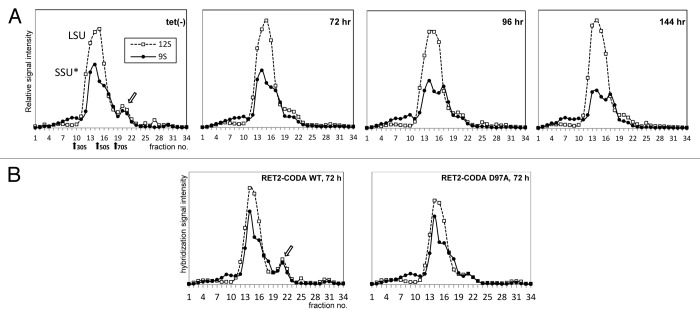
Denaturing gels allow for quantitation of full-length RNAs, but are less accurate in providing the overall RNA abundance. To independently corroborate the conclusions drawn from native gel analysis, we fractioned lysates from RET2 RNAi cell line induced for 0–144 h and RET2-CODA WT variants on sucrose gradients. RNA was extracted from each fraction and quantitated by dot-blot hybridization against synthetic 9S and 12S standards (Fig. 4). A discrete peak in the 80S region was observed for both 9S and 12S rRNAs in uninduced RET2 RNAi cells and RET2-CODA WT RNAi rescue; this particle was effectively reduced with progression of RET2 RNAi (Fig. 4A) or co-expression of inactive RET2-CODA D97A (Fig. 4B). In the major 45S-70S peak, the large subunit rRNA remained virtually unaffected while the small subunit RNA (45S region) declined in the RET2 RNAi more prominently than in the RET2-CODA D97A cell line. These findings further corroborate the existence of a monosome-mRNA 80S complex and indicate that under experimental conditions used in this study only a small (5–10%) fraction of mitochondrial monosomes in engaged in mRNA binding. Furthermore, this interaction apparently depends on functioning RNA editing pathway. We found no evidence of polysome formation in trypanosome mitochondrion.
Figure 4. Reduction of the 45S SSU-related complex (SSU*) and the 80S complex upon inactivation of RNA editing. (A) Ribosomal complexes in RET2 RNAi uninduced cells (tet[-]), as well as in cells upon induction of the RNAi at specified time points. Total cell lysates were fractionated in sucrose gradients; 9S and 12S rRNAs in gradient fractions were analyzed by slot-blot hybridization. (B) Ribosomal complexes in RET2-CODA WT and RET2-CODA D97A knock-in cells.
RNA editing is required for translation of edited and unedited mRNAs
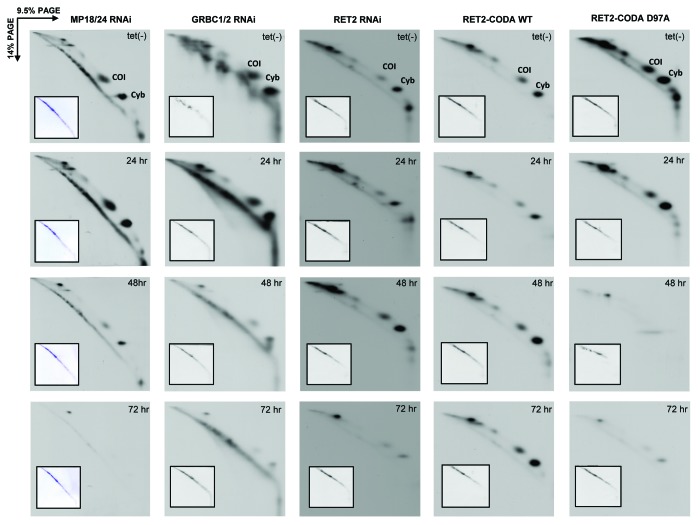
Interactions between RNA editing, polyadenylation, and translation machineries were initially suggested by mass spectrometric and immunochemical analyses of purified complexes.12,18,40 These contacts may be partially responsible for changes in unedited mRNA abundance caused by knockdown of seemingly unrelated editing proteins, e.g., increase in COI mRNA abundance in MP18/24 dual RNAi. Therefore, we analyzed mitochondrial translation in all RNA editing-deficient and RNA editing-restored genetic backgrounds described above. Among 18 predicted mitochondrially encoded proteins the de novo synthesis of only two polypeptides can be reliably monitored by metabolic cell labeling and two-dimensional PAGE (PAGE) in the presence of SDS.38,39 Encoded by unedited (COI) and edited (Cyb) mRNAs, these proteins migrate abnormally and present as off-diagonal spots in the second dimension. The edited Cyb mRNA was uniformly downregulated in all genetic backgrounds with impaired RNA editing (Figs. 1 and 2), which was reflected by a gradual decline of protein synthesis concomitant with RNAi induction time (Fig. 5). Restoration of RNA editing by co-expression of RET2-CODA WT provided for an effective complementation of protein synthesis (Fig. 5). Unedited COI mRNA underwent pleiotropic changes from overall increase in abundance (MP18/24 RNAi) to the loss of long A/U-tails (GRBC1/2 RNA) to insignificant difference with the control (RET2-CODA D97A) (Figs. 1 and 2). At the translation level, however, synthesis of COI polypeptide was uniformly reduced after 48 h and virtually eliminated at 72 h of RNAi induction in all cell lines lacking RNA editing. Restoration of editing in RET2-CODA WT cells, however, allowed for sustained COI synthesis (Fig. 5). To conclude, we found a strong correlation between the functional RNA editing pathway and translation of unedited mRNA.
Figure 5. Mitochondrial translation in parasites depleted of guide RNA binding complex (GRBC1/2), RNA editing core complex (MP18/24), RNA editing TUTase 2 (RET2), and knock-in cell lines expressing catalytically active (RET2-CODA WT) or inactive (RET2-CODA D97A) TUTases. RNAi and protein expression were induced for indicated periods of time and translation products were metabolically labeled with EasyTag mix (PerkinElmer Life Sciences) in isotonic buffer supplemented with 0.1 mg/ml of cycloheximide. Cells were collected by centrifugation, dissolved in SDS gel loading buffer, and fractionated by two-dimensional electrophoresis. Gels were stained with Coomassie brilliant blue R250 (inset panels) and exposed to X-ray film (large panels). Based on previous protein identifications,23 major spots represent COI and Cyb proteins encoded by unedited and edited mRNAs, respectively.
Edited A/U-tailed RPS12 mRNA is associated with ribosomes in both procyclic and bloodstream developmental stages of T. brucei
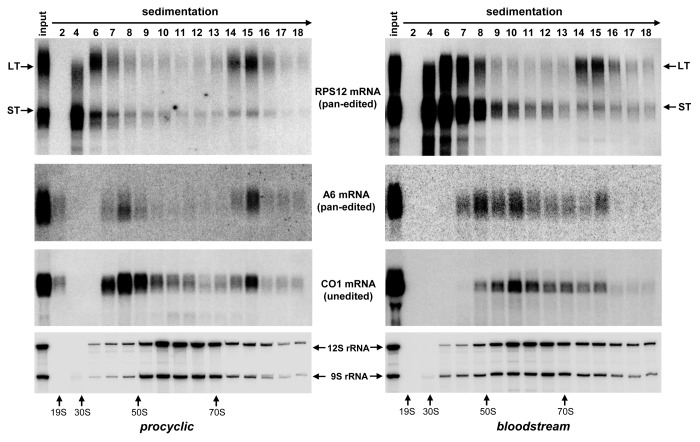
Predicted from the edited mRNA sequence, RSP12 protein is expected to be small (82 amino acids), cysteine-rich (six residues), and basic (9.04 isoelectric point). Despite numerous attempts and different mass spectrometric approaches, we and others41 have not been able to detect RPS12 protein in affinity-purified ribosomes, which precluded direct assessment of the RPS12 mRNA translation product. A large body of evidence suggests that pan-edited A6 mRNA provides a mitochondrially encoded protein essential for mitochondrial maintenance in the bloodstream form.15,16,42 It is held that energy requirements are fulfilled by an aerobic glycolysis where glucose is converted to pyruvate. The mitochondrion presumably lacks active respiratory chain while the membrane potential is maintained by reversing function of the F1F0 ATP synthase. We hypothesized that if RPS12 is universally required for mitochondrial translation, then translationally competent, i.e., long-tailed edited RPS12 and A6 mRNAs should be enriched in the 80S complex. On the contrary, subunits of respiratory complexes that are downregulated in BF, such as COI, should be excluded from 80S complexes. To test this hypothesis we compared sedimentation profiles of these transcripts in PF and BF parasites in reference to rRNAs (Fig. 6). In the procyclic form the long-tailed RPS12 mRNA was enriched in the 80S complex by ~3-fold vs. total RNA, which is consistent with our previous data18 while even higher (5-fold) enrichment of the LT mRNA population was observed in bloodstream parasites. Similarly, long-tailed A6 mRNA was detected in the 80S fractions in PF and BF parasites. In contrast, A/U-tailed COI mRNA was only present as discrete 80S peak in the procyclic, but not the bloodstream form. Edited Cyb mRNA was associated with the 80S complex in PF parasites, but could not be detected in BF trypanosomes by northern blotting (data not shown). These results show that the developmental downregulation of the mitochondrial translation occurring in BF trypanosomes does not include RPS12. In summary, our results indicate that loss of RNA editing impedes global mitochondrial translation in PF and BF cells.
Figure 6. Comparative sedimentation analysis of RPS12, A6 and COI mRNAs between procyclic and bloodstream developmental forms of T. brucei. Total cell lysates were obtained from parental cell lines routinely used for RNAi studies in procyclic (29–13) and bloodstream (Lister 427 “single marker”) parasites32 were fractioned on glycerol gradients. RNA was extracted from indicated fractions and separated on 5% polyacrylamide/8M urea gel (for RPS12 and rRNA detection) or 1.7% agarose/formaldehyde (for A6 and COI mRNA detection).
Discussion
In bacterial ribosome RPS12 protein participates in stabilizing the second base pair of the codon-anticodon duplex in the A site and is likely to be critical for the fidelity of decoding process.17 A similar role can be anticipated for this protein in mitochondrial ribosomes. Although this protein has not been detected by mass-spectrometry in T. brucei18,41 and L. tarentolae,43,44 a cryo-EM-3D reconstruction of the Leishmania mitoribosome45 indicated the presence of a protein mass in the fitting position within the small subunit. Apparently, the detection problem is caused by some unusual properties of this polypeptide, similar to the remaining mitochondrially encoded proteins, which proved to be refractory to identification by mass-spectrometry.
As the crucial ribosomal component, RPS12 protein is encoded in mitochondrial genomes across many taxa. In kinetoplastids, the RPS12 gene is among the 18 protein-coding genes retained on maxicircles of kDNA. A variable amount of U-editing, from numerous insertions/deletions to addition of just a few bases, is required for converting the original transcripts of 12 genes into translatable mRNAs. For a given gene, the level of required editing is not conserved among the kinetoplastid taxa24,25 and it was proposed that extensive editing (pan-editing) is being slowly eliminated or reduced by retroposition of edited or partially edited transcripts.24 Remarkably, the RPS12 gene represents a pan-edited cryptogene in all kinetoplastids so far examined, suggesting that the extent of editing per se may play an important role.
Pan-editing is performed by multi-protein editing complexes and sequential participation of multiple overlapping gRNAs. In this process, extension of the edited region by each next gRNA depends on completion of editing by a preceding gRNA.22 Modulation of this process by nuclear-encoded components provides a potential to regulate expression of the mitochondrial genes whose transcript require editing. At the same time, this seemingly straightforward scenario leaves out a question of coordinated shutdown or upregulation of edited and unedited mRNAs which often encode subunits of the same respiratory complexes. Such regulation becomes necessary during the differentiation of bloodstream (BF) trypanosomes into the insect stage (procyclic form, PF) that must be accompanied by a transition from mitochondrion lacking cytochrome-mediated electron transport chain into fully functional system of oxidative phosphorylation. This transformation is presumed to include translational activation of the mitochondrially encoded subunits of cytochrome c oxidase (COI, COII, COIII) and cytochrome bc1 (apocytochrome b). Conversely, their synthesis needs to be turned off during differentiation of PF into BF trypanosomes. This downregulation has to be selective, since BF cells still require the translation product of the A6 mRNA for functioning of F1F0 ATPase.
This work suggests the existence of a functional link between RNA editing and translation. The product of edited RPS12 mRNA translation represents an indispensable component of the mitoribosome's small subunit thereby providing a possible conduit for affecting translation of both edited and unedited mRNAs. Perturbations of the editing system caused by the RNAi repression of its structural (MP18/24), gRNA binding (GRBC1/2), or enzymatic (RET2) components resulted in a strong inhibition of the mitochondrial protein synthesis, affecting production of not only cyb protein, which is encoded by edited mRNA, but also the COI polypeptide produced from unedited template. This inhibition was most likely mediated by the decrease in the level of the long-tailed edited RPS12 mRNA and ablation of the COI mRNA association with the mitoribosomes (the 80S complex). These events essentially parallel a relative reduction of the 45S SSU* complexes (Maslov, unpublished) and significant reduction of the COI mRNA in the 80S complexes (this work) observed in BF trypanosomes. These results are also consistent with undetectable de novo synthesis of COI polypeptide in BF parasites.18,46 Albeit indirect, these observations lead us to a hypothesis of RPS12-mediated link between the editing and translation serving to align production of the polypeptides encoded by unedited and edited templates. The findings of this work show that translation of COI is sensitive to the decline in the edited RPS12 mRNA level. It can be envisioned that due to the existence of the positive feedback loop accelerating translation of the edited RPS12 mRNA, a relatively small modulation in the level of this mRNA will have an amplifying effect on abundance of the ribosomal small subunits. Since COI is among the most abundant translation products, its mRNA may represent a preferential translation template and a boost to the mitoribosomes may enhance the COI translation. It remains to be investigated whether or not this mechanism is employed during the developmental activation of mitochondrial translation in T. brucei.
Materials and Methods
Trypanosome culture, RNAi, and protein expression
RNAi plasmids were generated by cloning gene fragments in tandem into p2T7–177 vector for tetracycline-inducible expression.47 The following gene regions were targeted: MP18 (497–848); MP24 (53–490); GRBC1 (659–986), and GRBC2 (405–929). The constructs were transfected into procyclic 29–13 T. brucei strain.32 RNAi and RNAi-based RET2-CODA WT knock-ins were performed as described.12,35,36
Mitochondrial isolation, density gradients, and native gels
Mitochondrial fraction was isolated as described except omitting the Percoll density gradient.48 Fresh mitochondrial pellets were mixed with three parts (wet weight) of buffer containing 25 mM HEPES (pH 7.6), 125 mM KCl, 12 mM MgCl2, and 1.2% Nonidet P-40 (NP-40). After 15 min incubation on ice, the extract was clarified at 18 000 g for 10 min and fractionated on 10–30% glycerol gradient in SW41 rotor for 4 h at RCFavg = 178 000 x g (38 000 rpm). Fractions of 580 μl were collected from the top and 10 μl aliquot was further separated on 15-well 4–12% bis-Tris NuPAGE and transferred for Northern according to manufacturer’s protocol (Invitrogen). RNA was isolated from the remaining material by phenol/chloroform extraction. For sucrose gradients, approximately 108 cells were washed with 400 µL of the SoTE buffer solution (20 mM Tris-HCl, pH 7.5, 600 mM sorbitol, 2 mM EDTA) and were lysed in 400 µL of the DM buffer (1% dodecyl maltoside, 50 mM Tris-HCl, pH 7.5, 100 mM KCl, 10 mM Mg Cl2, 3 mM DTT, 0.1 mM EDTA) for 15–30 min on ice. RNA degradation was minimized by including 400 units of RNasin(R) Plus RNase Inhibitor (Promega) in the lysis buffer. The insoluble material was removed from lysates by centrifugation at 17 000 x g for 15 min. Mitochondrial lysates were fractionated in 7–30% sucrose gradients made with the SGB buffer (50 mM Tris-HCl, pH 7.5, 100 mM KCl, 10 mM Mg Cl2, 3 mM DTT, 0.1 mM EDTA, 0.05% dodecyl maltoside) at RCFavg = 35 670 x g (17 000 rpm, SW41 rotor) for 16 h.
RNA analysis
Total RNA isolation, qRT-PCR and northern blotting were performed as described.18 The change in relative abundance was calculated assuming the ratio between analyzed transcripts and loading control in mock-induced cells as 100%. Membranes and gels were exposed to phosphor storage screens and volume quantitation was performed with Quantity One software (BioRad). For dot-blot quantitation, RNA from gradient fractions was immobilized on BrightStar-Plus hybridization membranes (Ambion) using a Hoefer SlotBlot manifold followed by UV-crosslinking. The oligonucleotides
A304 (TGAACAATCA ATCATGGTAA TAAGTAGACG ATG) and
A504 (ACGGCTGGCA TCCATTTC) served as hybridization probes specific for the 12S rRNA and the 9S rRNA, respectively. The probes were 5′ labeled with [γ-32P]ATP (6000 Ci/mmol) and T4 polynucleotide kinase. Pre-hybridizations and hybridizations were performed at 42 °C in 6 x SSPE, 5 x Denhardt's solution, 0.5% SDS, and 0.2 mg/ml sheared denatured salmon sperm DNA. Results of the hybridizations were quantified by phosphorimaging. In vitro transcribed 9S-12S tandem RNA was used for the signal intensity normalization.
Mitochondrial translation
Cell labeling and gel separations were performed as described.18 In brief, procyclic trypanosomes (107 cells) were washed with SoTE and resuspended in the same buffer. Cytosolic translation was inhibited with 100 µg/ml cycloheximide and the products of mitochondrial translation were labeled with [35S]- EasyTag Express Protein Labeling Mix (Perkin Elmer) by incubation of the labeling mixture at 26 °C for 1 h with mild agitation. Labeled products were analyzed with 2D (9% vs 14% polyacrylamide) Tris-Glycine-SDS gels.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Marat Yusupov and members of our laboratories for discussions. This work was supported by NIH grants AI091914 to RA and AI088292 to DM.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/26733
References
- 1.Gray MW. Mitochondrial evolution. Cold Spring Harb Perspect Biol. 2012;4:a011403. doi: 10.1101/cshperspect.a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aphasizhev R, Aphasizheva I. Uridine insertion/deletion editing in trypanosomes: a playground for RNA-guided information transfer. Wiley Interdiscip Rev RNA. 2011;2:669–85. doi: 10.1002/wrna.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aphasizhev R, Aphasizheva I. Mitochondrial RNA processing in trypanosomes. Res Microbiol. 2011;162:655–63. doi: 10.1016/j.resmic.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Göringer HU. ‘Gestalt,’ composition and function of the Trypanosoma brucei editosome. Annu Rev Microbiol. 2012;66:65–82. doi: 10.1146/annurev-micro-092611-150150. [DOI] [PubMed] [Google Scholar]
- 5.Schneider A. Mitochondrial tRNA import and its consequences for mitochondrial translation. Annu Rev Biochem. 2011;80:1033–53. doi: 10.1146/annurev-biochem-060109-092838. [DOI] [PubMed] [Google Scholar]
- 6.Aphasizhev R, Aphasizheva I, Nelson RE, Gao G, Simpson AM, Kang X, Falick AM, Sbicego S, Simpson L. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 2003;22:913–24. doi: 10.1093/emboj/cdg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panigrahi AK, Schnaufer A, Ernst NL, Wang B, Carmean N, Salavati R, Stuart K. Identification of novel components of Trypanosoma brucei editosomes. RNA. 2003;9:484–92. doi: 10.1261/rna.2194603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blum B, Bakalara N, Simpson L. A model for RNA editing in kinetoplastid mitochondria: “guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990;60:189–98. doi: 10.1016/0092-8674(90)90735-W. [DOI] [PubMed] [Google Scholar]
- 9.Seiwert SD, Heidmann S, Stuart K. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell. 1996;84:831–41. doi: 10.1016/S0092-8674(00)81062-4. [DOI] [PubMed] [Google Scholar]
- 10.Aphasizhev R, Sbicego S, Peris M, Jang SH, Aphasizheva I, Simpson AM, Rivlin A, Simpson L. Trypanosome mitochondrial 3′ terminal uridylyl transferase (TUTase): the key enzyme in U-insertion/deletion RNA editing. Cell. 2002;108:637–48. doi: 10.1016/S0092-8674(02)00647-5. [DOI] [PubMed] [Google Scholar]
- 11.Aphasizhev R, Aphasizheva I, Simpson L. A tale of two TUTases. Proc Natl Acad Sci U S A. 2003;100:10617–22. doi: 10.1073/pnas.1833120100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weng J, Aphasizheva I, Etheridge RD, Huang L, Wang X, Falick AM, Aphasizhev R. Guide RNA-binding complex from mitochondria of trypanosomatids. Mol Cell. 2008;32:198–209. doi: 10.1016/j.molcel.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hannaert V, Bringaud F, Opperdoes FR, Michels PA. Evolution of energy metabolism and its compartmentation in Kinetoplastida. Kinetoplastid Biol Dis. 2003;2:11. doi: 10.1186/1475-9292-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schnaufer A, Panigrahi AK, Panicucci B, Igo RP, Jr., Wirtz E, Salavati R, Stuart K. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science. 2001;291:2159–62. doi: 10.1126/science.1058655. [DOI] [PubMed] [Google Scholar]
- 15.Schnaufer A, Clark-Walker GD, Steinberg AG, Stuart K. The F1-ATP synthase complex in bloodstream stage trypanosomes has an unusual and essential function. EMBO J. 2005;24:4029–40. doi: 10.1038/sj.emboj.7600862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown SV, Hosking P, Li J, Williams N. ATP synthase is responsible for maintaining mitochondrial membrane potential in bloodstream form Trypanosoma brucei. Eukaryot Cell. 2006;5:45–53. doi: 10.1128/EC.5.1.45-53.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. A new understanding of the decoding principle on the ribosome. Nature. 2012;484:256–9. doi: 10.1038/nature10913. [DOI] [PubMed] [Google Scholar]
- 18.Aphasizheva I, Maslov D, Wang X, Huang L, Aphasizhev R. Pentatricopeptide repeat proteins stimulate mRNA adenylation/uridylation to activate mitochondrial translation in trypanosomes. Mol Cell. 2011;42:106–17. doi: 10.1016/j.molcel.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhat GJ, Souza AE, Feagin JE, Stuart K. Transcript-specific developmental regulation of polyadenylation in Trypanosoma brucei mitochondria. Mol Biochem Parasitol. 1992;52:231–40. doi: 10.1016/0166-6851(92)90055-O. [DOI] [PubMed] [Google Scholar]
- 20.Maslov DA, Nawathean P, Scheel J. Partial kinetoplast-mitochondrial gene organization and expression in the respiratory deficient plant trypanosomatid Phytomonas serpens. Mol Biochem Parasitol. 1999;99:207–21. doi: 10.1016/S0166-6851(99)00028-6. [DOI] [PubMed] [Google Scholar]
- 21.Maslov DA, Simpson L. RNA editing and mitochondrial genomic organization in the cryptobiid kinetoplastid protozoan Trypanoplasma borreli. Mol Cell Biol. 1994;14:8174–82. doi: 10.1128/mcb.14.12.8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maslov DA, Sturm NR, Niner BM, Gruszynski ES, Peris M, Simpson L. An intergenic G-rich region in Leishmania tarentolae kinetoplast maxicircle DNA is a pan-edited cryptogene encoding ribosomal protein S12. Mol Cell Biol. 1992;12:56–67. doi: 10.1128/mcb.12.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nebohácová M, Maslov DA, Falick AM, Simpson L. The effect of RNA interference Down-regulation of RNA editing 3′-terminal uridylyl transferase (TUTase) 1 on mitochondrial de novo protein synthesis and stability of respiratory complexes in Trypanosoma brucei. J Biol Chem. 2004;279:7819–25. doi: 10.1074/jbc.M311360200. [DOI] [PubMed] [Google Scholar]
- 24.Simpson L, Maslov DA. RNA editing and the evolution of parasites. Science. 1994;264:1870–1. doi: 10.1126/science.8009214. [DOI] [PubMed] [Google Scholar]
- 25.Simpson L, Thiemann OH, Savill NJ, Alfonzo JD, Maslov DA. Evolution of RNA editing in trypanosome mitochondria. Proc Natl Acad Sci U S A. 2000;97:6986–93. doi: 10.1073/pnas.97.13.6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerasimov ES, Kostygov AY, Yan S, Kolesnikov AA. From cryptogene to gene? ND8 editing domain reduction in insect trypanosomatids. Eur J Protistol. 2012;48:185–93. doi: 10.1016/j.ejop.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Hashimi H, Zimmer SL, Ammerman ML, Read LK, Lukeš J. Dual core processing: MRB1 is an emerging kinetoplast RNA editing complex. Trends Parasitol. 2013;29:91–9. doi: 10.1016/j.pt.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo X, Carnes J, Ernst NL, Winkler M, Stuart K. KREPB6, KREPB7, and KREPB8 are important for editing endonuclease function in Trypanosoma brucei. RNA. 2012;18:308–20. doi: 10.1261/rna.029314.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tarun SZ, Jr., Schnaufer A, Ernst NL, Proff R, Deng J, Hol W, Stuart K. KREPA6 is an RNA-binding protein essential for editosome integrity and survival of Trypanosoma brucei. RNA. 2008;14:347–58. doi: 10.1261/rna.763308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Babbarwal VK, Fleck M, Ernst NL, Schnaufer A, Stuart K. An essential role of KREPB4 in RNA editing and structural integrity of the editosome in Trypanosoma brucei. RNA. 2007;13:737–44. doi: 10.1261/rna.327707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salavati R, Ernst NL, O’Rear J, Gilliam T, Tarun S, Jr., Stuart K. KREPA4, an RNA binding protein essential for editosome integrity and survival of Trypanosoma brucei. RNA. 2006;12:819–31. doi: 10.1261/rna.2244106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99:89–101. doi: 10.1016/S0166-6851(99)00002-X. [DOI] [PubMed] [Google Scholar]
- 33.Simpson L, Aphasizhev R, Lukes J, Cruz-Reyes J. Guide to the nomenclature of kinetoplastid RNA editing: a proposal. Protist. 2010;161:2–6. doi: 10.1016/j.protis.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aphasizheva I, Aphasizhev R. RET1-catalyzed uridylylation shapes the mitochondrial transcriptome in Trypanosoma brucei. Mol Cell Biol. 2010;30:1555–67. doi: 10.1128/MCB.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ringpis GE, Lathrop RH, Aphasizhev R. iCODA: RNAi-based inducible knock-in system in Trypanosoma brucei. Methods Mol Biol. 2011;718:23–37. doi: 10.1007/978-1-61779-018-8_2. [DOI] [PubMed] [Google Scholar]
- 36.Ringpis GE, Aphasizheva I, Wang X, Huang L, Lathrop RH, Hatfield GW, Aphasizhev R. Mechanism of U insertion RNA editing in trypanosome mitochondria: the bimodal TUTase activity of the core complex. J Mol Biol. 2010;399:680–95. doi: 10.1016/j.jmb.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ringpis GE, Stagno J, Aphasizhev R. Mechanism of U-insertion RNA editing in trypanosome mitochondria: characterization of RET2 functional domains by mutational analysis. J Mol Biol. 2010;399:696–706. doi: 10.1016/j.jmb.2010.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horváth A, Berry EA, Maslov DA. Translation of the edited mRNA for cytochrome b in trypanosome mitochondria. Science. 2000;287:1639–40. doi: 10.1126/science.287.5458.1639. [DOI] [PubMed] [Google Scholar]
- 39.Horváth A, Kingan TG, Maslov DA. Detection of the mitochondrially encoded cytochrome c oxidase subunit I in the trypanosomatid protozoan Leishmania tarentolae. Evidence for translation of unedited mRNA in the kinetoplast. J Biol Chem. 2000;275:17160–5. doi: 10.1074/jbc.M907246199. [DOI] [PubMed] [Google Scholar]
- 40.Etheridge RD, Aphasizheva I, Gershon PD, Aphasizhev R. 3′ adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria. EMBO J. 2008;27:1596–608. doi: 10.1038/emboj.2008.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zíková A, Panigrahi AK, Dalley RA, Acestor N, Anupama A, Ogata Y, Myler PJ, Stuart K. Trypanosoma brucei mitochondrial ribosomes: affinity purification and component identification by mass spectrometry. Mol Cell Proteomics. 2008;7:1286–96. doi: 10.1074/mcp.M700490-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zíková A, Schnaufer A, Dalley RA, Panigrahi AK, Stuart KD. The F(0)F(1)-ATP synthase complex contains novel subunits and is essential for procyclic Trypanosoma brucei. PLoS Pathog. 2009;5:e1000436. doi: 10.1371/journal.ppat.1000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maslov DA, Spremulli LL, Sharma MR, Bhargava K, Grasso D, Falick AM, Agrawal RK, Parker CE, Simpson L. Proteomics and electron microscopic characterization of the unusual mitochondrial ribosome-related 45S complex in Leishmania tarentolae. Mol Biochem Parasitol. 2007;152:203–12. doi: 10.1016/j.molbiopara.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maslov DA, Sharma MR, Butler E, Falick AM, Gingery M, Agrawal RK, Spremulli LL, Simpson L. Isolation and characterization of mitochondrial ribosomes and ribosomal subunits from Leishmania tarentolae. Mol Biochem Parasitol. 2006;148:69–78. doi: 10.1016/j.molbiopara.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 45.Sharma MR, Booth TM, Simpson L, Maslov DA, Agrawal RK. Structure of a mitochondrial ribosome with minimal RNA. Proc Natl Acad Sci U S A. 2009;106:9637–42. doi: 10.1073/pnas.0901631106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richterová L, Vávrová Z, Lukeš J. DEAD-box RNA helicase is dispensable for mitochondrial translation in Trypanosoma brucei. Exp Parasitol. 2011;127:300–3. doi: 10.1016/j.exppara.2010.06.036. [DOI] [PubMed] [Google Scholar]
- 47.Wickstead B, Ersfeld K, Gull K. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol Biochem Parasitol. 2002;125:211–6. doi: 10.1016/S0166-6851(02)00238-4. [DOI] [PubMed] [Google Scholar]
- 48.Pelletier M, Read LK, Aphasizhev R. Isolation of RNA binding proteins involved in insertion/deletion editing. Methods Enzymol. 2007;424:75–105. doi: 10.1016/S0076-6879(07)24004-2. [DOI] [PubMed] [Google Scholar]