Abstract
Simian Virus 40 (SV40) is a polyomavirus found in both monkeys and humans, which causes cancer in some animal models. In humans, SV40 has been reported to be associated with cancers but causality has not been proven yet. The transforming activity of SV40 is mainly due to its 94-kD large T antigen, which binds to the retinoblastoma (pRb) and p53 tumor suppressor proteins, and thereby perturbs their functions. Here we describe a 100 kD super T antigen harboring a duplication of the pRB binding domain that was associated with unusual high cell transformation activity and that was generated by a novel mechanism involving homologous RNA trans-splicing of SV40 early transcripts in transformed rodent cells. Enhanced trans-splice activity was observed in clones carrying a single point mutation in the large T antigen 5′ donor splice site (ss). This mutation impaired cis-splicing in favor of an alternative trans-splice reaction via a cryptic 5′ss within a second cis-spliced SV40 pre-mRNA molecule and enabled detectable gene expression. Next to the cryptic 5′ss we identified additional trans-splice helper functions, including putative dimerization domains and a splice enhancer sequence. Our findings suggest RNA trans-splicing as a SV40-intrinsic mechanism that supports the diversification of viral RNA and phenotypes.
Keywords: alternative splicing, trans-splicing, viral gene expression, SV40 100 kD super T antigen, cell transformation, molecular basis of diseases
Introduction
Trans-splicing of pre-mRNAs, the joining of sequences from distinct transcripts, represents an alternative form of splicing that contributes to the diversification of genotypes and phenotypes. Naturally occurring RNA trans-splicing was first detected in 1986 in trypanosomes and one year later also in the nematode Caenorhabditis elegans.1,2 In these organisms, a species-specific non-coding small RNA with a singular 5′ splice donor site, the so-called small leader RNA (SL RNA), is spliced to variable 3′ splice acceptor sites on separate pre-mRNA molecules. SL RNA trans-splicing, however, differs from regular cis-splicing, as the U1 small nuclear ribonucleoprotein (snRNP) containing the U1 small nuclear (sn)RNA is replaced by the SL snRNP, which carries the SL RNA.3 In the following five years, SL RNA trans-splicing was also detected in trematodes,4Euglena cells,5 and in tunicates,6 primitive chordates. Though also presumed to occur in vertebrates,7 SL RNA trans-splicing has not been detected as a natural event there yet. Natural RNA trans-splicing that differs from SL RNA trans-splicing and that resembles regular cis-splicing was also speculated to occur in vertebrate or mammalian cells. In silico analysis of cDNAs from a gene databank showed one percent of all sequenced mRNAs to be chimeric,8 some of which might be synthesized by RNA trans-splicing. In 1985 it was demonstrated that HeLa cell nuclear extracts support trans-splicing of artificial short RNA molecules containing extensive complementary antisense stretches in vitro.9,10 In the mid-’90s, the first experimental evidence was obtained that showed that mammalian cells can combine natural pre-mRNAs to new mRNA via RNA trans-splicing.11,12 Since then, several examples of naturally occurring mammalian RNA trans-splicing have been reported.13-21
Here we describe a novel mechanism that involves RNA trans-splicing of simian virus (SV40) early transcripts leading to a functional viral protein. SV40 is a polyomavirus of rhesus macaque origin that induces tumors in rodents and transforms many types of cells in culture including cells of human origin. SV40 can cause infections in humans and about 10–12% of men were found to be seropositive, independent of the administration of SV40-contaminated polio vaccines in the 1950s and ’60s.22 However, the immunological data are not definitive, as some of the SV40-reactive antibodies found in human serum are due to cross reaction with other polyomaviruses.23 SV40 has been reported to be associated with a variety of human cancers but causality still needs to be proven. SV40-mediated cellular transformation depends on the expression of the non-lytic viral early gene products and requires suppression of the lytic late gene expression. Late gene silencing occurs either in the event of integration of the viral DNA into the host genome or by antisense regulation of episomal SV40 gene expression as demonstrated in human mesothelial cells.24-26 The SV40 early region specifies two oncoproteins, the large T antigen (T-ag) and the small t antigen (t-ag), both of which are generated by alternative splicing of a common 2.7 knt precursor transcript. SV40 alternative splicing involves the use of a common T-ag and t-ag 3′ splice site (ss) and two different upstream 5′ss. The T-ag is capable of transforming a variety of mammalian cells by binding to the retinoblastoma protein (pRB) and p53, resulting in the inactivation of the two key tumor suppressor pathways.
In addition to the regular 94 kD T-ag, further variants of T-antigens (T-ags) have been described: truncated forms, species with similar but distinct molecular mass, and elongated forms of T-ag, so-called super T antigens (sT-ags).27,28 sT-ags encompass mainly three derivatives with molecular masses of 100, 115, and 145 kD. Whereas the 115 and 145 kD proteins are generated from genomic DNA rearrangements,29-32 the origin of the 100 kD sT-ag remains speculative. sT-ags of 100 kD have been reported in SV40 transformed cells for more than 30 years.27,28,33,34 Smith and colleagues describe a 100 kD sT-ag in transformed cells from rat and mouse, which exhibited a 35S-methionine fingerprint virtually identical to that of normal T-ag with neither additional nor missing methionine tryptic peptides.28 They speculated that a part of the T-ag might be duplicated and the mRNA coding for that protein might be the product of abnormal splicing of an RNA transcribed from genomic tandem integration. The 100 kD sT-ag described by Chen and colleagues33,34 in transformed mouse cells was found to be non-lytic, occurred independent of t-ag expression, was strongly associated with independence of anchorage requirement, was generated in cells transformed by molecularly cloned wild-type SV40 DNA, and required an intact SV40 origin of replication. The coding sequence of that 100 kD sT-ag was determined to include two separate partial repeats of the SV40 genome and the protein was speculated being generated from the spliced primary transcript.35 Finally in 1993, Kao and colleagues36 reported the generation of a sT-ag in transformed SV40-immortalized human uroepithelial cells. This 100 kD protein exhibited a 49 amino acid (aa) duplication from codons 83–132 of the T-ag, in which codon 132 was found just before the splice donor site and codon 83 representing the 5′ end of exon 2 of the SV40 T-ag just after the splice acceptor site, indicating the mRNA might have been generated by splicing. The origin of this sT-ag transcript in human cells was not clarified by further experiments but a double cis-splice reaction of a transcript generated from two integrated partial repeats of SV40 was discussed.
Here, we observed the formation of a 100 kD SV40 sT-ag under conditions which exclude the formation of the t-ag, i.e., when transforming cells with a SV40 construct deleterious of the t-ag 5′ss. This 100 kD protein was found to have superior cell transformation activity as compared with the regular 94 kD T-ag and was generated by an alternative RNA splice process involving trans-splicing. DNA sequencing revealed the formation of this 100 kD sT-ag was not associated with genomic SV40 DNA rearrangements, but instead supported by a single nucleotide mutation in the T-ag 5′ss. This mutation weakens this 5′ss, impairs its employment in cis-splicing to the downstream T-ag 3′ss, and favors the usage of an alternative cryptic 5′ss located downstream of the T-ag 3′ss. As one has to exclude splicing between a downstream (cryptic) 5′ss to an upstream 3′ss (that would generate a circular RNA), this cryptic 5′ss must be located on a second SV40 pre-mRNA molecule and the underlying splice process therefore constitutes a RNA trans-splice process. As compared with the T-ag mRNA, the sT-ag mRNA had gained 147 additional nucleotides coding for a duplicate of the N-terminal transforming pRb binding domain of the SV40 T-ag, which was presumably the reason for the aggressive growth of the sT-ag transformed cellular clones. It is possible but not proven that this 100 kD sT-ag resembles the specimen described previously by Kao and colleagues.36 The 100 kD sT-ag and its trans-spliced mRNA were detectable in the highly transformed clones containing the DNA with a mutation in the T-ag 5′ss but could also faintly be detected in one of the wild-type transformed clones. Thus, RNA trans-splicing is to be considered as a mechanism intrinsic and ubiquitous to the SV40 and possibly other viral systems increasing the diversity of viral sequences and phenotypes. This example further creates a link between RNA trans-splicing and the generation of highly transforming viral clones.
Results
In vitro transformation assay with rodent cells identifies highly transformed clones expressing a 100 kD SV40 sT-ag
To investigate the role of the 94 kD SV40 T-ag on cellular transformation in the absence of the 17 kD t-ag, which might have a helper function on cellular transformation,37 Rat-2 cells were transfected with the plasmid pSV-T/Δ5′t, which only expresses the T-ag. The 94 kD SV40 T-ag is encoded by two exons that are separated by a 346 nt intron (Fig. S1).38,39 Exon 1 codes for the first 82 amino acids and exon 2 codes for amino acids 83–708. Exon 2 contains two DNA sequences that bind and inactivate the tumor suppressor proteins p53 and pRb and thereby have a transforming effect in rodent cells. The pRb binding domain, essential for cellular transformation, is located between amino acids 103 and 107. Truncated SV40 T-ags with Met 109 in exon 2 as the first amino acid therefore are non-transforming, as shown before.11 The 17 kD t-ag is generated from the same pre-mRNA that codes for the T-ag but by an alternative splice process using a 5′ss 288 nt downstream the T-ag 5′ss. As this extended exon 1 of the t-ag contains a translation termination signal directly upstream of the t-ag 5′ss, the coding sequence of exon 2 is not used for 17 kD t-ag protein synthesis (Fig. S1). Therefore, the t-ag does not contain a pRb or p53 binding domain and does not have a transforming effect, although it may support the transforming activity of the T-ag.37,40 The transfection of Rat-2 cells with plasmid pSV-T/Δ5′t resulted in two cellular clones, clones 2 and 7, with a denser growth pattern compared with the other clones, displaying multilayer and superconfluent growth patterns in soft agar plates. This indicated a stronger transforming activity of the SV40 DNA in these clones possibly due to a spontaneous SV40 mutation or a DNA rearrangement. Analysis of immunoprecipitated SV40 T-ag proteins of these two Rat-2 cell clones indicated the presence of a 100 kD sT-ag in addition to the normal 94 kD T-ag (Fig. 1). Notably, a very faint 100 kD sT-ag signal was also present in the normally transformed clone 5.
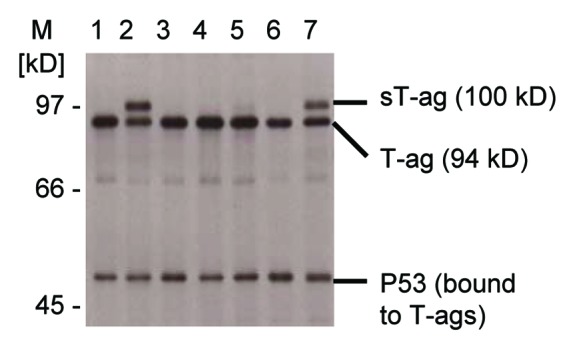
Figure 1. Detection of a 100 kD SV40 sT-ag in SV40-transfomed rat cell clones. SDS gel electrophoresis of immunoprecipitated SV40 T-ag-specific proteins from seven (lane 1 to 7) rat 2 cell lines that were transfected with SV40 DNA coding for the 94 kD T-ag. All cell clones expressed the regular 94 kD T-ag and co-immunoprecipitated T-ag-bound p53. Two highly transformed cell clones 2 and 7 strongly expressed the additional 100 kD sT-ag. A faint 100 kD sT-ag band was also detectable for the normally transformed cell clone 5.
sT-ag-expressing cell lines contain a transcript with a partial duplication of the T-ag exon 2 sequence
RT-PCR analysis of clones 2 and 7 revealed that the 100 kD sT-ag originated from a sT-ag mRNA that was about 150 nucleotides longer than the 462 nt amplicon relating to the normal 94 kD T-ag, as shown by an additional 609 nt RT-PCR product generated with primers g and w that bind to exon 1 and exon 2 of the T-ag RNA (Fig. 2). By using RT-PCR primers that exclusively bind to exon 1 or the distal part of exon 2 of the T-ag, however, this additional RT-PCR product was not observed. Sequencing of the isolated 609 nt RT-PCR product derived from this 100 kD sT-ag mRNA revealed a 147 nt duplication of the SV40 T-ag coding sequence between nucleotides 4571–4425, corresponding to a duplication of amino acids 83–131 (Fig. 2C). This region contains the binding region for pRb and is essential for transformation of rodent cells and growth of rodent cells in soft agar.11,41,42 We therefore hypothesized that the duplication of this domain was responsible for the more aggressive growth of the 100 kD sT-ag-expressing clones 2 and 7 as compared with the other cell lines, which predominantly expressed the 94 kD T-ag. Notably, a weak signal relating to the 100 kD sT-ag mRNA was also obtained from some normally transformed clones. That signal was most prominent in clone 5 (Fig. 2A).
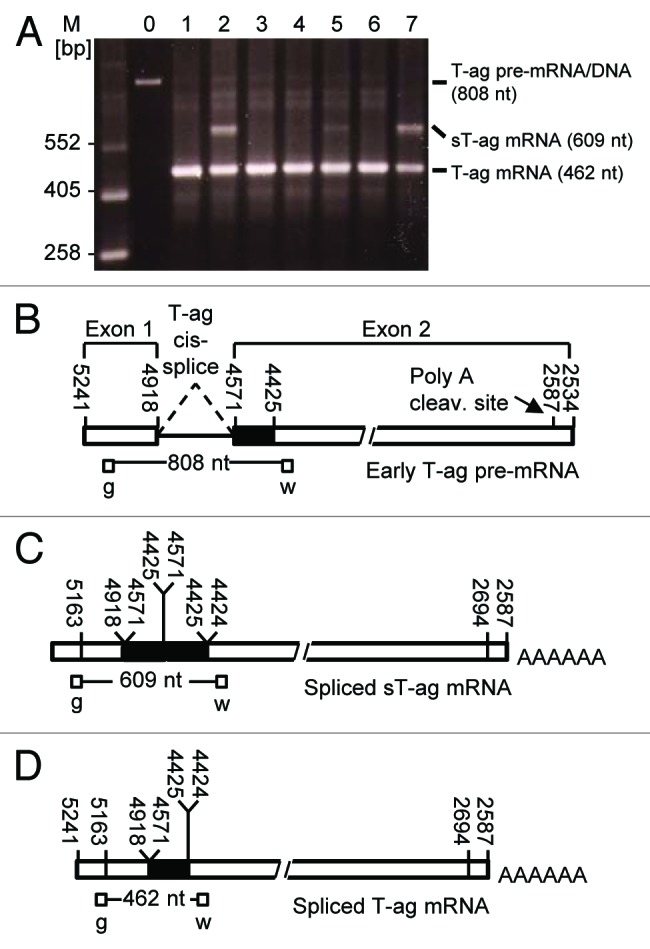
Figure 2. RT-PCR revealed sT-ag-expressing cell lines contained a transcript with a partial duplication of T-ag exon 2 sequences. (A) SV40-specific RT-PCR products of seven cell clones (lane 1 to 7) generated with the PCR-primers g and w that bind upstream and downstream the 346 nt T-ag intron. Lane 0 shows the 808 nt PCR product when the SV40 T-ag DNA was used as template—RT-PCR using the T-ag pre-mRNA as template generates the same PCR product. The T-ag mRNA (462 nt product) but not the T-ag pre-mRNA/genomic DNA was detected in all clones and an additional 609 nt band, but no 945 nt (609 nt + 346 nt) band, was detected in the sT-ag expressing clones. (B) Organisation and primer binding sites of the early T-ag pre-mRNA. (C) Organization and primer binding sites of the spliced sT-ag mRNA. (D) Organization and primer binding sites of the spliced T-ag pre-mRNA.
Sequence duplication in the sT-ag-associated transcript is not related to an equivalent DNA duplication
As SV40 DNA can rearrange in the mammalian host genome, partial SV40 DNA duplications may be generated that can code for sT-ags of sizes up to 145 kD in contrast to the regular 94 kD T-ag. The reported 145 kD30 or 115 kD SV40 sT-ags,27 for example, were generated by a partial 1212 or 573 bp SV40 DNA duplications coding for the additional 404 or 191 aa, respectively. Such a mechanism was also postulated to generate the 100 kD sT-ag.29,32 Therefore, we searched by PCR for a partial DNA duplication of about 150 bp in the genomic DNA that could be associated with the 100 kD sT-ag transcript using the primers g and w. However, in contrast to the RT-PCR analysis, the DNA of all clones, including clone 2 and 7, revealed a single PCR product of 808 nucleotides (Fig. 3), corresponding to the genomic distance of the primer binding sites (Fig. 2B). As an additional DNA PCR product of 955 (808 + 147) nt was not observed, a direct partial SV40 DNA duplication of 147 nt as the cause for sT-ag mRNA synthesis in clones 2 and 7 could be excluded.
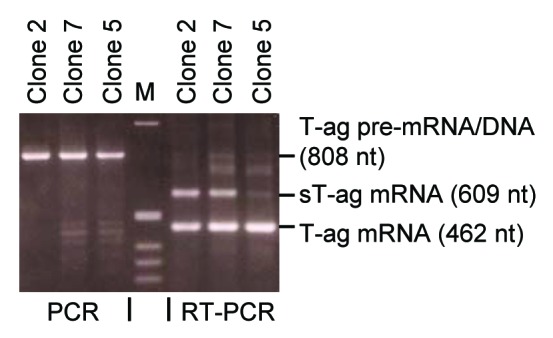
Figure 3. PCR on genomic DNA of sT-ag-positive cell clones revealed the sT-ag is not directly DNA-encoded. PCR / RT-PCR analyses of genomic DNA (lanes 1–3)/cellular RNA (lanes 4–6) of strongly (clones 2 and 7) and weakly (clone 5) expressing rat 2 cell clones. RT-PCR using primers g and w detected in all clones a 462 nt band resulting from the spliced T-ag mRNA as well as a 609 nt band related to the sT-ag mRNA, which was distinct in clones 2 and 7 and faint in clone 5. PCR from the genomic DNA detected in all clones a 808 nt band, which corresponds to the regular distance of the primer binding sites on the integrated SV40 DNA. A 955 nt (808 nt + 147 nt) PCR product relating to a direct genomic DNA template for sT-ag mRNA synthesis, e.g., as a result of a DNA rearrangement with a partial 147 nt SV40 DNA duplication, was not observed.
The 100 kD sT-ag-associated transcript is generated by a RNA splicing process using a cryptic 5′ss within the second T-ag exon
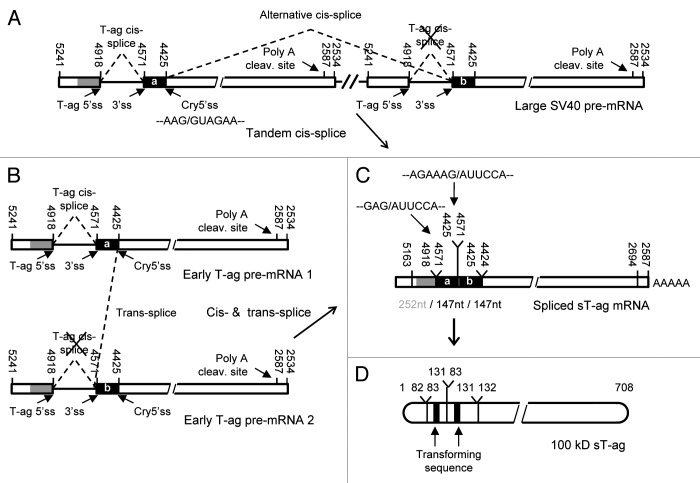
Sequence analysis of the 609 nt RT-PCR products derived from clones 2 and 7 revealed that these cDNAs contained the last 252 nts of exon 1 (SV40 nts 5169–4918, primer g starts at 5169) followed by a second segment with the first 147 nts of exon 2 (SV40 nts 4571–4425), which was followed by a third segment containing the first 210 nts of exon 2 (SV40 nts 4571–4362, primer w ends at 4362) of the T-ag (Fig. 4). Consequently, this cDNA sequences related to an RNA that was generated (1) by regular splicing of exon 1 to exon 2 of the T-ag and (2) a second splice process in which a cryptic splice site with the RNA sequence AAG/GUA GAA located 147 nts downstream of the proximal end of exon 2 was combined with the upstream T-ag 3′ss. As in this case a downstream 5′ss had been combined to an upstream 3′ss, this second splice process generating the spliced sequence ... AGA AAG/AUU CCA ... (/represents a splice junction) requires two T-ag pre-mRNA molecules or pre-mRNA portions for splicing. One explanation for such a result is that these two T-ag pre-mRNA portions lie on a common very large SV40-specific pre-mRNA molecule generated from a tail-to-head tandem SV40 DNA duplication integrated in the mammalian host genome. This SV40 DNA duplication then might generate a very large SV40-specific pre-mRNA, which contains two SV40 portions that might be directly linked, or be separated by a DNA linker of host cellular genomic DNA. The second splice generating the spliced sequence ... AGA AAG/AUU CCA ... would combine the cryptic 5′ss within exon 2 of the proximal SV40 pre-mRNA portion with the T-ag 3′ss of the distal SV40 pre-mRNA portion within this large pre-mRNA by conventional cis-splicing (Fig. 4A). To test the hypothesis of a tandem repeat integration of SV40 DNA into the host cell genome, a southern blot analysis was performed. For DNA cleavage we used the restriction enzymes BamHI and EcoRI, both of which cut once downstream of the HpaII-BamHI DNA segment in the multiple cloning site of the pSPT19 vector DNA but not within the early SV40 DNA HpaII–BamHI segment (Fig. 5A). A single copy insertion would give rise to a single SV40-specific fragment of 3.1 kb or longer whereas in case of a dual or multiple copy integration of SV40 DNA into the host genome two or more specific DNA fragments should be detected by the DNA probe. However, both enzymatic digestions generated only one SV40-specific DNA fragment (Fig. 5B). A dual or multiple integration of SV40 DNA within a distance of up to approximately 10 kb would also be detectable by PCR. Using PCR primer m, which binds at the distal part of exon 2 and elongates the DNA in a downstream direction, together with primer n, which binds at the proximal part of exon 1 and elongates the DNA in the upstream direction (Fig. 5C), and a primer extension time of 10 min, SV40 DNA tandem integrations in a distance of up to 10 kb were analysed, but no SV40-specific signal was detected. In two positive PCR-controls using circular viral wt 5243 bp SV40 DNA or the circular plasmid 6.1 kb psVT-t DNA as templates, primers m and n generated as expected a 2.8 kb (2798 bp) or 3.8 kb PCR fragment, respectively (data not shown). On the basis of both experiments, the southern hybridization and the PCR assay, a dual or multiple genomic integration of SV40 DNA as origin of the sT-ag transcript could be excluded. Notably, even if a SV40 DNA tandem repeat would have occurred, the generation of a common large pre-mRNA molecule containing two SV40 gene portions would be unlikely, as the early SV40 DNA HpaII–BamHI segment contains the signal sequences for RNA polyadenylation upstream and nearby the BamHI site (Fig. 4A). These polyadenylation signals would prevent a pre-mRNA elongation beyond the BamHI site of the upper and towards the lower SV40 DNA tandem repeat portion.
Figure 4. Schematic illustration of two mechanisms that can lead to the generation of the sT-ag mRNA and protein. (A) Alternative RNA cis-splicing of a long pre-mRNA transcript originating from genomic SV40 DNA tandem integration; this mechanism involves two cis-splice events, i.e., a regular splice from the T-ag 5′ss to the common 3′ss and a second alternative splice from the cryptic 5′ss related to the first SV40 genome to the 3′ss related to a second SV40 DNA tandem repeat. (B) RNA trans-splicing between two early T-ag pre-mRNAs; this mechanism involves a regular cis-splice of the first transcript from the T-ag 5′ss to the common 3′ss and a second alternative trans-splice from the cryptic 5′ss of the first transcript to the 3′ss of the second transcript which is not spliced in cis. (C) Structure of the identified spliced mRNA coding for the sT-ag, which contains a 147 nt partial duplication of exon 2 sequences. Linkages between exon 1 and exon 2 sequences as well as between the exon 2 repeats resemble the splice sites and are consistent with a regular splice from the T-ag 5′ss to the common 3′ss and a second alternative splice from the cryptic 5′ss to a duplicate of the 3′ss. a and b: identical sequence stretch being duplicated in the sT-ag mRNA. (D) Structure and aa numbering of the observed 100 kD sT-ag harboring a duplication of the transforming Rb binding domain (black rectangle).
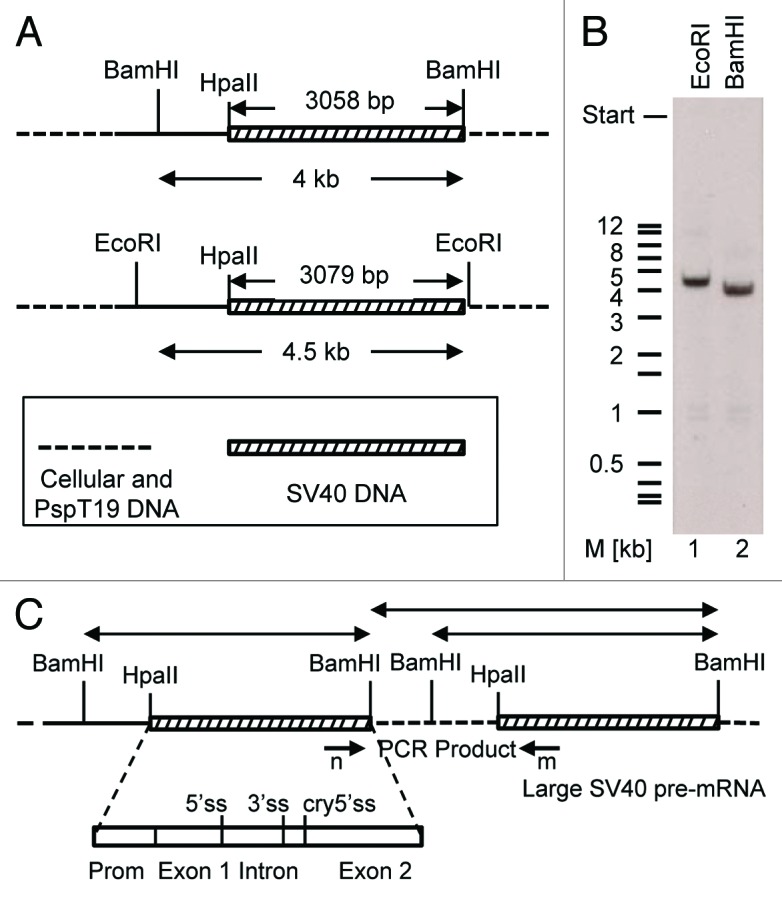
Figure 5. Sequence duplication in the sT-ag-associated transcript was not related to an equivalent DNA duplication. (A) Enzymatic cleavage scheme of genomic DNA for southern hybridization. The genomic DNA of a sT-ag producing clone was digested with EcoRI or BamHI. Both enzymes cut outside the SV40 DNA coding for T-ag: once in the multiple cloning site of the vector pspT19 that was used for cloning of the SV40 HpaII–BamHI early SV40 DNA segment reconstituting the BamHI site, and once upstream in a distance of about 3.1 kb (size of the HpaII-BamHI fragment) or more within the host genomic DNA. (B) Single bands of about 4.5 kb (EcoRI, lane 1) or 4 kb (BamHI, lane 2) indicated the integration of a single copy of the early SV40 DNA HpaII–BamHI segment into the genomic DNA of the host cell. (C) Hypothetical HpaII–BamHI SV40 DNA segment tandem duplication. BamHI (or EcoRI) digestion of the genomic DNA would generate two SV40-specific BamHI fragments. If the host-genomic DNA linker sequence does not contain a BamHI site, then the size of one fragment would indirectly indicate the distance of the two integrated HpaII–BamHI SV40 DNA segments in the genomic host DNA. A PCR from the genomic DNA with primers n and m, binding to the 3′end of exon 1 and the 5′end of exon 2, respectively, using conditions which favor the amplification of long DNA fragments, did not give any product. These results exclude a tandem integration of SV40 DNA into the genome of the sT-ag-producing host cell clone.
The 100 kD sT-ag-associated transcript is generated by intermolecular RNA trans-splicing between two separated early SV40 pre-mRNA molecules
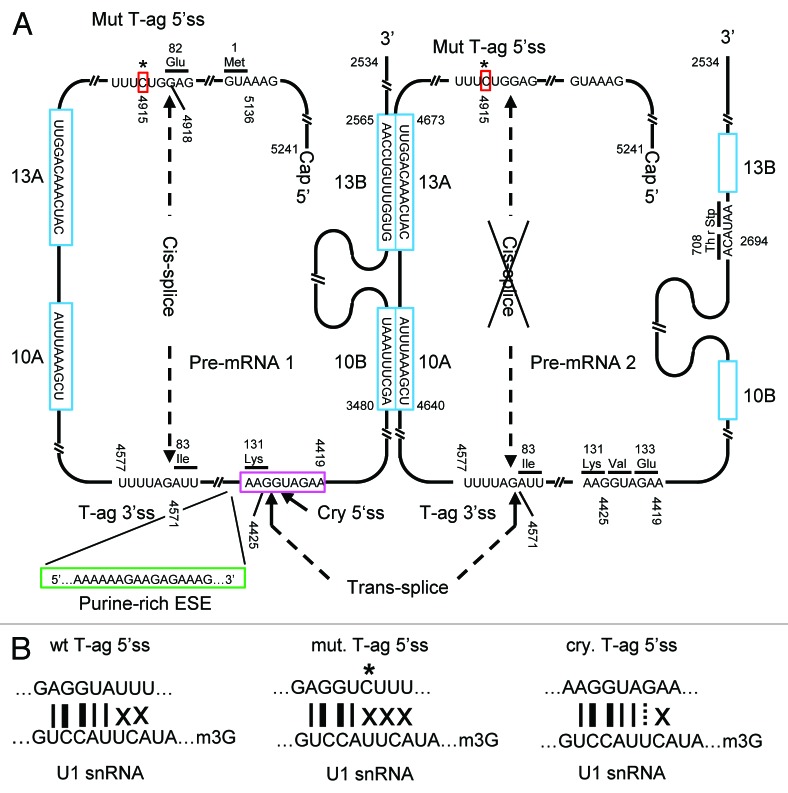
All the experiments described above indicate that a trans-splice reaction between two SV40 pre-mRNA molecules originating from a single copy genomic insertion of SV40 DNA remains to be the only plausible explanation for sT-ag formation in the cells of clones 2 and 7. Trans-splicing occurs between the cryptic 5′ss of the first SV40 pre-mRNA molecule and the T-ag 3′ss of the second transcript (Figs. 4B and 6). Notably, trans-splicing between these two splice sites of SV40 was reported earlier after microinjection of in vitro transcribed SV40 early transcripts into the nuclei of Rat-2 cells in the absence of genomic SV40 DNA.12 To generate the 100 kD sT-ag mRNA, the 346 nt T-ag intron of the first SV40 pre-mRNA molecule is additionally removed by conventional cis-splicing, either before or after trans-splicing has occurred.
Figure 6. Illustration of sT-ag mRNA synthesis by combining two T-ag pre-mRNA molecules via RNA trans-splicing. (A) The super T antigen mRNA is generated by combining the cryptic 5′ss (highlighted in magenta) of the first pre-mRNA molecule to the T-ag 3′ss of the second pre-mRNA molecule via RNA trans-splicing. Whereas the involvement of the T-ag 3′ss in trans-splicing abrogates cis-splicing of the second pre-mRNA, the first pre-mRNA is additionally spliced in cis using the T-ag 5′- and 3′ss. Besides the presence of a strong cryptic 5′ss, which is in 6 nts complementary to the U1 snRNA, trans-splicing is supported by several helper functions: First, two pairs of potential RNA dimerization domains (highlighted in cyan) of 10 (10A + 10B) and 13 (13A + 13B) complementary nucleotides in length which can generate steric proximity between the trans-splice sites; second, a putative purine-rich exonic splice enhancer sequence (highlighted in green) directly upstream of the cryptic 5′ss. In the strongly sT-ag expressing clones, trans-splicing is further supported by a A to C point mutation (highlighted in red) within the T-ag 5′ss, which weakens this ss by reducing the complementarity to the U1 snRNA from 5 to 4 nt in favor of the use of the cryptic 5′ss. (B) Complementarity between the wild-type T-ag 5′ss, the mutated T-ag 5′ss, and the cryptic 5′ss. Thin lines, A = U bp; thick lines, G ≡ C bp; dashed lines, GסU wobble bp; X, mismatch.
Finally, we investigated why trans-splicing and subsequent generation of the 100 kD sT-ag was much more efficient in the Rat-2 cell line clones 2 and 7 as compared with other clones. Sequencing of the SV40 DNA revealed a single A to C point mutation within the T-ag 5′ss of both highly transformed cell lines, but not in normally transformed clones; i.e., the T-ag 5′ss wt sequence GAG/GTA TTT was found to be mutated to the sequence GAG/GTC TTT only in the highly transformed clones.
Discussion
We describe the occurrence and the formation pathway of an aberrant 100 kD SV40 sT-ag in a fraction of highly transformed rodent cells stably transfected with a gene coding for the T-ag of SV40. The 100 kD sT-ag was found to contain a duplication of the transforming Rb binding domain of the T-ag encoded by a 147 nt RNA duplication and sequencing revealed that the dual arrangement was obviously the result of a splice process (Fig. 4). 100 kD sT-ags have been repeatedly reported to regularly occur in SV40 transformed rodent and in human cells. It is likely that all these reports describe the same protein, which can be generated in two different ways: either by alternative tandem-cis-splicing36 or by RNA trans-splicing as evidenced in this work. That both alternative splice mechanisms can generate identical proteins was demonstrated earlier for a truncated SV40 T-ag.43 Here we excluded a tandem-cis-splice as possible cause by southern hybridization and PCR analysis. Thus, a trans-splicing reaction between an unspliced and a cis-spliced variant of the T-ag pre/mRNA is the only explanation for the emergence of the aberrant transcript and its gene product.
In principle, intra-molecular cis-splicing, in which splice donor and splice acceptor sites are covalently linked, is favored compared with intermolecular trans-splicing. However, alternative cis- and trans-splicing can be triggered when regular 5′- or 3′ss cis-splices sites are missing or weakend by mutations.44 In our example, the T-ag 3′ss favors the use of an alternative cryptic 5′ss in trans because the regular T-ag 5′ cis ss was weakened by a single A to C point mutation (Fig. 6B). Consequently, only four instead of 5 nts of the 9 nt 5′ splice donor can base-pair to the U1 snRNA of the spliceosomal U1 snRNP, likely impeding its recruitment to this 5′ss. The alternatively used cryptic 5′ss can bind with 6 of 9 nts to the U1 snRNA and, hence, represents a much stronger splice donor site (Fig. 6B). RNA secondary structure predictions further indicated that the T-ag 5′ss point mutation renders this ss less accessible for U1 snRNA binding suggesting that this mutation disturbs both, the thermodynamics and the kinetics of U1 snRNA annealing (Fig. S2). In contrast to the mutated T-ag 5′ss, the alternative cryptic 5′ss is closely situated downstream of the purine-rich sequence 5′ … AAAAAAGAAG AGAAAG ... 3′ of exon 2 (Fig. 6A). Purine-rich RNA sequences can enhance the recruitment of the U1 snRNP to a downstream 5′ss thus, functioning as exonic splice enhancer (ESE).45,46 A similar purine-rich sequence 5′ ... GAAGAAG ... 3′ was shown to act as ESE in trans-splice reactions involving the rat carnithine octanoyltransferase RNA.13 Together, the alternative cryptic 5′ss represents an excellent site for U1snRNP binding and splicing.
A more tight steric ss fixation in trans can be achieved by complementary antisense binding domains, which were shown to strongly support bimolecular RNA-RNA trans-splice reactions in vitro.9,10 It remains to be tested if, analogous to antisense RNA-mediated inhibition of gene expression,47 RNA trans-splicing is a kinetically controlled process, i.e., depends on the on-rate of binding between splice-donor and -acceptor RNA. We identified two-plus-two potential RNA dimerization domains with a respective length of 10 (10A+10B) and 13 (13A+13B) complementary nucleotides on the SV40 T-ag pre-mRNA molecule, which appear to be suitable to promote trans-splicing (Fig. 6A). However, we haven’t confirmed this postulated RNA::RNA dimerization of the SV40 T-ag pre-mRNA yet. Dimerization domain 13B is located downstream of the SV40 polyA cleavage site and, hence, can only be functional if dimerization occurs faster than and anticipates 3′ end cleavage and polyadenylation. However, U1 snRNP binding could protect the unspliced transcript from cleavage and polyadenylation and, thus, favor trans-splicing.48 Basically, RNA tri- or multi-merization could even lead to tri- or multi-partite mRNA and larger sT-ag variants, which were not observed (Fig. S3). Exclusively trans-spliced transcripts would generate the 17 kD t-ag only but no T- or sT-ag. The resulting clones would not be transformed and could not be selected in soft agar. Thus, a trans-splice in the absence of an additional cis-splice may have occurred but was not detectable under our assay conditions that highlights that the combination of cis- and trans-splicing triggers a growth advantage by generating a highly transforming 100 kD sT-ag. The cis-splice likely occurs prior to or even supports the trans-splice, which would be consistent with the observation that cis-splicing of a trans-splicing RNA enhances trans-splicing activities.49
Intron-like bridging of splice sites via intermolecular RNA::RNA antisense base-pairing may favor, but is not an absolute prerequisite for RNA trans-splicing as demonstrated by in vitro trans-splicing experiments where RNA::RNA-annealing regions were omitted on the corresponding pre-mRNA substrate molecules.9,50,51 In that case splice sites are assumed to combine via “bridging”-interactions between the spliceosomal proteins and RNAs bound before to the separated 5′ and 3′ss.52
Many mutations of the SV40 virus were described.53 Since the cells in our assays were transfected with plasmid DNA originating from a single clone, one has to assume that the mutation within the T-ag 5′ss was created by the transformed cells rather than by the investigator. The generation of this mutation found in two highly transformed cellular clones, was likely supported by the ability of the SV40 ori-containing plasmid DNA to replicate endogenously in the presence of the T-ag, giving rise to high DNA turnover and error-prone duplication. A dependence of 100 kD sT-ag formation on an active origin of replication was evidenced previously.33
As to the SV40 virus, which is not being considered a human tumor virus so far, one cannot exclude that spontaneously occurring mutations triggering the formation of the 100 kD sT-ag or other gene products, convert it into a highly transforming virus exhibiting higher pathogenic potential than the wild-type virus or as originally thought and, thus, being involved in human tumorigenesis.
RT-PCR and less pronounced immunoprecipitation indicate the formation of the sT-ag transcript and protein and, hence, trans-splicing even in wild-type transformed clone 5, which does not carry the 5′ donor ss mutation (Figs. 1–3). Therefore, the 100 kD sT-ag may be formed without viral mutagenesis under conditions which support trans-splicing, e.g., if, after genomic integration, transcription is controlled by a strong promoter giving rise to high endogenous concentrations of SV40 primary transcripts, which support the kinetics of their dimerization as a prerequisite for intermolecular RNA trans-splicing. As opposed to tandem integration-based generation of sT-ags, trans-splicing-based generation is independent of genomic integration. Thus, even replicating episomal SV40 DNA may give rise to the generation of the highly transforming non-lytic 100 kD sT-ag and thereby contribute to the high reported rate of malignant transformation associated with episomal SV40 in human mesothelial cells.54
In the homologous trans-splice reaction described here, two pre-mRNAs coding for the regular 94 kD T-ag (containing each a single copy of the transforming region) combine to a single transcript coding for the 100 kD sT-ag (including two copies of the transforming region). That is, the total copy number of the transforming Rb binding region is independent of the splicing mechanism indicating that the molecular duplication rather than the overall abundance of the transforming region is associated with the elevated transforming activity of the sT-ag, though we cannot exclude that transformation depends on the coexistence of the T-ag. The T-ag vs. sT-ag cDNA (Fig. 3, clones 2 and 7) levels point toward an approximate 2:1 ratio of exclusively cis-spliced vs. trans-spliced (50% of which are cis-spliced as well) transcripts. A similar ratio was observed for the related gene products (Fig. 1, lanes 2 and 7). Assuming comparable rates of processing and RNA stability of the normally cis-spliced and the trans-spliced transcript, a 2:1 ratio suggests that about half of the pre-mRNA transcripts are spliced in trans. That is much compared with rates in the single-digit percentage area reported for artificial trans-splicing RNA that is expressed from an integrated construct.55 In the example reported here, trans-splicing is supported by a variety of molecular features, some of which may be explored toward the design of therapeutic trans-splicing RNA: (1) mutation-based weakening of the regular 5′ cis ss, (2) the absence of an alternative 5′ cis ss upstream, and (3) the presence of an alternative cryptic 5′ss downstream of the 3′ss, whereby the use of that cryptic 5′ss is likely supported by (4) splice enhancer sequences and (5) two pairs of rather short putative RNA dimerization domains.
The detection of RNA trans-splicing in the presence/absence of the T-ag 5′ss as described in this work/previously11 together with the ubiquitous occurrence of the 100 kD sT-ag, suggests trans-splicing and, thus, generated gene products as common process intrinsic to SV40 infections of mammalian, including human cells. The alternative cryptic T-ag 5′ss was shown being recruited for trans-splicing with the T-ag 3′ss even under conditions where the regular t-ag 5′ss (but not the T-ag 5′ss) was present and not mutated.43 Inhibition of competitive cis-splicing by deletion of the t-ag 5′ss, a situation which corresponds to the experimental setup of this study, supported trans-splicing, which was not observed at all when the cryptic T-ag 5′ss was mutated.12 Thus, trans-splicing of SV40 transcripts occurs inescapably once the cryptic T-ag 5′ss and the common T-ag/t-ag 3′ss are available. We hypothesize homologous trans-splicing as a novel mechanism to increase protein variation in SV40 and possibly other viruses as well.
This hypothesis is supported by previous descriptions of heterologous trans-splicing that all involved viral transcripts, such as between the HIV-nef pre-mRNA,56 the adenoviral major late transcript,17 or a HCV transgenic transcript57 and cellular sequences or between pre-mRNAs from SV40 and HIV.56 Functions have not been assigned to the resulting fusion proteins. Notably, all these trans-splicing events involved cryptic viral ss and were not associated with mutations of the regular cis ss. Thus, the concurrence of potential trans-splicing helper functions identified in this work likely is not coincidental but instead these functions constitute intrinsic characteristics of the viral sequences leading them to trans-splicing. It is not surprising that viruses as the smallest replicating genetic specimen use the mechanism of trans-splicing, which doesn’t enlarge the genomic size, for the generation of viral proteins and phenotypes.
This first example of the generation of a highly transforming tumor antigen creates a link between RNA trans-splicing and oncogenesis even beyond the scope of SV40 or other viral infections. Estimated 15% of all diseases with genetic origin are caused by point mutations that impede the use of regular ss through different mechanisms thereby fostering the activation of alternative new or cryptic ss and leading to the formation of aberrant mRNAs and gene products.58 Cryptic splice sites, in particular 3′ss in 5′ terminal exons and 5′ss in 3′ terminal exons as in the example reported here, seem to be most suitable for trans-splicing as they are not involved in and cannot get lost by regular cis-splicing. In cancer cells, alternative splicing generates a variety of aberrant transcripts, many of which are directly or indirectly involved in carcinogenesis.59-63 Since mammalian cells are generally trans-splicing competent,13 one may speculate that a group of human diseases of unknown molecular biological genesis including cancers are attributed to alternative RNA trans-splicing.64
Materials and Methods
SV40 DNA plasmid constructs
To analyze the transforming activity of SV40 oncoproteins in different mammalian cell types, the entire early SV40 3058 bp HpaII–BamHI DNA segment, which also includes the origin of replication, was cloned into the AccI/BamHI cloning sites of the 3.15 kb pSPT19 plasmid DNA (Pharmacia), generating plasmid pSV-T/t as described previously.11 This DNA contains the early SV40 T-ag promoter and the coding sequence for both, the t-ag and T-ag, which are generated from a common pre-mRNA by an alternative splice process using a common 3′ss but two different 5′ss. In order to investigate the transforming activity of the 94 kD T-ag in the absence of the 17 kD t-ag, which under some conditions might enhance cellular transformation, t-ag formation was anticipated by mutational inactivation of the t-ag 5′ss G/GTA sequence at nt 4638–4635, which was converted to the sequence G/CGC by primer directed mutagenesis. The resulting plasmid pSV-T/Δ5′t expresses the T-ag but not the t-ag.
Cells and DNA transfer
Thymidine kinase (TK) negative Rat-2 cells were cultivated in Dulbecco’s modified Eagle medium (DMEM) supplemented with 5% fetal calf serum. Using calcium phosphate transfection standard protocols, the SV40 pSPT19 DNAs (pSV-T/t or pSV-T/Δ5′t) were co-transfected with plasmid pHSV106, which expresses the herpes simplex virus thymidine kinase gene. Successfully transfected Rat-2 cell clones were selected in HAT medium (hypoxanthin, aminopterin, and thymidine) and transformed clones were subsequently selected in soft agar.
SV 40 T antigen protein preparation
About 107 Rat-2 cells were incubated for 1 h at 37 °C in a methionine-free DMEM, and afterwards labeled for 3 h with 10 µCi [35S] methionine. Cells were washed with PBS and lysed with 1% Nonidet P40 (NP 40), 150 mM NaCl, 10 mM β-mercaptoethanol, and 20 mM Tris-HCl (pH 8.6) at 4 °C for 30 min. After lysis the cell debris was pelleted and the supernatant was used for immunoprecipitation with the SV40-specific antibody Ab2 (Oncogene Science), which recognizes an epitope within the T-ag amino acid sequence position 83–108.12 Proteins were separated on a 7.5% SDS-polyacrylamide gel, and visualized by autoradiography.
Isolation of cellular DNA and RNA
The genomic DNA or RNA from 1–5 × 107 cells was isolated using Trisolv reagent (Biotecx Laboratories) according to manufacturer’s recommendations. The RNA was further purified from contaminating DNA by digestion with RQ-1 RNase-free DNase (Promega) in 50 mM Tris, 10 mM NaCl, 6 mM MgCl2, and 10 mM CaCl2 (pH 8.0) at 37 °C for 60 min. Successful elimination of SV40-specific DNA in total RNA was controlled and confirmed by PCR using SV40-specific primers.
Southern blot analysis
Genomic DNA (10 µg) was digested with 2 U EcoRI or BamHI per µg DNA at 37 °C for 6 h. DNA fragments were separated on a 1% agarose gel, transferred to a nylon filter (Hybond, Amersham) and UV cross-linked. The hybridization was performed for 12 h at 65 °C in a buffer containing 1% BSA, 7% SDS, 0.5 M sodium phosphate (pH 7.3), and 32P-labeled SV40 DNA, using the 3.06 kb HpaII-BamHI DNA segment. The filter membrane was washed with 1% SDS, 40 mM sodium phosphate (pH 7.3) at 65 °C three times for 30 min. The blots were exposed for 1–7 d at -70 °C to a X-ray film (Kodak).
cDNA synthesis
Total cellular RNA (5 µg) was converted into single-stranded cDNA using either 0.5 µg oligo-dT15 primer or 0.1 µg of SV 40-specific primers, 0.5 mM of each dNTP, and 200 U M-MLV reverse transcriptase (BRL) in 20 µl enzyme buffer at 37 °C (oligo-dT15 primer) or 45 °C (SV40-specific primers) for 2 h.
PCR and SV40-specific primers
PCR was performed as described in standard protocols either with 1/10 of the volume (2 µl) of the cDNA preparation or 1 µg isolated genomic DNA, 0.2 mM of each dNTP, 0.5 µg of a pair of SV40-specific primers, and 1–2 U of Taq DNA polymerase (Perklin Elmer) or proof reading Vent DNA polymerase (NEB) in 100 µl reaction buffer. The temperature profile used in 30 or 35 cycles hot start PCR amplification was 1 min 94 °C, 1 min 52 °C, and 3 min 72 °C. The SV40-specific antisense primers used for PCR or first strand cDNA synthesis were primers n (5′-CTTTATCCAT CTTTGCAAAG-3′, 5′end at SV40 nt position 5154), r (5′-GCTTTAAATC TCTGTAGG-3′, 5′end at SV40 nt position 4644), w (5′-ACTAAACACA GCCATGACT-3′, 5′end at SV40 nt position 4362), k (5′-CTCTGCTTCT TCTGGGTT-3′, 5′end at SV40 nt position 4029), and o (5′-CCAGACATGA TAAGATTAC-3′, 5′end at SV40 nt position 2537). The sense primers used for PCR were primers z (5′-TATTCCAGAA GTAGTGAG-3′, 5′end at SV40 nt position 5226), g (5′-GCAAAGATGG ATAAAGTTT-3′, 5′end at SV40 nt position 5169), and m (5′-CCTGAACCTG AAACATAA-3′, 5′end at SV40 nt position 2708). The t-ag 5′ss mutation in plasmid pSV-T/Δ5′t was generated using primer × (5′-TCTAAGCGCA ATATAAAATT TTTAAGTG-3′, 5′end at SV 40 nt position 4643) with three altered nucleotides (underlined) compared with the SV40 wt sequence. Standard RT-PCR protocols to detect mRNA splicing patterns used the primer combination g and w. To analyze a possible SV40 DNA tandem integration in the genomic DNA of the sT-ag-expressing Rat-2 cell clone 7, the genomic DNA extracted from these cells was PCR analyzed using primers m and n or m and w and a primer extension time of 10 min. With these primer combinations it would have been possible to detect a second HpaII-BamHI segment genomic insertion in up to 10 kb distance of the detected HpaII-BamHI segment.
Cloning and sequencing of PCR products
PCR products used for subsequent sequencing were purified by the Qiagen PCR purification method, 5′-phosphorylated using T4 polynucleotide kinase (BRL), and cloned in the dephosphorylated blunt-ended SmaI site of pUC18 (Pharmacia) prior to transformation of E. coli NM522. Clones were analyzed by restriction enzyme digestions and analytical PCR. Plasmid DNA of positive clones was directly sequenced using the Taq Terminator Cycle sequencing kit (ABS). Dye-terminators were used to label 0.5–1 µg double-stranded DNA for sequence analysis in ABS model 373A.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by NUS-Cambridge Start-up grant R-182-000-163-646 from the National University of Singapore and by New Investigator Grant 1058/2011 from the National Medical Research Council of Singapore, both to VP. We thank E Guhl for technical assistance and S Poddar as well as N Yamamoto for critically reading the manuscript and helpful comments.
Glossary
Abbreviations:
- SV40
simian virus 40
- pRB
retinoblastoma protein
- ss
splice site
- t-ag
small T antigen
- T-ag
large T antigen
- sT-ag
super T antigen
- SL
small leader
- snRNP
small nuclear ribonucleoprotein
- mfe
minimum free energy
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/rnabiology/article/26707
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/26707
References
- 1.Sutton RE, Boothroyd JC. Evidence for trans splicing in trypanosomes. Cell. 1986;47:527–35. doi: 10.1016/0092-8674(86)90617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krause M, Hirsh D. A trans-spliced leader sequence on actin mRNA in C. elegans. Cell. 1987;49:753–61. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hannon GJ, Maroney PA, Nilsen TW. U small nuclear ribonucleoprotein requirements for nematode cis- and trans-splicing in vitro. J Biol Chem. 1991;266:22792–5. [PubMed] [Google Scholar]
- 4.Rajkovic A, Davis RE, Simonsen JN, Rottman FM. A spliced leader is present on a subset of mRNAs from the human parasite Schistosoma mansoni. Proc Natl Acad Sci U S A. 1990;87:8879–83. doi: 10.1073/pnas.87.22.8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tessier LH, Keller M, Chan RL, Fournier R, Weil JH, Imbault P. Short leader sequences may be transferred from small RNAs to pre-mature mRNAs by trans-splicing in Euglena. EMBO J. 1991;10:2621–5. doi: 10.1002/j.1460-2075.1991.tb07804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vandenberghe AE, Meedel TH, Hastings KE. mRNA 5′-leader trans-splicing in the chordates. Genes Dev. 2001;15:294–303. doi: 10.1101/gad.865401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dandekar T, Sibbald PR. Trans-splicing of pre-mRNA is predicted to occur in a wide range of organisms including vertebrates. Nucleic Acids Res. 1990;18:4719–25. doi: 10.1093/nar/18.16.4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romani A, Guerra E, Trerotola M, Alberti S. Detection and analysis of spliced chimeric mRNAs in sequence databanks. Nucleic Acids Res. 2003;31:e17. doi: 10.1093/nar/gng017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konarska MM, Padgett RA, Sharp PA. Trans splicing of mRNA precursors in vitro. Cell. 1985;42:165–71. doi: 10.1016/S0092-8674(85)80112-4. [DOI] [PubMed] [Google Scholar]
- 10.Solnick D. Trans splicing of mRNA precursors. Cell. 1985;42:157–64. doi: 10.1016/S0092-8674(85)80111-2. [DOI] [PubMed] [Google Scholar]
- 11.Eul J, Graessmann M, Graessmann A. Experimental evidence for RNA trans-splicing in mammalian cells. EMBO J. 1995;14:3226–35. doi: 10.1002/j.1460-2075.1995.tb07325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eul J, Graessmann M, Graessmann A. In vitro synthesized SV40 cRNA is trans-spliced after microinjection into the nuclei of mammalian cells. FEBS Lett. 1996;394:227–32. doi: 10.1016/0014-5793(96)00957-X. [DOI] [PubMed] [Google Scholar]
- 13.Caudevilla C, Serra D, Miliar A, Codony C, Asins G, Bach M, Hegardt FG. Natural trans-splicing in carnitine octanoyltransferase pre-mRNAs in rat liver. Proc Natl Acad Sci U S A. 1998;95:12185–90. doi: 10.1073/pnas.95.21.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caudevilla C, Codony C, Serra D, Plasencia G, Román R, Graessmann A, Asins G, Bach-Elias M, Hegardt FG. Localization of an exonic splicing enhancer responsible for mammalian natural trans-splicing. Nucleic Acids Res. 2001;29:3108–15. doi: 10.1093/nar/29.14.3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finta C, Zaphiropoulos PG. Intergenic mRNA molecules resulting from trans-splicing. J Biol Chem. 2002;277:5882–90. doi: 10.1074/jbc.M109175200. [DOI] [PubMed] [Google Scholar]
- 16.Flouriot G, Brand H, Seraphin B, Gannon F. Natural trans-spliced mRNAs are generated from the human estrogen receptor-alpha (hER alpha) gene. J Biol Chem. 2002;277:26244–51. doi: 10.1074/jbc.M203513200. [DOI] [PubMed] [Google Scholar]
- 17.Kikumori T, Cote GJ, Gagel RF. Naturally occurring heterologous trans-splicing of adenovirus RNA with host cellular transcripts during infection. FEBS Lett. 2002;522:41–6. doi: 10.1016/S0014-5793(02)02878-8. [DOI] [PubMed] [Google Scholar]
- 18.Takahara T, Kasahara D, Mori D, Yanagisawa S, Akanuma H. The trans-spliced variants of Sp1 mRNA in rat. Biochem Biophys Res Commun. 2002;298:156–62. doi: 10.1016/S0006-291X(02)02419-1. [DOI] [PubMed] [Google Scholar]
- 19.Tasic B, Nabholz CE, Baldwin KK, Kim Y, Rueckert EH, Ribich SA, Cramer P, Wu Q, Axel R, Maniatis T. Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol Cell. 2002;10:21–33. doi: 10.1016/S1097-2765(02)00578-6. [DOI] [PubMed] [Google Scholar]
- 20.Jehan Z, Vallinayagam S, Tiwari S, Pradhan S, Singh L, Suresh A, Reddy HM, Ahuja YR, Jesudasan RA. Novel noncoding RNA from human Y distal heterochromatic block (Yq12) generates testis-specific chimeric CDC2L2. Genome Res. 2007;17:433–40. doi: 10.1101/gr.5155706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brooks YS, Wang G, Yang Z, Smith KK, Bieberich E, Ko L. Functional pre- mRNA trans-splicing of coactivator CoAA and corepressor RBM4 during stem/progenitor cell differentiation. J Biol Chem. 2009;284:18033–46. doi: 10.1074/jbc.M109.006999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jafar S, Rodriguez-Barradas M, Graham DY, Butel JS. Serological evidence of SV40 infections in HIV-infected and HIV-negative adults. J Med Virol. 1998;54:276–84. doi: 10.1002/(SICI)1096-9071(199804)54:4<276::AID-JMV7>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 23.Viscidi RP, Rollison DE, Viscidi E, Clayman B, Rubalcaba E, Daniel R, Major EO, Shah KV. Serological cross-reactivities between antibodies to simian virus 40, BK virus, and JC virus assessed by virus-like-particle-based enzyme immunoassays. Clin Diagn Lab Immunol. 2003;10:278–85. doi: 10.1128/CDLI.10.2.278-285.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ozer HL, Banga SS, Dasgupta T, Houghton J, Hubbard K, Jha KK, Kim SH, Lenahan M, Pang Z, Pardinas JR, et al. SV40-mediated immortalization of human fibroblasts. Exp Gerontol. 1996;31:303–10. doi: 10.1016/0531-5565(95)00024-0. [DOI] [PubMed] [Google Scholar]
- 25.Gazdar AF, Butel JS, Carbone M. SV40 and human tumours: myth, association or causality? Nat Rev Cancer. 2002;2:957–64. doi: 10.1038/nrc947. [DOI] [PubMed] [Google Scholar]
- 26.Carbone M, Pannuti A, Zhang L, Testa JR, Bocchetta M. A novel mechanism of late gene silencing drives SV40 transformation of human mesothelial cells. Cancer Res. 2008;68:9488–96. doi: 10.1158/0008-5472.CAN-08-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tegtmeyer P. Altered patterns of protein synthesis in infection by SV40 mutants. Cold Spring Harb Symp Quant Biol. 1975;39:9–15. doi: 10.1101/SQB.1974.039.01.004. [DOI] [PubMed] [Google Scholar]
- 28.Smith AE, Smith R, Paucha E. Characterization of different tumor antigens present in cells transformed by simian virus 40. Cell. 1979;18:335–46. doi: 10.1016/0092-8674(79)90053-9. [DOI] [PubMed] [Google Scholar]
- 29.May E, Jeltsch JM, Gannon F. Characterization of a gene encoding a 115 K super T antigen expressed by a SV40-transformed rat cell line. Nucleic Acids Res. 1981;9:4111–28. doi: 10.1093/nar/9.16.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.May E, Lasne C, Prives C, Borde J, May P. Study of the functional activities concomitantly retained by the 115,000 Mr super T antigen, an evolutionary variant of simian virus 40 large T antigen expressed in transformed rat cells. J Virol. 1983;45:901–13. doi: 10.1128/jvi.45.3.901-913.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clayton CE, Lovett M, Rigby PW. Functional analysis of a simian virus 40 super T-antigen. J Virol. 1982;44:974–82. doi: 10.1128/jvi.44.3.974-982.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lovett M, Clayton CE, Murphy D, Rigby PWJ, Smith AE, Chaudry F. Structure and synthesis of a simian virus 40 super T-antigen. J Virol. 1982;44:963–73. doi: 10.1128/jvi.44.3.963-973.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen S, Verderame M, Lo A, Pollack R. Nonlytic simian virus 40-specific 100K phosphoprotein is associated with anchorage-independent growth in simian virus 40-transformed and revertant mouse cell lines. Mol Cell Biol. 1981;1:994–1006. doi: 10.1128/mcb.1.11.994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen S, Grass DS, Blanck G, Hoganson N, Manley JL, Pollack RE. A functional simian virus 40 origin of replication is required for the generation of a super T antigen with a molecular weight of 100,000 in transformed mouse cells. J Virol. 1983;48:492–502. doi: 10.1128/jvi.48.2.492-502.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen S, Pollack R. DNA rearrangements and the role of viral origin in SV40-transformed mouse cells. DNA Tumor Viruses, Cold Spring Harbour Laboratory. Cancer Cells. 1986;4:381–6. [Google Scholar]
- 36.Kao C, Hauser P, Reznikoff WS, Reznikoff CA. Simian virus 40 (SV40) T-antigen mutations in tumorigenic transformation of SV40-immortalized human uroepithelial cells. J Virol. 1993;67:1987–95. doi: 10.1128/jvi.67.4.1987-1995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bikel I, Montano X, Agha ME, Brown M, McCormack M, Boltax J, Livingston DM. SV40 small t antigen enhances the transformation activity of limiting concentrations of SV40 large T antigen. Cell. 1987;48:321–30. doi: 10.1016/0092-8674(87)90435-1. [DOI] [PubMed] [Google Scholar]
- 38.Sáenz Robles MT, Pipas JM. T antigen transgenic mouse models. Semin Cancer Biol. 2009;19:229–35. doi: 10.1016/j.semcancer.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng J, DeCaprio JA, Fluck MM, Schaffhausen BS. Cellular transformation by simian virus 40 and murine polyoma virus T antigens. Semin Cancer Biol. 2009;19:218–28. doi: 10.1016/j.semcancer.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montano X, Millikan R, Milhaven JM, Newsom DA, Ludlow JW, Arthur AK, Fanning E, Bikel I, Livingston DM. Simian virus 40 small tumor antigen and an amino-terminal domain of large tumor antigen share a common transforming function. Proc Natl Acad Sci U S A. 1990;87:7448–52. doi: 10.1073/pnas.87.19.7448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Figge J, Webster T, Smith TF, Paucha E. Prediction of similar transforming regions in simian virus 40 large T, adenovirus E1A, and myc oncoproteins. J Virol. 1988;62:1814–8. doi: 10.1128/jvi.62.5.1814-1818.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen S, Paucha E. Identification of a region of simian virus 40 large T antigen required for cell transformation. J Virol. 1990;64:3350–7. doi: 10.1128/jvi.64.7.3350-3357.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eul J, Graessmann M, Graessmann A. Trans-splicing and alternative-tandem-cis-splicing: two ways by which mammalian cells generate a truncated SV40 T-antigen. Nucleic Acids Res. 1996;24:1653–61. doi: 10.1093/nar/24.9.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahara T, Tasic B, Maniatis T, Akanuma H, Yanagisawa S. Delay in synthesis of the 3′ splice site promotes trans-splicing of the preceding 5′ splice site. Mol Cell. 2005;18:245–51. doi: 10.1016/j.molcel.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 45.Dirksen WP, Hampson RK, Sun Q, Rottman FM. A purine-rich exon sequence enhances alternative splicing of bovine growth hormone pre-mRNA. J Biol Chem. 1994;269:6431–6. [PubMed] [Google Scholar]
- 46.Tanaka K, Watakabe A, Shimura Y. Polypurine sequences within a downstream exon function as a splicing enhancer. Mol Cell Biol. 1994;14:1347–54. doi: 10.1128/mcb.14.2.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patzel V, Sczakiel G. In vitro selection supports the view of a kinetic control of antisense RNA-mediated inhibition of gene expression in mammalian cells. Nucleic Acids Res. 2000;28:2462–6. doi: 10.1093/nar/28.13.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–8. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mansfield SG, Clark RH, Puttaraju M, Kole J, Cohn JA, Mitchell LG, Garcia-Blanco MA. 5′ exon replacement and repair by spliceosome-mediated RNA trans-splicing. RNA. 2003;9:1290–7. doi: 10.1261/rna.5101903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chew SL, Liu HX, Mayeda A, Krainer AR. Evidence for the function of an exonic splicing enhancer after the first catalytic step of pre-mRNA splicing. Proc Natl Acad Sci U S A. 1999;96:10655–60. doi: 10.1073/pnas.96.19.10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boukis LA, Liu N, Furuyama S, Bruzik JP. Ser/Arg-rich protein-mediated communication between U1 and U2 small nuclear ribonucleoprotein particles. J Biol Chem. 2004;279:29647–53. doi: 10.1074/jbc.M313209200. [DOI] [PubMed] [Google Scholar]
- 52.Warf MB, Berglund JA. Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem Sci. 2010;35:169–78. doi: 10.1016/j.tibs.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Butel JS, Lednicky JA. Cell and molecular biology of simian virus 40: implications for human infections and disease. J Natl Cancer Inst. 1999;91:119–34. doi: 10.1093/jnci/91.2.119. [DOI] [PubMed] [Google Scholar]
- 54.Carbone M, Pannuti A, Zhang L, Testa JR, Bocchetta M. A novel mechanism of late gene silencing drives SV40 transformation of human mesothelial cells. Cancer Res. 2008;68:9488–96. doi: 10.1158/0008-5472.CAN-08-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mansfield SG, Chao H, Walsh CE. RNA repair using spliceosome-mediated RNA trans-splicing. Trends Mol Med. 2004;10:263–8. doi: 10.1016/j.molmed.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 56.Caudevilla C, Da Silva-Azevedo L, Berg B, Guhl E, Graessmann M, Graessmann A. Heterologous HIV-nef mRNA trans-splicing: a new principle how mammalian cells generate hybrid mRNA and protein molecules. FEBS Lett. 2001;507:269–79. doi: 10.1016/S0014-5793(01)02957-X. [DOI] [PubMed] [Google Scholar]
- 57.Desai MM, Tumurbataar B, Zhang Y, Chan LN, Sun J, Chan TS. Aberrant transcription and post-transcriptional processing of hepatitis C virus non-structural genes in transgenic mice. Transgenic Res. 2011;20:1273–84. doi: 10.1007/s11248-011-9494-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J, Smith PJ, Krainer AR, Zhang MQ. Distribution of SR protein exonic splicing enhancer motifs in human protein-coding genes. Nucleic Acids Res. 2005;33:5053–62. doi: 10.1093/nar/gki810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brinkman BMN. Splice variants as cancer biomarkers. Clin Biochem. 2004;37:584–94. doi: 10.1016/j.clinbiochem.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 60.Srebrow A, Kornblihtt AR. The connection between splicing and cancer. J Cell Sci. 2006;119:2635–41. doi: 10.1242/jcs.03053. [DOI] [PubMed] [Google Scholar]
- 61.Pettigrew CA, Brown MA. Pre-mRNA splicing aberrations and cancer. Front Biosci. 2008;13:1090–105. doi: 10.2741/2747. [DOI] [PubMed] [Google Scholar]
- 62.Pio R, Montuenga LM. Alternative splicing in lung cancer. J Thorac Oncol. 2009;4:674–8. doi: 10.1097/JTO.0b013e3181a520dc. [DOI] [PubMed] [Google Scholar]
- 63.Miura K, Fujibuchi W, Sasaki I. Alternative pre-mRNA splicing in digestive tract malignancy. Cancer Sci. 2011;102:309–16. doi: 10.1111/j.1349-7006.2010.01797.x. [DOI] [PubMed] [Google Scholar]
- 64.Li H, Wang J, Ma X, Sklar J. Gene fusions and RNA trans-splicing in normal and neoplastic human cells. Cell Cycle. 2009;8:218–22. doi: 10.4161/cc.8.2.7358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.