Abstract
Staphylococcus aureus is a major mastitis-causing pathogen in dairy cows. The latex agglutination-based Staphaurex test allows bovine S. aureus strains to be grouped into Staphaurex latex agglutination test (SLAT)-negative [SLAT(−)] and SLAT-positive [SLAT(+)] isolates. Virulence and resistance gene profiles within SLAT(−) isolates are highly similar, but differ largely from those of SLAT(+) isolates. Notably, specific genetic changes in important virulence factors were detected in SLAT(−) isolates. Based on the molecular data, it is assumed that SLAT(+) strains are more virulent than SLAT(−) strains. The objective of this study was to investigate if SLAT(−) and SLAT(+) strains can differentially induce an immune response with regard to their adhesive capacity to epithelial cells in the mammary gland and in turn, could play a role in the course of mastitis. Primary bovine mammary epithelial cells (bMEC) were challenged with suspensions of heat inactivated SLAT(+) (n = 3) and SLAT(−) (n = 3) strains isolated from clinical bovine mastitis cases. After 1, 6, and 24 h, cells were harvested and mRNA expression of inflammatory mediators (TNF-α, IL-1β, IL-8, RANTES, SAA, lactoferrin, GM-CSF, COX-2, and TLR-2) was evaluated by reverse transcription and quantitative PCR. Transcription (ΔΔCT) of most measured factors was induced in challenged bMEC for 6 and 24 h. Interestingly, relative mRNA levels were higher (P<0.05) in response to SLAT(+) compared to SLAT(−) strains. In addition, adhesion assays on bMEC also showed significant differences between SLAT(+) and SLAT(−) strains.
The present study clearly shows that these two S. aureus strain types cause a differential immune response of bMEC and exhibit differences in their adhesion capacity in vitro. This could reflect differences in the severity of mastitis that the different strain types may induce.
Introduction
Mastitis has a profound impact on dairy production causing considerable economic losses [1], and affecting animal welfare [2]. Its predominant cause is the invasion of bacteria that enter the udder via the teat canal [3], [4]. The virulence of the pathogen and the immune response of the cow determine the clinical outcome of mastitis [5], [6].
According to several in vivo and in vitro studies [7], distinct bacterial species affect the cow's immune response differently: E. coli and its cell wall component LPS induce a stronger and qualitatively different immune reaction in the mammary gland and in bovine mammary epithelial cells (bMEC) than S. aureus or LTA from S. aureus. Moreover, it was shown that different strains of a bacterial species have varying effects on the immune response [5], [8], [9].
S. aureus belongs to one of the most important etiological agents of bovine mastitis and is referred to as a major udder pathogen. The Staphaurex latex agglutination test (Remel, Oxoid, Pratteln, Switzerland) is a diagnostic instrument widely used to confirm putative S. aureus isolates through detection of characteristic S. aureus surface proteins. Latex particles coated with human IgG and fibrinogen interact with the bacterial target proteins SpA (staphylococcal protein A), ClfA/B (clumping factor A/B), and FnbA/B (fibronectin-binding protein A/B), mediating a rapid agglutination reaction visible to the naked eye. While the Staphaurex latex agglutination test exhibits high specificity (99.5%) and sensitivity (99.8%) when applied to S. aureus strains obtained from humans, Stutz et al. [10] reported that 54% of S. aureus isolates obtained from cases of bovine mastitis yield negative test results. The latex agglutination-negative phenotypes are due to sequence polymorphisms leading to impaired functionality of one or several of the targeted virulence factors Spa, ClfA/B, and FnbA/B. According to Moser et al. [11], all Staphaurex latex agglutination test (SLAT)-negative [SLAT(−)] strains belong to clonal complex (CC) 151, whereas SLAT-positive [SLAT(+)] strains can be assigned to various CCs. Furthermore, DNA microarray profiles for virulence and resistance genes were highly similar among SLAT(−) isolates, but differed largely from those of SLAT(+) isolates. Based on all this molecular data, it is assumed that SLAT(+) strains exceed SLAT(−) strains in virulence [11].
The bMEC lining the inner surface of the mammary gland are crucial for the early defense against intramammary pathogens. They constitute a physical barrier, and they produce, concomitantly to somatic cells in milk, several antimicrobial substances and inflammatory mediators that enhance effector functions of innate immunity and stimulate adaptive immunity [6], [12], [13].
Pathogens that enter the lumen of the mammary gland are sensed via Toll-like receptors (TLR) that recognize pathogen-associated molecular patterns (PAMPs) [14]–[16]. Finally, signaling pathways that eventually activate transcription factors for genes that promote protective and inflammatory responses are induced. Important factors of the mammary immune response include the pro-inflammatory cytokines tumor necrosis factor-α (TNF-α) and interleukin (IL)-1β, known to be expressed in the early immune response of the mammary gland [1]. They induce other immune factors like granulocyte-macrophage colony stimulating factor (GM-CSF) which mediates differentiation of hematopoietic progenitors into granulocytes and monocytes [17]. Chemokines such as IL-8 and RANTES (Regulated and on activation, normal T cell expressed and secreted) attract leukocytes from the blood to the site of infection, which is reflected by an increase of somatic cell count (SCC) in milk [1]. Furthermore, antibacterial proteins including lactoferrin (Lf) and serum amyloid A (SAA), as well as cyclooxygenase-2 (COX-2), an enzyme involved in the prostaglandin synthesis, are secreted by bMEC [12], [18], [19].
As it is assumed that SLAT(+) are more virulent than SLAT(−) and the immune response of the mammary gland to bacteria of different virulence is diverse, the aim of the present study was to compare the immune response of bMEC and their adhesion capacity to different S. aureus strains, belonging either to the SLAT(+) or SLAT(−) group.
According to the current study, there is clear evidence that the SLAT phenotype affects the immune response of bMEC and the adhesion capacity in vitro.
Materials and Methods
S. aureus strains
The six S. aureus strains (Table 1) used in this study were isolated from bovine mastitis milk samples collected from different cows [11]. The milk samples from mastitis were collected after the diagnosis of acute mastitis by the attending veterinarian, including a positive result of the California Mastitis Test. Bacterial cultures were maintained in peptone with 20% glycerol at −70°C. Prior to experiments, isolates were grown overnight on tryptic soy agar (TSA; Difco, Becton Dickinson Diagnostics, Mississauga, ON, Canada) at 37°C. Individual S. aureus strains were then inoculated in 10 ml cultures of brain heart infusion (BHI; Oxoid, Pratteln, Switzerland) broth and grown overnight at 37°C. Colony counts were determined for each strain (∼109 CFU/ml).
Table 1. S. aureus strains used in this study.
| Strain | Clonal complex | Staphaurex test phenotype | Anamestic data |
| 692 | CC8 | positive | acute mastitis |
| 1130 | CC97 | positive | acute mastitis |
| 1989 | CC20 | positive | acute mastitis |
| 1586 | CC705 | negative | acute mastitis |
| 1904 | CC705 | negative | acute mastitis |
| 2071 | CC705 | negative | acute mastitis |
Reference: [11].
Treatment of bMEC with different S. aureus strains
Primary cultures of mammary gland epithelial cells of two Holstein dairy cows with clinically healthy udders (SCC<105 cells/ml) were developed as previously described [20]. Mammary tissue was removed directly after slaughter with permission of the slaughterhouse Marmy SA, Estavayer-le-Lac, Switzerland. Cells in passage 2 were cryopreserved in DMEM/F12 (Sigma-Aldrich, Munich, Germany) containing 20% fetal bovine serum (FBS, Sigma-Aldrich) and 10% dimethyl sulfoxide (DMSO, Sigma-Aldrich) and stored in aliquots at −80°C until the experiment. Cells from both cows were thawed and cultured in growth medium consisting of DMEM/F12 supplemented with 10% FBS, penicillin G (500 units, Sigma-Aldrich), streptomycin (100 µg/ml, Sigma-Aldrich), and ITS (0.5 mg/ml insulin, 0.5 mg/ml apo-transferrin, 0.5 µg/ml sodium selenite; Sigma-Aldrich). After two further passages, cells were seeded at a concentration of 3×105 cells/well on BD Falcon™ 6-well cell culture plates (BD Biosciences, San Jose, CA, USA). On the following day, growth medium was replaced by DMEM/F12 supplemented with 5% FBS and ITS.
Suspensions of heat inactivated bacteria (inactivated by heating for 20 min at 80°C) were diluted to a concentration of 1×109 CFU/ml in BHI broth. The cells were challenged in triplicate with 150 µl of the bacterial suspension. Assuming that 3×105 cells/well after 24 h of incubation and a confluence of about 70% represent approximately 1×106 cells/well, this corresponds to a multiplicity of infection (MOI) of 150. As a positive control, cells were challenged with 150 MOI of Escherichia coli in duplicate. Cells incubated in DMEM/F12, 5% FBS, and ITS only served as negative controls. Cells were incubated at 37°C with 5% CO2.
Total RNA extraction and reverse transcription
After 1, 6, or 24 h of incubation, cells were harvested with 0.5 ml peqGOLD Trifast™ (PEQLAB Biotechnologie GmbH, Erlangen, Germany) and total RNA was extracted according to the manufacturer's protocol. Total RNA yield and purity were determined by absorbance at 260 nm and 280 nm using a NanoDrop-2000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). Finally, 500 ng of total RNA was reverse transcribed by 200 units of Moloney Murine Leukemia Virus Reverse Transcriptase (M-MLV RT; Promega Corp., Madison, WI, USA) using 100 pmol of random hexamer primers (Invitrogen, Leek, The Netherlands).
Quantitative real-time PCR
Quantitative real-time PCR analysis was performed with the Sensimix DNA Kit (Quantace, Biolabo, Châtel St. Denis, Switzerland) on a Rotor-Gene 6000 (Corbett Research, Sydney, Australia). One reaction mixture contained 2 µl of cDNA equivalent to 25 ng of total RNA, 0.8 µl RNase-free water (Qiagen, Hilden, Germany), 1 µl (5 pmol) of forward primer, 1 µl (5 pmol) of reverse primer, and 5.2 µl of 2x SensiMix plus SYBR-Green (1 mM MgCl2). Primers for the housekeeping (GAPDH and ubiquitin) and target genes were synthesized commercially (Microsynth, Balgach, Switzerland) using previously published sequences [12], [18], [21], [22], or designed using the open source primer design software Primer 3 (primer sequences are listed in Table 2).
Table 2. Sequences, accession numbers, annealing temperature of the PCR primers, and length of the PCR products.
| Gene1 | Sequence 5′→3′ | GenBank accession no. | Annealing temperature (°C) | Length (bp) | |
| IL-1β | for | AGT GCC TAC GCA CAT GTC TTC2 | M37211 | 60 | 114 |
| rev | TGC GTC ACA CAG AAA CTC GTC2 | ||||
| TNF-α | for | CCA CGT TGT AGC CGA CAT C2 | NM173966 | 60 | 155 |
| rev | CCC TGA AGA GGA CCT GTG AG2 | ||||
| IL-8 | for | ATG ACT TCC AAG CTG GCT GTT G2 | AF232704 | 60 | 149 |
| rev | TTG ATA AAT TTG GGG TGG AAA G2 | ||||
| RANTES | for | GCC AAC CCA GAG AAG AAG TG2 | BC102064 | 60 | 119 |
| rev | CTG CTT AGG ACA AGA GCG AGA2 | ||||
| SAA | for | CCT GGG CTG CTA AAG TGA TC3 | AF540564 | 57 | 184 |
| rev | TAC TTG TCA GGC AGG CCA G3 | ||||
| Lf | for | GGC CTT TGC CTT GGA ATG TAT4 | L08604 | 62 | 338 |
| rev | ATT TAG CCA CAG CTC CCT GGA G4 | ||||
| GM-CSF | for | TTC TCC GCA CCT ACT CGC | NM174027 | 62 | 195 |
| rev | GTT CTT GTA CAG CTT CAG GCG | ||||
| COX-2 | for | TCC TGA AAC CCA CTC CCA ACA5 | NM174445 | 62 | 242 |
| rev | TGG GCA GTC ATC AGG CAC AG5 | ||||
| TLR2 | for | CAT TCC CTG GCA AGT GGA TTA TC2 | NM174197 | 62 | 201 |
| rev | GGA ATG GCC TTC TTG TCA ATG G2 | ||||
| GAPDH | for | GTC TTC ACT ACC ATG GAG AAG G2 | NM001034034 | 60 | 197 |
| rev | TCA TGG ATG ACC TTG GCC AG2 | ||||
| Ubiquitin | for | AGA TCC AGG ATA AGG AAG GCA T2 | NM174133 | 62 | 198 |
| rev | GCT CCA CCT CCA GGG TGA T2 | ||||
for = forward, rev = reverse
IL-1β = interleukin-1β; TNF-α = tumor necrosis factor-α; IL-8 = interleukin-8; RANTES = regulated on activation, normal T cell expressed and secreted; SAA = serum amyloid A; Lf = lactoferrin; GM-CSF = granulocyte-macrophage colony-stimulating factor; COX-2 = cyclooxygenase-2; TLR2 = toll-like receptor 2; GAPDH = glyceraldehyde-3-phosphate dehydrogenase
The following 3-step PCR program was used: initial denaturation for 10 min at 95°C, followed by 40 cycles with denaturation for 15 s at 95°C, 30 s at primer-specific annealing temperature, and elongation for 20 s at 72°C. Fluorescence was acquired at 72°C after each cycle, and a dissociation melt curve of the PCR product was determined at the end of each run to verify the specificity of the PCR reactions.
Cycle threshold (Ct) values were determined by the Rotor-Gene software version 1.7.75, and the relative mRNA expression was calculated with the comparative Ct method [23] using the following equation:
ΔCt = Ct target gene – Ct endogenous control (arithmetic mean of housekeeping genes).
To visualize the impact of SLAT(+) and SLAT(−) strains on the immune response of bMEC, data are presented as ΔΔCt± SEM, where:
ΔΔCt = ΔCt sample (1, 6, or 24 h; treated) −ΔCt negative control (1, 6, or 24 h; untreated).
Adhesion Assay
The adhesion assay was performed as previously described [24]. Bovine mammary epithelial cells derived from the same cows mentioned above were grown on sterile plastic coverslips (13 mm diameter, Bibby Sterilin, Stone, UK) coated with rat tail collagen (BD Biosciences, Allschwil, Switzerland). The bMEC on coverslips were cultured at 37°C for 24 h in 24-well plates (24-well flat-bottom cell culture plate with Low-Evaporation Lid, TPP, Trasadingen, Switzerland) until they reached a confluency of 70–80%. Suspensions of live bacteria were used at a concentration of 150 MOI to infect coverslips in duplicate. Infected bMEC cells of both cows were incubated for 3 h at 37°C. Uninfected bMECs were incubated in parallel and used as negative controls. After 3 h post infection, cell monolayers were washed five times with Dulbecco's phosphate buffered saline (DPBS) (GIBCO, Invitrogen, Carlsbad, CA, USA) and fixed with absolute methanol (−20°C) for 10 min. After the staining with May-Grünwald-Giemsa (Fluka, Buchs SG, Switzerland), the coverslips were examined by oil immersion light microscopy at a magnification of 1000×.
Adhesion affinity of the SLAT(+) and SLAT(−) S. aureus strains on bMEC was assessed by counting of 200 cells per coverslip and the presence of adhered bacteria. Each cell with at least one firmly adhered bacterium was counted as positive. Mean percentage of positive cells were compared between the three SLAT(+) and the three SLAT(−) S. aureus strains, respectively.
Statistical Analysis
Data are presented as means ± SEM. Statistical analysis of the inflammatory response data was performed with ANOVA using a MIXED procedure of SAS (Release 9.2; SAS Institute Inc., Cary, NC, USA). The model included strain, type, and their interaction as fixed effects. Results of the triplicates were nested within cow. Statistical analysis of the adhesion experiments was performed using an unpaired t-test. Differences were considered significant if P<0.05.
Results
Relative mRNA expression of immune factors
Cells challenged with E. coli (positive control) induced an increase in mRNA expression of IL-1β, TNF-α, IL-8, and GM-CSF after 1 h of stimulation. Furthermore, E. coli induced mRNA expression of all factors after 6 and 24 h of stimulation (data not shown).
Cells challenged with S. aureus for only 1 h had no significant effect on the relative mRNA expression of all measured factors in bMEC, with the exception of IL-1β and IL-8. IL-1β mRNA expression was significantly upregulated in response to most of the tested S. aureus strains, regardless of SLAT phenotype. IL-8 mRNA expression was significantly induced in bMEC challenged for 1 h with SLAT(+) strains (Table 3).
Table 3. Changes of mRNA abundance (Mean ± SEM ΔΔCt1) of immune factors in bMEC stimulated with six heat-inactivated S. aureus strains for 1, 6, or 24 h.
| SLAT(+)3 | SLAT(−)3 | Analysis of variance (P-value) | ||||||||
| Gene2 | Time (h) | 692 | 1130 | 1989 | 1586 | 1904 | 2071 | Strain(Type) | Cow | Strain*Cow |
| IL-1β | 1 | 0.5±0.3 ac | 1.6±0.2 b * | 1.3±0.3 bc * | 1.3±0.5 bc * | 1.5±0.4 b * | 0.5±0.3 a | 0.01 | <0.01 | 0.03 |
| 6 | 8.3±0.2 a * | 8.7±0.2 a * | 8.3±0.3 a * | 3.5±0.1 b * | 5.7±0.5 c * | 5.3±0.2 c * | <0.01 | <0.01 | 0.01 | |
| 24 | 5.1±0.3 ab * | 5.7±0.2 b * | 4.8±0.1 a * | 0.4±0.4 c | 1.8±0.3 d * | 1.6±0.3 d * | <0.01 | <0.01 | 0.03 | |
| TNF-α | 1 | 0.7±0.2 a | 0.8±0.1 a | 0.3±0.3 a | 0.2±0.6 ab | 05±0.5 a | −0.6±0.3 b | 0.07 | <0.01 | 0.02 |
| 6 | 5.4±0.4 a * | 5.1±0.2 a * | 5.1±0.4 a * | 1.3±0.6 b * | 3.4±0.3 c * | 2.9±0.4 c * | <0.01 | <0.01 | 0.75 | |
| 24 | 4.1±0.2 a * | 4.3±0.2 a * | 4.1±0.1 a * | 1.1±0.2 b * | 2.0±0.4 c * | 1.6±0.1 bc * | 0.11 | 0.06 | 0.23 | |
| IL-8 | 1 | 2.0±0.5 a * | 2.1±0.4 a * | 1.5±0.2 ab * | 0.6±0.4 b | 0.8±0.3 b | 0.9±0.5 b | 0.73 | <0.01 | 0.07 |
| 6 | 7.8±0.2 a * | 7.1±0.2 b * | 7.0±0.4 b * | 4.7±0.1 c * | 6.1±0.3 d * | 5.4±0.3 e * | <0.01 | <0.01 | 0.01 | |
| 24 | 7.1±0.3 a * | 7.0±0.2 a * | 7.1±0.1 a * | 3.5±0.4 b * | 4.8±0.2 c * | 4.1±0.6 c * | <0.01 | <0.01 | 0.01 | |
| RANTES | 1 | −0.2±0.2 ab | −0.6±0.3 ab | 0.1±0.5 a | −0.6±0.4 ab | −0.5±0.4 ab | −0.8±0.5 b | 0.56 | <0.01 | 0.72 |
| 6 | 3.9±0.2 a * | 4.3±0.2 a * | 2.9±0.3 b * | 0.0±0.2 c | 1.8±0.4 d * | 1.0±0.3 e * | <0.01 | <0.01 | 0.69 | |
| 24 | 3.2±0.3 a * | 4.0±0.3 b * | 3.0±0.3 a * | −0.1±0.5 c | 1.1±0.2 d * | 0.6±0.4 d | <0.01 | <0.01 | 0.06 | |
| SAA | 1 | −1.0±0.5 a | −0.6±0.4 ab | 0.8±1.0 c | −0.9±0.9 a | 0.7±0.8 bc | −0.9±0.6 a | 0.01 | <0.01 | 0.50 |
| 6 | 5.6±0.6 a * | 4.9±0.5 ab * | 4.7±0.4 b * | 0.8±0.4 c | 3.6±0.3 d * | 1.6±0.3 e * | <0.01 | <0.01 | <0.01 | |
| 24 | 5.3±0.4 a * | 5.3±0.6 a * | 6.3±0.5 b * | 1.9±0.5 c * | 3.3±0.4 d * | 3.0±0.6 d * | <0.01 | <0.01 | 0.47 | |
| Lf | 1 | 0.7±0.2 a | 0.1±0.3 bc | −0.1±0.2 d | 0.4±0.4 ab | −0.7±0.4 d | −0.6±0.3 cd | 0.01 | <0.01 | 0.53 |
| 6 | 2.6±0.4 a * | 2.1±0.1 ab * | 1.9±0.4 b * | 0.0±0.5 c | 1.6±0.2 b * | 0.6±0.2 d | <0.01 | <0.01 | 0.02 | |
| 24 | 3.9±0.5 a * | 3.6±0.5 a * | 4.1±0.5 a * | 1.9±0.8 b * | 2.4±0.4 b * | 1.6±0.7 b * | 0.36 | <0.01 | 0.03 | |
| GM-CSF | 1 | 0.6±0.2 a | 0.0±0.4 ab | −0.5±0.5 b | 0.3±0.1 a | 0.4±0.2 a | 0.5±0.3 a | 0.07 | 0.01 | 0.16 |
| 6 | 3.1±0.1 a * | 3.9±0.2 b * | 3.0±0.4 a * | 0.0±0.3 c | 1.6±0.2 d * | 1.9±0.3 d * | <0.01 | 0.71 | 0.04 | |
| 24 | 0.4±0.6 ab | 1.7±0.4 c * | 0.5±0.4 a | −1.4±0.3 d | −0.7±0.4 bd | −1.0±0.6 d | 0.06 | <0.01 | 0.39 | |
| COX-2 | 1 | 0.5±0.3 | 0.3±0.2 | −0.1±0.4 | 0.6±0.3 | −0.3±0.5 | 0.0±0.1 | 0.27 | 0.08 | 0.03 |
| 6 | 1.8±0.2 a * | 1.8±0.4 ab * | 1.5±0.3 abd * | 0.2±0.1 c | 1.1±0.1 bd * | 1.0±0.4 d | 0.06 | <0.01 | 0.01 | |
| 24 | 0.1±0.5 ab | 0.8±0.2 b | 0.8±0.2 b | −0.6±0.3 a | −0.3±0.2 a | −0.4±0.4 a | 0.58 | 0.59 | <0.01 | |
| TLR2 | 1 | 0.0±0.3 a | 0.6±0.3 a | 0.3±0.9 a | 0.6±0.3 a | −0.3±0.3 a | −0.2±0.5 a | 0.57 | 0.05 | 0.95 |
| 6 | 2.7±0.3 a * | 1.9±0.2 b * | 2.0±0.3 b * | 0.4±0.4 c | 1.7±0.2 bd * | 1.1±0.3 d * | <0.01 | <0.01 | <0.01 | |
| 24 | 2.1±0.4 ab * | 2.2±0.6 a * | 2.6±0.5 a * | 0.9±0.4 c | 1.5±0.3 bc * | 1.1±0.6 c * | 0.23 | <0.01 | 0.51 | |
Means within a row without common superscript letters differ (P<0.05).
Means differ significantly from negative control.
ΔΔCt values are normalized to negative controls and corrected for the two reference genes, GAPDH and ubiquitin.
IL-1β = interleukin-1β; TNF-α = tumor necrosis factor-α; IL-8 = interleukin-8; RANTES = regulated on activation, normal T cell expressed and secreted; SAA = serum amyloid A; Lf = lactoferrin; GM-CSF = granulocyte-macrophage colony-stimulating factor; COX-2 = cyclooxygenase-2; TLR2 = toll-like receptor 2.
SLAT(−) = S. aureus latex agglutination test negative; SLAT(+) = S. aureus latex agglutination test positive.
The relative mRNA expression of IL-1β, TNF-α, and IL-8 was significantly increased after 6 and 24 h in response to all S. aureus strains, except for IL-1β mRNA expression after challenge with SLAT(−) strain 1586 for 24 h. After 24 h of challenge, mRNA levels of IL-1β, TNF-α, and IL-8 were equal or lower than after 6 h (Table 3).
All SLAT(+) strains induced increased mRNA levels of RANTES in bMEC challenged for 6 or 24 h. Within the SLAT(−) group, only strain 1904 induced elevated mRNA levels after 6 and 24 h of challenge. For all strains, similar RANTES mRNA levels were obtained in cells challenged for either 6 or 24 h (Table 3). For IL-1β, TNF-α, IL-8, and RANTES the relative mRNA abundances were significantly higher in cells challenged for 6 and 24 h with SLAT(+) than with SLAT(−) strains (Table 4, Figure 1).
Table 4. Effect of SLAT type on changes of mRNA expression (Mean ± SEM ΔΔCt1) of immune factors in bMEC stimulated for 1, 6, or 24 h.
| Type3 | Analysis of variance (P-value) | ||||
| Gene2 | Time (h) | SLAT(+) | SLAT(-) | Type | Cow |
| IL-1β | 1 | 1.1±0.2 a * | 1.1±0.2 a * | 0.87 | 0.01 |
| 6 | 8.4±0.1 a * | 4.8±0.3 b * | <0.01 | 0.03 | |
| 24 | 5.2±0.1 a * | 1.3±0.2 b * | <0.01 | <0.01 | |
| TNF-α | 1 | 0.6±0.1 a | 0.0±0.3 b | 0.03 | <0.01 |
| 6 | 5.2±0.2 a * | 2.5±0.3 b * | <0.01 | 0.02 | |
| 24 | 4.2±0.1 a * | 1.6±0.2 b * | <0.01 | 0.08 | |
| IL-8 | 1 | 1.9±0.2 a * | 0.8±0.2 b | <0.01 | <0.01 |
| 6 | 7.3±0.2 a * | 5.2±0.2 b * | <0.01 | <0.01 | |
| 24 | 7.1±0.1 a * | 4.1±0.3 b * | <0.01 | <0.01 | |
| RANTES | 1 | −0.2±0.2 a | −0.7±0.2 a | 0.09 | <0.01 |
| 6 | 3.7±0.2 a * | 0.9±0.2 b | <0.01 | 0.04 | |
| 24 | 3.4±0.2 a * | 0.6±0.2 b | <0.01 | <0.01 | |
| SAA | 1 | −0.2±0.4 a | −0.4±0.5 a | 0.52 | <0.01 |
| 6 | 5.1±0.3 a * | 2.0±0.3 b * | <0.01 | <0.01 | |
| 24 | 5.6±0.3 a * | 2.7±0.3 b * | <0.01 | <0.01 | |
| Lf | 1 | 0.2±0.2 a | −0.3±0.2 b | 0.05 | <0.01 |
| 6 | 2.2±0.2 a * | 0.7±0.2 b * | <0.01 | <0.01 | |
| 24 | 3.9±0.3 a * | 2.0±0.4 b * | <0.01 | <0.01 | |
| GM-CSF | 1 | 0.0±0.2 a | 0.4±0.1 a | 0.09 | 0.01 |
| 6 | 3.3±0.2 a * | 1.2±0.2 b | <0.01 | 0.79 | |
| 24 | 0.9±0.3 a | −1.0±0.3 b | <0.01 | <0.01 | |
| COX-2 | 1 | 0.2±0.2 a | 0.1±0.2 a | 0.67 | 0.08 |
| 6 | 1.7±0.2 a * | 0.7±0.2 b | <0.01 | 0.01 | |
| 24 | 0.6±0.2 a | −0.4±0.2 b | <0.01 | 0.58 | |
| TLR2 | 1 | 0.3±0.3 a | 0.0±0.2 a | 0.45 | 0.04 |
| 6 | 2.2±0.2 a * | 1.1±0.2 b | <0.01 | 0.01 | |
| 24 | 2.3±0.3 a * | 1.2±0.3 b * | <0.01 | <0.01 | |
Means within a row without common superscript letters differ (P<0.05).
Means differ significantly from negative control.
ΔΔCt values are normalized to negative controls and corrected for the two reference genes, GAPDH and ubiquitin.
IL-1β = interleukin-1; TNF-α = tumor necrosis factor-α; IL-8 = interleukin-8; RANTES = regulated on activation, normal T cell expressed and secreted; SAA = serum amyloid A; Lf = lactoferrin; GM-CSF = granulocyte-macrophage colony-stimulating factor; COX-2 = cyclooxygenase-2; TLR2 = toll-like receptor 2.
SLAT(−) = S. aureus latex agglutination test negative; SLAT(+) = S. aureus latex agglutination test positive.
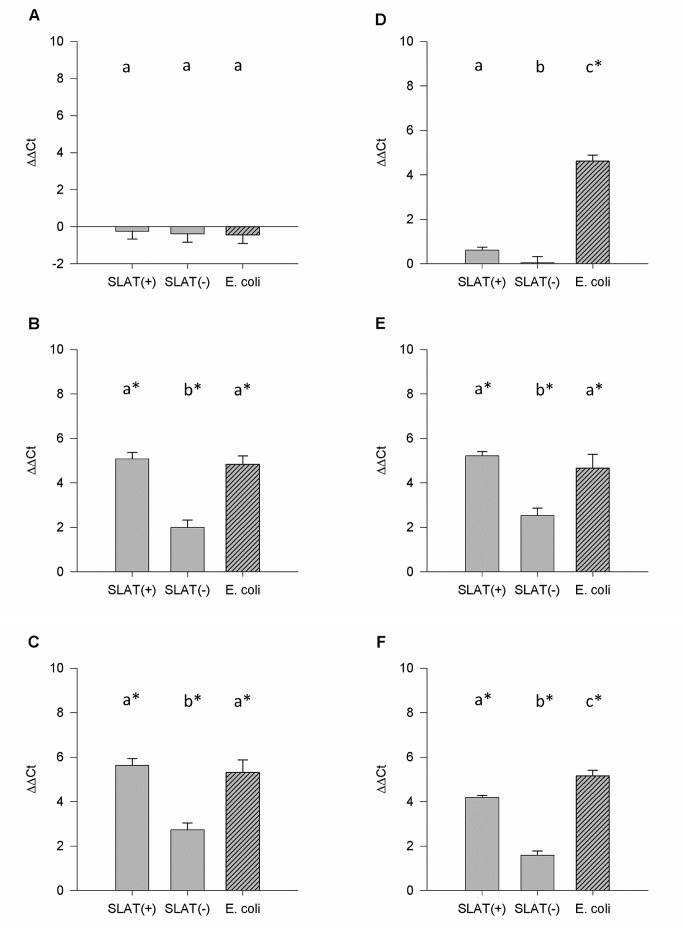
Figure 1. Relative SAA and TNF-α mRNA expression.
Relative mRNA expression (ΔΔCt) of SAA in bMEC stimulated for (A) 1 h, (B) 6 h, or (C) 24 h with SLAT(+), SLAT(−), or E. coli. Relative mRNA expression (ΔΔCt) of TNF-α in bMEC stimulated for (D) 1 h, (E) 6 h, or (F) 24 h with SLAT(+), SLAT(−) or E. coli. Data are presented as Means ± SEM. a,bMeans without common superscript letters differ (P<0.05).
Lactoferrin and SAA were upregulated in bMEC challenged for 6 and 24 h with SLAT(+) strains, whereas the increase was equal or higher after 24 h (Table 3). Two of the SLAT(−) strains increased SAA mRNA expression after 6 and 24 h of stimulation (Table 3). Lactoferrin mRNA expression was significantly increased after 24 h of stimulation in response to all SLAT(−) strains, whereas after 6 h only one of the SLAT(−) strains induced Lf transcription (Table 3). SLAT(+) significantly induced higher relative mRNA levels of SAA and Lf than SLAT(−) strains (Table 4, Figure 1).
In response to all SLAT(+) strains, GM-CSF mRNA expression was significantly increased after 6 h of challenge, but decreased again between 6 and 24 h. The challenge of bMEC with SLAT(+) for 6 h caused significantly higher GM-CSF mRNA expression compared to SLAT(−) strains (Tables 3 & 4). Two SLAT(−) strains affected GM-CSF mRNA expression in bMEC stimulated for 6 h (Table 3).
Expression levels of COX-2 mRNA were affected after stimulation for 6 h with all SLAT(+), as well as with one SLAT(−) strain (Table 3). Although the SLAT(+)-induced response was significantly stronger after 6 h for COX-2 compared to SLAT(−) (Table 4), individual strain differences were not consistently significant between both groups (Table 3).
In response to all S. aureus strains, except for SLAT(−) 1586, relative mRNA levels of TLR2 were significantly increased in bMEC stimulated for 6 and 24 h. The mRNA levels were similar in bMEC stimulated for 6 or 24 h (Table 3). After 6 and 24 h, the expression of TLR2 mRNA was more pronounced in bMEC stimulated with SLAT(+) than with the SLAT(−) type (Table 4).
Although SLAT(+) significantly induced higher mRNA expression levels than SLAT(−) for all measured factors (Table 4), mRNA expression differed between individual strains for most time points (Table 3). In addition, there were significant strain by cow interactions in half of the measurements, and the cells from differing cows had a significant impact on mRNA expression of all factors at most time points (Table 3). A similar expression pattern of most immune factors was seen in SLAT(+) and E. coli challenged bMEC after 6 and 24 h (data not shown).
Adhesion Assay
After 3 h post infection, bMEC were morphologically unaltered irrespective of the S. aureus strain and when compared to the negative controls (confluency of 70–80%). All six S. aureus strains showed a variable degree of adhesion (10–55%) to bMEC. Clusters of more than 20 adherent bacteria were frequently observed in SLAT(+) but not in SLAT(−) S. aureus strains. There was a statistically significant difference in adhesion affinity between the SLAT(+) and SLAT(−) S. aureus strains in cells of both cows (Table 5).
Table 5. Degree of adhesion (%) of SLAT(+) and SLAT(−) strains to bMEC.
| bMEC | SLAT(+) | SLAT(−) |
| cow 1 | 36.1±6.1 a | 20.6±2.8 b |
| cow 2 | 36.1±6.1 a | 21.0±3.6 b |
| total (cow 1 & 2) | 36.3±4.9 a | 20.8±3.1 b |
Means within a row without common superscript letters differ (P<0.05).
Discussion
Cultured primary bMEC responded to the challenge with heat inactivated bacteria by upregulation of genes relevant in the immune reaction of the mammary gland. This was highly reproducible and verifies the suitability of the model for comparison of the innate immune response to different bacterial strains. By using cells from two different cows, the biological reproducibility was proven. With some exceptions, the factor ‘cow’ had a significant effect on the results. Differences are most likely attributed to the genotype of the cows, as it was shown that bMEC from cows with genetic differences in mastitis susceptibility can show a difference in magnitude of transcription of immune factors in response to bacterial challenge [25].
Escherichia coli was chosen as a positive control as it often causes acute mastitis. Heat inactivated E. coli isolated from acute mastitis cases induced a strong immune response characterized by highly induced mRNA expression of important immune factors in mammary epithelial cells in culture [12]. This is in agreement with the present study, as the mRNA expression for all measured factors was strongly upregulated after at least 6 h post challenge.
Three different periods of challenge were used in the present study to capture the change of different immune factors that are expressed in the early and later phases of the immune response. Griesbeck-Zilch et al. [12] found that mRNA levels of IL-1β, TNF-α, and IL-8 were already increased in bMEC after 1 h of challenge with the heat inactivated S. aureus strain M60. Comparable results were seen in the present study with several S. aureus strains, which indicates that bMEC respond directly to contact with heat inactivated S. aureus, regardless of the bacterial type, i.e. SLAT(+) or SLAT(−). Interestingly for IL-8, a stronger increase of transcription was detectable after 1 h of challenge for SLAT(+) strains compared to SLAT(−) strains. It is apparent for the other factors that 1 h of challenge was not long enough to induce measurable changes in transcription of the selected factors. However, after challenging the cells for a longer period of time, the induction of mRNA expression of all measured immune factors was more pronounced by SLAT(+) compared to SLAT(−) strains.
In clinical mastitis caused by S. aureus, IL-1β and TNF-α are expressed in the early stages of the infection period, soon followed by a considerable decrease [4], [26]. It is assumed that the duration of enhanced transcription of these pro-inflammatory cytokines is not sufficient to eliminate S. aureus from the mammary gland, and therefore may often lead to a chronic outcome [1]. In contrast, E. coli typically causes a more sustained increase of these cytokines [4]. E. coli intramammary infections are often characterized by acute and severe clinical manifestations, but can, if not leading to death, be cured within a few days [1], [27]. According to the review by Oviedo-Boyso et al. [1], in the present study, S. aureus-induced mRNA expression of IL-1β and TNF-α was increased at least after 6 h of stimulation and decreased again until 24 h post challenge. The observation that SLAT(+) and SLAT(−) strains induced a different expression of these pro-inflammatory cytokines in bMEC may indicate that these strains induce different severities of mastitis. However, the severity of mastitis that these strains induced is not known.
The stronger induction of the mRNA expression pattern of the chemokines IL-8 and RANTES in bMEC may suggest earlier and stronger leukocyte recruitment during the innate immune response after an intramammary infection with SLAT(+) compared to infections with SLAT(−) strains. The immediate recruitment of somatic cells from the blood into the udder is essential for effective elimination of intramammary pathogens [28]. Thus, a deviating time point and magnitude of leukocyte recruitment, which is reflected in the chemokine expression, might influence the clinical course of mastitis. Since no clinical data including somatic cell count were available for the tested S. aureus strains in the present study, conclusions cannot be drawn on the clinical manifestation of the infection.
The acute phase protein SAA is known to be expressed in bMEC and is upregulated during mastitis [29]. Wellnitz et al. [21] reported similarly increased mRNA levels of SAA in cells challenged for 6 h with S. aureus or E. coli. In the present study, SLAT(+) induced a stronger SAA mRNA expression compared to SLAT(−) S. aureus strains. However, mRNA levels induced by E. coli were equal or higher during the whole study than in response to S. aureus, depending on the SLAT type. As SAA is also a chemoattractant [30], [31], pathogens that upregulate SAA transcription could be associated with an increase of SCC in the mammary gland. This effect could, as described for IL-8 above, have an effect on the progress of the mastitis.
Epithelial cells from the mammary gland are the major source of the iron-binding protein Lf that increases in milk during bovine clinical mastitis [28]. Griesbeck-Zilch et al. [12] showed a more pronounced Lf mRNA expression in bMEC by S. aureus than by E. coli challenge. In the present study, E. coli induced comparable Lf mRNA levels to SLAT(+) but higher levels compared to SLAT(-). However, neither the SLAT phenotype of S. aureus strains, nor details on the E. coli strain used in the study of Griesbeck-Zilch et al. [12] are known, which could account for a deviating Lf expression pattern in bMEC. Comparing the Lf mRNA expression between S. aureus strains in the present study clearly shows that SLAT(+) strains induced higher levels than SLAT(-), which again confirms a deviating immune response of the mammary gland to SLAT(+) and SLAT(−) strains. Only SLAT(−) strain 1904 caused Lf mRNA levels that were not significantly lower than in response to two of the SLAT(+) strains. Thus, the induction of Lf mRNA expression in bMEC might follow different strain-dependent mechanisms.
Another cytokine that was stronger induced on the mRNA level by SLAT(+) compared to SLAT(−) S. aureus strains in bMEC is GM-CSF. Since GM-CSF is responsible for an appropriate supply of leukocytes, the effector cells of the innate immune system, it is likely that high levels of GM-CSF expression represent a strong inflammatory response in the mammary gland. As a consequence, it is possible that SLAT(+) cause more severe forms of mastitis.
The transient upregulation of GM-CSF in response to all SLAT(+), as well as to the majority of SLAT(−) strains indicates that the role of bMEC in stimulating phagocyte differentiation is rather short term. This is consistent with an in vivo study, where mRNA expression of somatic cells upon S. aureus infection was evaluated [32].
The observation that TLR2 mRNA, a receptor for cell wall components of gram-positive bacteria, was induced in response of the majority of the S. aureus strains, confirms the involvement of this pattern recognition receptor (PRR) [12]. SLAT(+) S. aureus strains showed a stronger induction of TLR2 mRNA than SLAT(−) strains. Interestingly, one strain, SLAT(−) 1586, apparently did not induce TLR2 mRNA expression in bMEC. One possible reason could be that this strain has an altered PAMP, i.e. LTA or peptidoglycan that is normally recognized by this PRR. To confirm this, further investigations are necessary. Even though most studies describe TLR2 as a primary receptor for gram-positive bacteria, it is also known that this PPR is responsive to gram-negative bacteria [33]. This point also explains the induced TLR2 expression by E. coli.
Prostaglandins are further inflammatory mediators in the bovine mammary gland with chemotactic activity [34]. COX-2 is one of the enzymes involved in prostaglandin synthesis that is transiently upregulated during inflammation [35], [36]. These findings are reflected by this study. COX-2 was only transiently upregulated in bMEC in response to all S. aureus strains, except to one SLAT(−) strain. Compared to the other measured immune factors, the SLAT(+) group is not as strong of an inducer of COX-2 and TLR2 compared to SLAT(−), since differences were not consistently significant between the SLAT groups.
Conclusion
The results of this study clearly indicate a different immunological response of bMEC to SLAT(+) and SLAT(−) strains. Although individual differences within SLAT groups and the cow seem to influence the immune response, SLAT(+) S. aureus strains induce a more pronounced transcription of several important immune factors compared to SLAT(−) strains in mammary epithelial cells. Furthermore, results obtained from adhesion assays indicate that SLAT(+) S. aureus strains show an increased affinity to adhere to bMEC than SLAT(−) strains. These findings together with molecular data on the S. aureus strains might support the hypothesis that SLAT(+) exceed SLAT(−) strains in virulence. The influence of these effects on the varying course and severity of mastitis can be suggested and should be further investigated in vivo.
Funding Statement
This work was financially supported by the Swiss Federal Veterinary Office (BVET). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Oviedo-Boyso J, Valdez-Alarcón JJ, Cajero-Juárez M, Ochoa-Zarzosa A, López-Meza JE, et al. (2007) Innate immune response of bovine mammary gland to pathogenic bacteria responsible for mastitis. J Infect 54: 399–409. [DOI] [PubMed] [Google Scholar]
- 2. Le Maréchal C, Thiery R, Vautor E, Le Loir Y (2011) Mastitis impact on technological properties of milk and quality of milk products-a review. Dairy Sci Technol 91: 247–282. [Google Scholar]
- 3. Kerro Dego O, van Dijk JE, Nederbragt H (2002) Factors involved in the early pathogenesis of bovine Staphylococcus aureus mastitis with emphasis on bacterial adhesion and invasion. A review. Vet Q 24: 181–198. [DOI] [PubMed] [Google Scholar]
- 4. Bannerman DD, Paape MJ, Lee JW, Zhao X, Hope JC, et al. (2004) Escherichia coli and Staphylococcus aureus elicit differential innate immune responses following intramammary infection. Clin Diagn Lab Immunol 11: 463–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lahouassa H, Moussay E, Rainard P, Riollet C (2007) Differential cytokine and chemokine responses of bovine mammary epithelial cells to Staphylococcus aureus and Escherichia coli. Cytokine 38: 12–21. [DOI] [PubMed] [Google Scholar]
- 6. Bannerman DD (2009) Pathogen-dependent induction of cytokines and other soluble inflammatory mediators during intramammary infection of dairy cows. J Anim Sci 87: 10–25. [DOI] [PubMed] [Google Scholar]
- 7. Wellnitz O, Bruckmaier RM (2012) The innate immune response of the bovine mammary gland to bacterial infection. Vet J 192: 148–152. [DOI] [PubMed] [Google Scholar]
- 8. Zecconi A, Cesaris L, Liandris E, Daprà V, Piccinini R (2006) Role of several Staphylococcus aureus virulence factors on the inflammatory response in bovine mammary gland. Microb Pathog 40: 177–183. [DOI] [PubMed] [Google Scholar]
- 9. Wellnitz O, Berger U, Schaeren W, Bruckmaier R (2012) Mastitis severity induced by two Streptococcus uberis strains is reflected by the mammary immune response in vitro. Schweiz Arch Tierheilkd 154: 317–323. [DOI] [PubMed] [Google Scholar]
- 10. Stutz K, Stephan R, Tasara T (2011) SpA, ClfA, and FnbA genetic variations lead to Staphaurex test-negative phenotypes in bovine mastitis Staphylococcus aureus isolates. J Clin Microbiol 49: 638–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moser A, Stephan R, Corti S, Johler S (2013) Comparison of genomic and antimicrobial resistance features of latex agglutination test-positive and latex agglutination test-negative Staphylococcus aureus isolates causing bovine mastitis. J Dairy Sci 96: 329–334. [DOI] [PubMed] [Google Scholar]
- 12. Griesbeck-Zilch B, Meyer HH, Kühn CH, Schwerin M, Wellnitz O (2008) Staphylococcus aureus and Escherichia coli cause deviating expression profiles of cytokines and lactoferrin messenger ribonucleic acid in mammary epithelial cells. J Dairy Sci 91: 2215–2224. [DOI] [PubMed] [Google Scholar]
- 13. Wellnitz O, Arnold ET, Bruckmaier RM (2011) Lipopolysaccharide and lipoteichoic acid induce different immune responses in the bovine mammary gland. J Dairy Sci 94: 5405–5412. [DOI] [PubMed] [Google Scholar]
- 14. Goldammer T, Zerbe H, Molenaar A, Schuberth HJ, Brunner RM, et al. (2004) Mastitis increases mammary mRNA abundance of beta-defensin 5, toll-like-receptor 2 (TLR2), and TLR4 but not TLR9 in cattle. Clin Diagn Lab Immunol 11: 174–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Strandberg Y, Gray C, Vuocolo T, Donaldson L, Broadway M, et al. (2005) Lipopolysaccharide and lipoteichoic acid induce different innate immune responses in bovine mammary epithelial cells. Cytokine 31: 72–86. [DOI] [PubMed] [Google Scholar]
- 16. Petzl W, Zerbe H, Günther J, Yang W, Seyfert HM, et al. (2008) Escherichia coli, but not Staphylococcus aureus triggers an early increased expression of factors contributing to the innate immune defense in the udder of the cow. Vet Res 39: 18. [DOI] [PubMed] [Google Scholar]
- 17. Kehrli ME, Cullor JS, Nickerson SC (1991) Immunobiology of hematopoietic colony-stimulating factors: potential application to disease prevention in the bovine. J Dairy Sci 74: 4399–4412. [DOI] [PubMed] [Google Scholar]
- 18. Pfaffl MW, Wittmann SL, Meyer HH, Bruckmaier RM (2003) Gene expression of immunologically important factors in blood cells, milk cells, and mammary tissue of cows. J Dairy Sci 86: 538–545. [DOI] [PubMed] [Google Scholar]
- 19. Weber A, Weber AT, McDonald TL, Larson MA (2006) Staphylococcus aureus lipotechoic acid induces differential expression of bovine serum amyloid A3 (SAA3) by mammary epithelial cells: Implications for early diagnosis of mastitis. Vet Immunol Immunopathol 109: 79–83. [DOI] [PubMed] [Google Scholar]
- 20. Wellnitz O, Kerr DE (2004) Cryopreserved bovine mammary cells to model epithelial response to infection. Vet Immunol Immunopathol 101: 191–202. [DOI] [PubMed] [Google Scholar]
- 21. Wellnitz O, Reith P, Haas SC, Meyer HHD (2006) Immune relevant gene expression of mammary epithelial cells and their influence on leukocyte chemotaxis in response to different mastitis pathogens. Vet Med (Praha) 51: 125–132. [Google Scholar]
- 22. Takagi M, Yamamoto D, Ogawa S, Otoi T, Ohtani M, et al. (2008) Messenger RNA expression of angiotensin-converting enzyme, endothelin, cyclooxygenase-2 and prostaglandin synthases in bovine placentomes during gestation and the postpartum period. Vet J 177: 398–404. [DOI] [PubMed] [Google Scholar]
- 23. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 24. Hensen SM, Pavicić MJ, Lohuis JA, Poutrel B (2000) Use of bovine primary mammary epithelial cells for the comparison of adherence and invasion ability of Staphylococcus aureus strains. J Dairy Sci 83: 418–429. [DOI] [PubMed] [Google Scholar]
- 25. Griesbeck-Zilch B, Osman M, Kühn C, Schwerin M, Bruckmaier RM, et al. (2009) Analysis of key molecules of the innate immune system in mammary epithelial cells isolated from marker-assisted and conventionally selected cattle. J Dairy Sci 92: 4621–4633. [DOI] [PubMed] [Google Scholar]
- 26. Alluwaimi AM, Leutenegger CM, Farver TB, Rossitto PV, Smith WL, et al. (2003) The cytokine markers in Staphylococcus aureus mastitis of bovine mammary gland. J Vet Med B Infect Dis Vet Public Health 50: 105–111. [DOI] [PubMed] [Google Scholar]
- 27. Hogan J, Smith KL (2003) Coliform mastitis. Vet Res 34: 507–519. [DOI] [PubMed] [Google Scholar]
- 28. Rainard P, Riollet C (2006) Innate immunity of the bovine mammary gland. Vet Res 37: 369–400. [DOI] [PubMed] [Google Scholar]
- 29. Molenaar AJ, Harris DP, Rajan GH, Pearson ML, Callaghan MR, et al. (2009) The acute-phase protein serum amyloid A3 is expressed in the bovine mammary gland and plays a role in host defence. Biomarkers 14: 26–37. [DOI] [PubMed] [Google Scholar]
- 30. Badolato R, Wang JM, Murphy WJ, Lloyd AR, Michiel DF, et al. (1994) Serum amyloid A is a chemoattractant: induction of migration, adhesion, and tissue infiltration of monocytes and polymorphonuclear leukocytes. J Exp Med 180: 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. He R, Sang H, Ye RD (2003) Serum amyloid A induces IL-8 secretion through a G protein-coupled receptor, FPRL1/LXA4R. Blood 101: 1572–1581. [DOI] [PubMed] [Google Scholar]
- 32. Lee JW, Bannerman DD, Paape MJ, Huang MK, Zhao X (2006) Characterization of cytokine expression in milk somatic cells during intramammary infections with Escherichia coli or Staphylococcus aureus by real-time PCR. Vet Res 37: 219–229. [DOI] [PubMed] [Google Scholar]
- 33. Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, et al. (1999) Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity 11: 443–451. [DOI] [PubMed] [Google Scholar]
- 34. Craven N (1986) Chemotactic factors for bovine neutrophils in relation to mastitis. Comp Immunol Microbiol Infect Dis 9: 29–36. [DOI] [PubMed] [Google Scholar]
- 35. Smith WL, Garavito RM, DeWitt DL (1996) Prostaglandin endoperoxide H synthases (cyclooxygenases)-1 and -2. J Biol Chem 271: 33157–33160. [DOI] [PubMed] [Google Scholar]
- 36.Crofford LJ (1997) COX-1 and COX-2 tissue expression: implications and predictions. J Rheumatol Suppl 49: 15–19. [PubMed]