Abstract
In Sweden, mosquitoes are considered the major vectors of the bacterium Francisella tularensis subsp. holarctica, which causes tularaemia. The aim of this study was to investigate whether mosquitoes acquire the bacterium as aquatic larvae and transmit the disease as adults. Mosquitoes sampled in a Swedish area where tularaemia is endemic (Örebro) were positive for the presence of F. tularensis deoxyribonucleic acid throughout the summer. Presence of the clinically relevant F. tularensis subsp. holarctica was confirmed in 11 out of the 14 mosquito species sampled. Experiments performed using laboratory-reared Aedes aegypti confirmed that F. tularensis subsp. holarctica was transstadially maintained from orally infected larvae to adult mosquitoes and that 25 % of the adults exposed as larvae were positive for the presence of F. tularensis-specific sequences for at least 2 weeks. In addition, we found that F. tularensis subsp. holarctica was transmitted to 58 % of the adult mosquitoes feeding on diseased mice. In a small-scale in vivo transmission experiment with F. tularensis subsp. holarctica-positive adult mosquitoes and susceptible mice, none of the animals developed tularaemia. However, we confirmed that there was transmission of the bacterium to blood vials by mosquitoes that had been exposed to the bacterium in the larval stage. Taken together, these results provide evidence that mosquitoes play a role in disease transmission in part of Sweden where tularaemia recurs.
Introduction
Francisella tularensis, the etiological agent of the zoonotic disease tularaemia, is a Gram-negative coccobacillus. At present, F. tularensis is divided into four subspecies (F. tularensis subsp. tularensis, F. tularensis subsp. holarctica, F. tularensis subsp. novicida and F. tularensis subsp. mediasiatica) [1, 2]. However, the taxonomic boundaries of Francisella novicida have recently been debated [3, 4]. Two subspecies are of clinical importance: type A (F. tularensis subsp. tularensis) and type B (F. tularensis subsp. holarctica) [2]. Subspecies F. tularensis strains are limited to North America, whilst F. tularensis subsp. holarctica strains exist throughout the Northern Hemisphere [2]. The clinical expression of tularaemia largely depends on the route of entry of the infectious agent [5].
The ecology of F. tularensis and how the bacterium persists between outbreaks is not clear; indeed, the historic focus on the epidemiology of tularaemia may have skewed our views [6]. Numerous wildlife species and several potential vectors (i.e. fleas, flies, lice, midges, mites, mosquitoes and ticks) have been associated with transmission of the bacteria [5, 7]. Outbreaks and occurrences of tularaemia are often patchy, i.e. it frequently occurs within natural foci in geographically restricted areas, typically in association with just one or a few key mammalian and arthropod species. It has also been suggested that subspecies of F. tularensis differ with respect to their ecological niches [5]. Historically, the distribution of human tularaemia caused by F. tularensis subsp. holarctica has been associated with close proximity to water [8]. There is also evidence that F. tularensis subsp. holarctica strains persist in watercourses [9, 10], possibly in association with protozoa [11–14]. Even though mosquitoes are considered to be mechanical vectors of F. tularensis [7], it is generally thought that they are the major route of transmission for human cases of tularaemia in Sweden [15, 16], and the occurrence of infected mosquitoes (Aedes cinereus) was reported as early as 1942 [17]. Recent detection of F. tularensis subsp. holarctica deoxyribonucleic acid (DNA) in adult mosquitoes, reared from larvae collected in an endemic area, implies a novel transmission cycle originating in the aquatic habitats of mosquito larva [18]. However, the dynamics of outbreaks of tularaemia remain unknown.
The aim of this study was to evaluate further the role of mosquitoes as vectors of F. tularensis subsp. holarctica by investigating the occurrence of the bacterium in mosquito populations in a Swedish area with endemic tularaemia. Further, experimental exposure and transmission were investigated using a fully virulent F. tularensis subsp. holarctica strain in a laboratory colony of the tropical mosquito Aedes aegypti.
Methods
Occurrence of F. tularensis subsp. holarctica in Field-Sampled Mosquitoes
Study Area, Mosquito Sampling and Species Identification
In 2003 tularaemia re-emerged in Örebro County (59° 16′ 0″ N, 15° 12′ 0″ E, a population of 281,100, 2011). Before 2000, only a handful of cases had been reported from the county; however, between 2003 and 2005, 229 human cases of tularaemia were reported (http://www.smi.se/in-english/statistics/tularaemia/). Mosquito sampling points were chosen based on the information supplied by local physicians about the geographical distribution of human tularaemia cases.
Adult mosquitoes were sampled using the Centre for Disease Control (CDC) miniature light traps with carbon dioxide as an additional attractant, as previously described [19]. From June to September in 2004, mosquitoes were sampled on eight sampling occasions, each comprising three successive nights. Sampling was performed at four locations (Ormesta, Karlslund, Vattenparken and Kårsta) within the Örebro Region, and three traps were used at each location. The CDC traps were placed in trees approximately 1.5 m above the ground and were activated early in the evening and run for 12–14 h. Trapped mosquitoes were anaesthetized with carbon dioxide, separated into three random groups, placed in plastic ampoules (each containing 1 to 500 mosquitoes) and euthanized by freezing on dry ice. The three different groups were used for the following: (1) an initial PCR screening for F. tularensis, (2) mosquito species identification and (3) attempts to cultivate the bacteria. Ampoules containing mosquitoes were stored at −70 °C until further analysis.
During species identification, the mosquitoes were kept cold on a chill table, illuminated by a cold light lamp and identified to species based on their morphology [20]. Mosquitoes identified to species were sorted into ampoules according to species, capture date and trap and were returned to the −70 °C freezer.
Molecular and Culture-Based Methods for F. tularensis Subsp. holarctica Detection in Field-Sampled Mosquitoes
Prior to DNA extraction from mosquitoes collected in the field, 250 μl distilled water and 12.5 μl 2.8 M NH4OH were added to the ampoules of a maximum of 50 mosquitoes. The mixture was incubated at room temperature (RT) for 10 min. The samples were vortexed for 20 s and, thereafter, bead beaten to disrupt the mosquito bodies and release intracellular bacteria (450 mg each of 1 and 0.1 mm silica beads for 60 s, Mini-BeadBeater-8, BioSpec). Bead-beaten samples were incubated at 95 °C for 30 min before centrifugation at 800×g for 3 min. DNA was extracted using phenol (pH 8)–chloroform–isoamyl alcohol (25:24:1) and subsequently precipitated using ethanol. The pellet was resuspended in 40 μl of distilled water and frozen if not immediately subjected to PCR analysis.
The screening of ampoules of mosquitoes for the presence of F. tularensis was performed using a real-time PCR probe-based assay for detection of the F. tularensis-specific lpnA gene, using iQFt1 F/R primers and probe as previously described [13] (Table 1). Positive samples were subjected to a F. tularensis subsp. holarctica-specific PCR based on the 30-bp deletion region and fragment size analysis according to the literature [18, 21]. Each reaction consisted of 1 μl template, 1× AmpliTaq Gold PCR buffer, 40 μM each of the primers FtM19InDel (Table 1), 2.6 mM MgCl2, 1 M betaine, 0.2 mM dNTP, 0.5 U of AmpliTaq Gold Polymerase and Milli-Q water to a total volume of 25 μl. An initial denaturation at 94 °C for 2 min was followed by 50 cycles of 94 °C for 30 s, 60 °C for 30 s and 72 °C 30 s, followed by final incubation at 72 °C for 5 min on MyCycler (Bio-Rad Laboratories, Hercules, CA, USA). Positive control reactions using DNA from F. tularensis and negative control reactions without a template were included in each PCR run. Genomic rat control DNA and internal control primers IQFPrat36B4 and IQRPrat36B4 were used as previously described [13] (Table 1). The resulting amplicons were sized by capillary electrophoresis on the CEQ™ 8800 Genetic Analysis System (Beckman Coulter, Inc., Fullerton, CA, USA). A volume of 1 μl PCR products was mixed with CEQ DNA Size Standard Kit 400 in a sample loading solution according to the manufacturer's manual; a fragment size of 100 bp corresponds to F. tularensis subsp. holarctica.
Table 1.
Primers used in this study
| Primer | Target | Sequence | Ref |
|---|---|---|---|
| iQFt1 F | lpnA | 5′-CGCAGGTTTAGCGAGCTGTT-3′ | [13] |
| iQFt1 R | 5′-GCAGCTTGCTCAGTAGTAGCTGTCT-3′ | ||
| iQFt1 p | FAM 5′-CATCATCAGAGCCACCTAACCCTA-3′ | ||
| lpnA2F | lpnA | 5′-CGCAGGTTTAGCGAGCTGTT-3′ | [18] |
| lpnA2R | 5′-GAGCAGCAGCAGTATCTTTAGC-3′ | ||
| FtM19InDel F | 30-bp deletion | Wellred-PA 5′ CCAGTACAAACTCAATTTGGTTATCATC-3′ | [9] |
| FtM19InDel R | 5′-GTTTCAGAATTCATTTTTGTCCGTAA-3′ | ||
| iQFPrat36B4 | Internal control | 5′-GCCCAGAGGTGCTGGACAT-3′ | [13] |
| iQRPrat36B4 | 5′-ATTGCGGACACCCTCTAGGA-3′ | ||
| iQrat36B4 p | TET 5′-ACAGAGCAGGCCCTGCACACTCG-3′ | ||
| ITS1_F338 | Mosquito | CGCTCGGACGCTCGTAC | [28] |
| ITS1_R427 | CTTCGAGCTTCGACGACACA |
A subset of the third group of the mosquitoes collected in the field was homogenized and spread on selective and non-selective cysteine heart agar plates supplemented with sheep blood, CHAB-PACCV and CHAB [22]. Selective and non-selective agar medium according to the literature [23] was also used in parallel. The selective supplements used were 600,000 IU of penicillin, 1,000,000 IU of polymyxin B and 100 mg of cycloheximide. All agar plates were incubated at 37 °C in 5 % CO2. Culture plates were checked for potential Francisella colonies after 2 days of incubation. Potential Francisella colonies were picked and analyzed with PCR using the MicroSeq 500 16S rRNA Bacterial Identification Kit (Applied Biosystems).
The infection rate (IR) and confidence intervals of F. tularensis in field-sampled mosquitoes were calculated using bias-corrected likelihood methods and skew-corrected score [24].
A Laboratory Mosquito Model for Studies of Transstadial Maintenance and Transmission of Virulent F. tularensis Subsp. holarctica
Francisella Strains and Constructs
F. tularensis subsp. holarctica FSC 200 [25] was grown on modified Thayer–Martin agar plates [26] at 37 °C in 5 % CO2. The amino acid sequence of Discosoma sp. red fluorescent protein, DsRed-Express (Clontech Laboratories, Inc., Mountain View, CA, USA) was converted into a DNA sequence with optimized codon usage for protein expression in F. tularensis and chemically synthesized by GenScript, NJ, USA. The 684-bp sequence was introduced into the NdeI-EcoRI site of pKK289Km-gfp [11] under control of the LVS GroESL promoter, after removal of the gfp gene. The resulting plasmid pKK289Km-DsRed was then introduced to F. tularensis subsp. holarctica FSC200 by cryotransformation [27]. The resulting plasmid-carrying strain was denoted FSC 849. All experiments using FSC 200 and FSC 849 were performed under BSL 3 conditions.
Rearing of Mosquito A. aegypti
Eggs of the tropical mosquito A. aegypti, kindly provided by Oxitec (Oxitec Ltd., Oxford, England), were hatched in deionized water. Larvae, approximately 20 per container (Mosquito Breeder, BioQuip Products, Rancho Dominguez, CA, USA), were maintained in tap water at 26 °C and fed crushed fish flakes. When they developed, pupae were removed from the larval containers and placed in clean tap water. Adult mosquitoes were fed from cotton wool soaked in 10 % dextrose solution.
Exposure of Mosquito Larvae to F. tularensis Subsp. holarctica and Transstadial Maintenance
Uptake of F. tularensis subsp. holarctica by the mosquito larvae and transstadial maintenance to the resulting pupae and adult mosquitoes was studied using DsRed-labelled F. tularensis subsp. holarctica (FSC 849) (Fig. 1). Mosquito larvae (second instar) were transferred to tap water containing bacteria at a concentration of 107 colony-forming units (cfu) per ml. After a 24-h period of exposure to bacteria, the larvae were washed three times in water and transferred to fresh tap water where they were kept until harvested (by freezing −70 °C for 5 min). Harvesting was performed at one of the three developmental stages: larva (fourth instar), pupa or adult mosquito. In addition, haemolymph from pupae and adults were obtained through perfusing the thorax using capillary tubes. All samples (larva, pupa, adult and haemolymph) were stored at −70 °C until further analysis for the presence of F. tularensis using real-time PCR.
Fig. 1.
Experimental set-up for investigating the uptake and transstadial maintenance of F. tularensis subsp. holarctica in the mosquito A. aegypti. Percent of F. tularensis-positive mosquitoes during different developmental stages (fourth instar larva, pupa and adult mosquito). Data are summarized from three separate experiments involving between 59 and 68 mosquitoes in each run. Transmission of the transstadially maintained F. tularensis subsp. holarctica was investigated after mosquito feeding from an artificial source of blood. The mosquitoes that were exposed to F. tularensis subsp. holarctica as larvae were allowed to feed from blood vials. The results are presented as percent of blood vials positive for F. tularensis DNA after mosquito feeding
Mosquitoes and Artificial Blood Meal Experiments
Adult A. aegypti mosquitoes, exposed to F. tularensis subsp. holarctica as larvae (as described above), were presented with an artificial blood meal (Fig. 1). A small vial containing sheep, guinea pig or mouse blood was covered with parafilm and placed upside down on the mosquito container, representing an artificial source of blood. Transmission of bacteria from the mosquito to the blood was investigated using F. tularensis-specific real-time PCR with lpnA2 primers (Table 1), culturing and immunofluorescence microscopy.
In another experiment, nine batches (one to eight individuals) of uninfected adult mosquitoes were allowed to feed on vials of mouse blood spiked with approximately 106 cfu per ml (FSC 849). Mosquitoes were harvested within 48 h after feeding. In both experiments, blood-fed mosquitoes were harvested by freezing, as described above, and analyzed for the presence of F. tularensis using real-time PCR.
Mosquitoes and Transmission Experiments Using Rodent Host
In order to study potential uptake and transmission of F. tularensis subsp. holarctica by mosquitoes to susceptible hosts, C57Bl/6 mice were used (in-house bred). Mice were housed under conventional conditions and allowed to acclimatize for at least 7 days before infection. The study was approved by the Local Ethical Committee on Laboratory Animals in Umeå, Sweden. With the aim of determining the optimal time for mosquitoes to feed on blood of infected mice, bacterial counts in mouse blood during F. tularensis subsp. holarctica infection were investigated. For these studies, five mice were subcutaneously (s.c.) infected with F. tularensis subsp. holarctica (FSC 200, infection dose 27 cfu). Bacterial counts in mouse blood were monitored daily. Blood from infected mice was bead beaten, serially diluted and spread on Thayer–Martin agar plates [26] incubated at 37 °C in 5 % CO2. Bacterial growth was recorded by counting cfu. All transmission experiments with infected mosquitoes and mice were performed under BSL 3 conditions.
Prior to exposure to the mosquitoes in the transmission study, mice were anesthetized by intra-peritoneal injection of ketamine (Ketalar® vet) (75 mg/kg) and medetomidine (Domitor® vet) (1 mg/kg) and placed on top of a mosquito container to allow the mosquitoes to feed on mouse blood. All mice were removed from the mosquito container when either ten mosquitoes had fed (as observed by extended blood-filled abdomen) or 30 min had passed. The experiment evaluated the possibility that A. aegypti could transstadially transmit F. tularensis subsp. holarctica. Here, 184 female mosquitoes exposed to F. tularensis subsp. holarctica at the larval stage (as described above) were allowed to feed on 14 naïve mice (Fig. 2(a)). As negative controls, non-infected mosquitoes fed on five naïve mice. Mice exposed to mosquitoes were monitored for signs of disease for 25 days.
Fig. 2.
Experiment to investigate the ability of the mosquito A. aegypti to transstadially and directly transmit F. tularensis subsp. holarctica to susceptible hosts. a Transstadial transmission of F. tularensis subsp. holarctica: mosquitoes exposed to the bacterium as larvae were allowed to feed on the blood of naïve mice. Chance of transmission is the number of mosquitoes that took a blood meal and were later identified as positive for F. tularensis. Transmission is the number of mice positive for F. tularensis at the end of the experiment. b Mosquito-mediated direct transmission of F. tularensis subsp. holarctica between hosts: mosquitoes were allowed to feed on the blood of mice infected with F. tularensis subsp. holarctica. Five days later, the mosquitoes were allowed to feed on blood of naïve mice in order to establish whether there was direct transmission. Uptake is the number of mosquitoes that took blood from infected mice and were later identified as positive for F. tularensis. In both experiments, mice were monitored for signs of disease for up to 25 days, and no traces of F. tularensis were detected in mouse spleen or blood samples analyzed using real-time PCR and culture methods (transmission). Mosquitoes were analyzed for the presence of F. tularensis and mouse DNA using PCR (uptake and chance of transmission)
Another experiment was designed to evaluate the ability of A. aegypti to transmit F. tularensis subsp. holarctica between susceptible hosts. Here, 33 non-exposed female A. aegypti divided into five batches were allowed to feed on two mice (Fig. 2(b)). The mice had been infected with F. tularensis subsp. holarctica s.c. (FSC 849, infection dose 27) 3 days prior to the mosquito blood meal. Four days after the mosquitoes fed on the infected mice, they were allowed to feed on five naïve mice (Fig. 2(b)). Mice were observed for symptoms of the disease for 21 days. Mosquitoes from the two experiments were harvested after the blood meal and analyzed for the presence of the bacteria using F. tularensis-specific real-time PCR and lpnA2 primers (Table 1).
Detection of F. tularensis Subsp. holarctica in the Laboratory Mosquito Model
DNA Extraction
Tris–EDTA (TE) buffer (20–100 μl) was added to mosquito larvae, pupae and haemolymph samples and boiled for 15 min. DNA from adult mosquitoes was extracted using the SoilMaster DNA Extraction Kit (Epicentre Biotechnologies, Madison, WI, USA) according to the manufacturer's instructions. The resulting DNA pellet was resuspended in 50 μl TE buffer. Blood samples (50 μl) were prepared using the Qiagen Tissue Kit, adding 150 μl G2 and 2 μl proteinase K, and were incubated at 65 °C for 15 min and then extracted using the EZ1 Advanced PrepRobot (Qiagen, Venlo, Netherlands) and EZ1 Tissue Kit (Qiagen). The elution volume was 50 μl.
PCR Analysis
Samples from the mosquito experiments were screened for the presence of F. tularensis using a modified real-time PCR SYBR-based assay for detection of the F. tularensis-specific lpnA gene, as previously described [18]. Each reaction volume consisted of 1–5 μl template, 10 μl 1× SsoFast EVA Green (Bio-Rad), 400 nmol/l each of the lpnA2R/F primer (Table 1) and Milli-Q up to 20 μl. An initial denaturation at 98 °C for 2 min was followed by 45 cycles of 98 °C for 5 s and 60 °C for 5 s and a melting curve of 60–95 °C on CFX96 (Bio-Rad). In addition, an internal control was included using a construct of a non-coding part of the pSKluxCDABE (ATG: Biosynthetics) in plasmid pKK214tet, flanked by the lpnA2 primer pair sequence generating a 93-bp fragment (CGCAGGTTTAGCGAGCTGTTTAGATTTCGAGTTGCAGCGAGGCG GCAAGTGAACGAATCCCCAGGAGCATAGCTAAAGATACTGCTGCTGCTC).
In order to confirm the material from A. aegypti in samples, specific primers ITS1-F338 and ITS1-R427 (Table 1) were used as previously described [28].
To test the detection limit for the real-time PCR assay and to generate a standard curve for assessing target DNA concentrations in the matrixes analyzed, DNA extraction was performed in triplicate on samples of larvae, pupae, haemolymph and adults spiked with F. tularensis subsp. holarctica concentrations ranging from 1 × 100 to 1 × 106 cells per ml.
Culture of F. tularensis Subsp. holarctica
Cultures from laboratory-reared A. aegypti mosquitoes and from vials of blood used for the artificial blood meals were produced from samples that had been homogenized in screw-capped microtubes with integral o-ring seals using 1-mm silica beads and the Mini-BeadBeater-8 (BioSpec Products, Inc., Bartlesville, OK, USA). The resulting samples were spread on selective T4 agar plates [29] and incubated at 37 °C in 5 % CO2. Bacterial growth was recorded after 3 to 12 days of incubation depending on the degree of contamination. Potential Francisella colonies were picked and confirmed using the PCR SYBR-based assay for detection of the F. tularensis-specific lpnA gene described above.
Fluorescence Microscopy
To confirm transmission of F. tularensis to the vials of blood used to feed artificially the mosquitoes exposed to F. tularensis subsp. holarctica in the larval stage, fluorescence microscopy was performed using the Leica DMR (UV light, 330–380 nm). Blood was smeared on glass slides and allowed to dry before analysis either by direct detection of fluorescence from DsRed (using FSC849) or by incubation with a primary rabbit anti-F. tularensis antibody (Grünow, R.) (1:200) for 2 h at RT. The glass slides were thereafter rinsed five times in dH2O before incubation with a secondary FITC-labelled mouse anti-rabbit antibody (1:1,000) for 1 h at RT. The glass slides were then rinsed five times with dH2O before microscopy.
Results
Mosquito Abundance and Occurrence of F. tularensis in Wild-Caught Mosquitoes
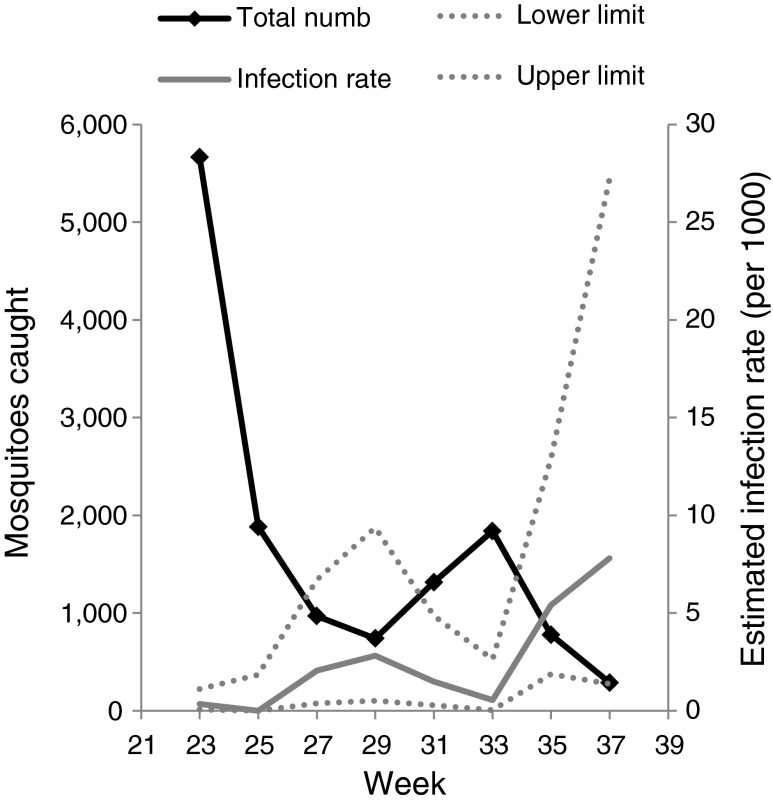
The first group of mosquitoes collected in the field was used for the initial PCR screening for the presence of F. tularensis. This material comprised approximately 13,500 mosquitoes in 188 mixed species ampoules (Table 2). The number of mosquitoes caught at the four locations on each sampling occasion peaked twice (Fig. 3). The peak in June (week 23) corresponds to the emergence of snow-pool mosquito species (mosquito functional group 2a), while the peak in August (week 33) corresponds to the emergence of flood water species (mosquito functional group 2b) [19].
Table 2.
The first group of the mosquitoes collected from Örebro, an area of Sweden with endemic tularaemia, was analyzed for the presence of the F. tularensis-specific sequence lpnA. Samples were collected from four locations (Ormesta, Karlslund, Vattenparken and Kårsta) on eight sampling occasions (June to September)
| Sampling occasion | Total ampoules (individuals) | F. tularensis-positive ampoules (individuals) |
|---|---|---|
| W 23 | 41 (5,666) | 2 (66) |
| Ormesta | 19 (3,810) | n.d. |
| W 25 | 27 (1,882) | n.d. |
| Ormesta | 8 (960) | n.d. |
| W 27 | 21 (968) | 2 (70) |
| Ormesta | 8 (549) | 1 (60) |
| W 29 | 19 (740) | 3 (125) |
| Ormesta | 6 (376) | 1 (70) |
| W 31 | 22 (1,316) | 2 (47) |
| Ormesta | 7 (635) | n.d. |
| W 33 | 25 (1,839) | 1 (70) |
| Ormesta | 6 (465) | n.d. |
| W 35 | 19 (778) | 4 (143) |
| Ormesta | 5 (98) | 3 (63) |
| W 37 | 14 (287) | 2 (85) |
| Ormesta | 5 (126) | 2 (85) |
| Total | 188 (13,476) | 16 |
The numbers refer to the number of ampoules containing mosquitoes that were analyzed, and those within brackets refer to the estimate of the total number of individuals within the ampoules. Results of the analysis of material from all four locations are presented for each sampling week (W 23 to 37). Results of analysis of the material from Ormesta are presented separately for each sampling week (Ormesta) since they were the basis for selecting dates and locations for further analysis
n.d. not detected
Fig. 3.
The first group of the mosquitoes collected in the field that was subjected to an initial PCR screening for the presence of F. tularensis. The number of mosquitoes caught on each sampling occasion in relation to the estimated IR per 1,000 mosquitoes collected. Dotted lines represent the calculated upper and lower IR limits. The F. tularensis infection rate were highest at 2 to 6 weeks after the peak in total number of mosquitoes
F. tularensis-specific sequences (lpnA) were detected in mosquito ampoules collected throughout the summer season and at all four sampling locations (Table 2). The F. tularensis IR in the mixed species mosquito pools increased approximately 4–6 weeks after the peak occurrence of snow-pool mosquitoes and, again, approximately 2–4 weeks after the peak occurrence of flood water mosquitoes (Fig. 3). This demonstrates that the infection rate of F. tularensis in the mosquito population varies over the summer, and it increases towards the end of summer with the declining mosquito population comprising an increasing proportion of flood water mosquitoes.
F. tularensis Subsp. holarctica in Several Species of Wild-Caught Mosquitoes
The second group of mosquitoes collected in the field was used for species identification and comprised originally of approximately 9,000 individuals in 181 ampoules. The material was identified to species and sorted by sampling occasion, location and species, resulting in 859 ampoules (data not shown). In order to investigate the presence of F. tularensis subsp. holarctica in wild-caught mosquitoes and any potential association between the bacterium and a specific mosquito species, a subset of the species sorted sample was analyzed further. The subset was selected in order to maximize the chance of finding F. tularensis-positive mosquitoes. The selection was performed on the basis of the initial PCR screening of the first group of mosquitoes collected in the field, during which high frequencies of F. tularensis positives were detected in the mixed species mosquito ampoules from the Ormesta area on four sampling occasions (i.e. weeks 27, 29, 35 and 37, see Table 2). This subset of species identified mosquitoes comprised of 791 mosquitoes in 89 single-species ampoules (Table 3). Twenty of the 89 mosquito ampoules were positive for F. tularensis-specific sequences (lpnA, Table 3). Eighteen of the 20 lpnA-positive ampoules were also positive in the screening for F. tularensis subsp. holarctica using the FtM19InDel primer pair that targets the F. tularensis subsp. holarctica-specific 30-bp deletion region (resulting in a 100-bp amplicon) (Table 3). Of the 14 mosquito species identified in the subset, 11 were found to be positive for F. tularensis subsp. holarctica sequences. These 11 mosquito species belonged to the four mosquito genera: Aedes, Anopheles, Coquillettidia and Culex (Table 3).
Table 3.
The second group of the field-sampled mosquito material was species identified. A subset of this material was subjected for further analysis for the presence of F. tularensis subsp. holarctica. The subsample consisted of ampoules of mosquitoes captured in the Ormesta area on four sampling occasions W 27, 29, 35 and 37 (chosen based on results presented in Table 2, as it was found that samples from Ormesta collected during these weeks had the highest frequency of F. tularensis-positive ampoules)
| Mosquito species | Total ampoules (individuals) | F. tularensis positive | F. tularensis subsp. holarctica positive |
|---|---|---|---|
| A. cinereus | 18 (410) | 4 (146) | 4 |
| Aedes sticticus | 10 (33) | 3 (11) | 3+ |
| Aedes vexans | 5 (5) | 1 (1) | 1 |
| Anopheles claviger | 6 (8) | 2 (3) | 1 |
| Anopheles maculipennis | 7 (16) | 2 (5) | 1 |
| Coquillettidia richiardii | 10 (153) | 3 (45) | 3 |
| Culiseta morsitans | 4 (7) | n.d. | – |
| Culex pipiens/Culex torrentium | 6 (28) | 1 (1) | 1 |
| Aedes annulipes | 5 (11) | 1 (1) | 1 |
| Aedes cantans | 9 (86) | 1 (13) | 1 |
| Aedes communis | 4 (11) | n.d. | – |
| Aedes intrudens | 3 (19) | 1 (1) | 1 |
| Aedes leucomelas | 1 (2) | 1 | 1+ |
| Aedes punctor | 1 (2) | n.d. | – |
| Total | 89 (791) | 20 | 18 |
The numbers refer to the number of ampoules containing mosquitoes that were analyzed, and those within brackets refer to the estimate of the total number of individuals within the ampoules. The presence of F. tularensis-specific sequences was determined, and positive samples were further investigated for the presence of the clinically relevant subspecies F. tularensis subsp. holarctica using fragment size analysis of a 30-bp deletion region unique to F. tularensis subsp. holarctica (FtM19Indel); + indicates the presence of F. tularensis subsp. holarctica and other F. tularensis subspecies
n.d. not detected
Attempts to culture Francisella performed on the third group of mosquitoes collected in the field did not result in any identification of Francisella colonies.
Transstadial Maintenance of F. tularensis Subsp. holarctica in Laboratory-Reared A. aegypti
A. aegypti mosquitoes exposed to F. tularensis subsp. holarctica as larvae were harvested at different developmental stages (larvae, pupae and adults) and analyzed for the presence of F. tularensis using molecular methods (i.e. the lpnA PCR assay, Fig. 1). Spiking experiments showed that real-time PCR detection of the F. tularensis lpnA gene (lpnA2 primers) was possible for concentrations of >200 cells per ml in all matrixes (F. tularensis subsp. holarctica in mosquito larvae, pupae and adults). The range of linearity for all matrixes was determined to be between 103 and 106 F. tularensis subsp. holarctica cfu/ml.
Real-time PCR analysis of mosquito larvae exposed to F. tularensis subsp. holarctica showed that 5 days after infection, 69 ± 27 % of the larvae (total of 59 analyzed) were positive for F. tularensis (Fig. 1). Approximately 1 week later, the remaining exposed larvae developed into pupae. In the PCR analysis of a subset of the pupae, 34 ± 1 % of the 62 pupae examined were positive for the presence of the bacterium. From the remaining pupae, adult mosquitoes emerged (n = 68). Approximately 14–16 days after the original exposure of mosquito larva, 25 ± 5 % of adult mosquitoes were positive for F. tularensis. These results are based on three independent experiments, each starting with 59 to 68 mosquito larvae.
In addition, haemolymph from pupae and adult mosquitoes was sampled and analyzed in order to confirm the presence of F. tularensis inside the mosquitoes. Approximately 29 ± 4 % of the haemolymph from pupae (n = 62) and 19 ± 2 % of the haemolymph from adult mosquitoes (n = 68) were positive for F. tularensis.
In order to investigate the possibility that mosquitoes take up and subsequently maintain viable F. tularensis subsp. holarctica derived from a blood meal, nine batches of previously uninfected mosquitoes (one to eight mosquitoes in each batch) were allowed to feed from vials containing mouse blood spiked with F. tularensis subsp. holarctica (approximately 106 cfu per ml). Mosquitoes from five out of the nine batches were, when newly fed (analyzed within 48 h of the blood meal), found positive for F. tularensis using real-time PCR. However, attempts to culture the bacterium from mosquitoes were unsuccessful.
Transstadial Transmission of F. tularensis Subsp. holarctica to Blood After Artificial Mosquito Feeding
Batches of approximately 20 mosquitoes exposed to F. tularensis subsp. holarctica in the larval stage were allowed to feed from vials of blood from three different sources (sheep, guinea pig and mouse). A total of 1–11 mosquitoes were observed to feed from each vial of blood. Mosquito feeding was confirmed by PCR on blood from the vials using ITS1 primers targeting a mosquito specific sequence. Real-time PCR analysis of blood indicated the presence of F. tularensis in one out of six sheep blood vials, in five out of 25 guinea pig blood vials and in five out of 20 mouse blood vials. Overall, F. tularensis subsp. holarctica was transmitted to 20 ± 4 % of the blood vials as a result of feeding by mosquitoes exposed to F. tularensis subsp. holarctica as larvae (Fig. 1). Transmission results were confirmed by fluorescence microscopy, which directly detected F. tularensis subsp. holarctica (DsRed) in the post-feeding blood vials (data not shown). The numbers of bacteria recorded were low, and efforts to cultivate the bacteria from mosquitoes and blood samples were unsuccessful.
Mosquito Transmission of F. tularensis Subsp. holarctica to Susceptible Hosts
Transstadial Transmission
A total of 184 female A. aegypti, exposed to F. tularensis subsp. holarctica during the larval stage, were divided into 14 batches and allowed to feed on naïve mice (n = 14) (Fig. 2(a)). In total, we observed 58 mosquito blood meals, 11 of which were by mosquitoes that were later shown to be positive for F. tularensis based on the presence of both mouse and F. tularensis DNA in the mosquito. The mice were monitored for a minimum of 15 days and a maximum of 25 days for clinical signs of disease. None of the mice developed tularaemia, and no traces of F. tularensis were detected in spleen or blood samples collected from the mice after they had been euthanized and analyzed using real-time PCR and culturing.
We speculated a potential requirement for prior activation of the bacteria, resulting from a blood meal, for mosquito transmission of F. tularensis subsp. holarctica. Thus, five of the mosquito batches (a total of 37 mosquitoes exposed to F. tularensis subsp. holarctica in the larval stage) were allowed a first blood meal on naïve mice (n = 5) 5 days prior to a second blood meal. Thirteen mosquitoes were fed during this first run. None of the mice developed tularaemia, and no bacteria were detected in blood or spleen samples from these mice.
The negative controls, four mice bitten by non-infected mosquitoes, displayed no signs of illness during the entire experiment (21 days). The positive controls, five mice infected (s.c.) with F. tularensis subsp. holarctica, were euthanized on day 3 or 4 after infection due to severe symptoms.
Direct Transmission
The possibility of mosquitoes as vectors for F. tularensis subsp. holarctica transmission between diseased and naïve hosts was investigated. Counts of F. tularensis subsp. holarctica in the blood during mouse infection were observed to increase from 5 ± 4 × 102 cfu/ml blood 2 days after infection to 4 ± 4 × 1010 bacteria/ml blood 4.5 days after infection when the experiment was terminated due to clinical signs of the disease (Fig. 4). In order to investigate the possibility of direct transmission, the mosquitoes were allowed to feed on mice on the 3rd day of F. tularensis subsp. holarctica infection. Thirty-three uninfected female mosquitoes divided into five batches were allowed to blood feed on mice (n = 2) infected with F. tularensis subsp. holarctica (27 cfu of FSC 849, Fig. 2(b)). At the time of mosquito feeding, bacterial counts in the blood of infected mice were determined to be 4 ± 0.5 × 105 cfu/ml, similar to what is observed during mouse infection with wild-type F. tularensis subsp. holarctica (Fig. 4). Out of 12 observed blood meals, seven lead to transmission of the bacteria to the feeding mosquito (infection rate of 58 %). Mosquitoes that acquired F. tularensis subsp. holarctica through blood feeding maintained the bacteria for at least 2 weeks. However, transmission of the disease was not observed upon a second blood meal on five naïve mice, where 14 out of the 33 mosquitoes were observed to take a blood meal. Four of the mosquitoes that took a second blood meal and fed on the naïve mice were later found to be positive for F. tularensis.
Fig. 4.
Bacterial counts of F. tularensis subsp. holarctica in blood during mouse infection. Colony-forming units (cfu) of the bacterium F. tularensis subsp. holarctica in the mouse blood following subcutaneous injection (infectious dose of 27 cfu/ml)
Discussion
F. tularensis subsp. holarctica circulates in the mosquito population in Örebro, a Swedish area with endemic tularaemia. Sequences specific for F. tularensis (lpnA) were detected in mosquitoes from all sampling locations in the area and throughout the summer season (June to September). Almost all F. tularensis (lpnA)-positive samples subjected for further analysis were also positive for the F. tularensis subsp. holarctica-specific region (FtM19InDel). Thus, sequences specific to the clinically relevant F. tularensis subsp. holarctica were found in mosquitoes of several different species. Considering recent reports of a high diversity of Francisella in environmental samples [9, 30–32], these results indicate a restriction of Francisella diversity through persistence in mosquitoes. In line with this, F. novicida has been shown not to be transstadially maintained during mosquito development [33]. This suggests selectivity associated with the pathogenic F. tularensis subsp. holarctica persistence in mosquitoes.
Traditionally, mosquitoes have mainly been considered to function as mechanical vectors for transmission of F. tularensis subsp. holarctica between hosts [7]. We have previously shown that the bacterium can be transstadially maintained, from larvae to adult, in mosquitoes of several species collected as larvae in the field and reared to adults in the laboratory [18]. Here, a model for mosquito exposure, where A. aegypti larvae were feed on planktonic F. tularensis subsp. holarctica, was established in order to investigate the role of a potential mosquito vector in the life cycle of the bacterium. Our results show that F. tularensis subsp. holarctica is transferred to mosquito larvae from its aquatic habitat and transstadially maintained in 25 % of the adult mosquitoes. Further, the bacterium is transmissible to the blood when mosquitoes feed on artificial sources of blood (i.e. blood in vials). However, none of the mammals exposed to the F. tularensis-positive mosquitoes were infected during the transmission experiment. It is very likely, however, that transmission of tularaemia by mosquitoes in nature occurs at much lower frequencies than could be detected in this study. Here, out of the 72 mosquitoes that fed on the blood of naïve mice, a total of 14 were later confirmed to be positive for F. tularensis. We speculate that the frequency of F. tularensis subsp. holarctica transmission by mosquitoes in the environment is low and probably depends on species-specific interaction alternative that transmission is mechanical and requires inoculation via smashing of vector onto the skin. There was no association between F. tularensis lpnA and any specific mosquito species in the material collected in the field. Mosquito species positive for F. tularensis included both snow-pool species (four positive out of 23 ampoules, infection rate of 0.17) and flood water species (eight positives out of 33 ampoules, infection rate of 0.24). The initial screening of the first group of mosquitoes collected in the field revealed an increasing IR of F. tularensis with declining mosquito populations in the late August and September. This coincides largely with human cases of tularaemia, which were reported in the area from August to October 2004 (a total of 50 cases, http://www.smi.se/in-english/statistics/tularaemia/?y=2004#statistics-nav). The relevance of using A. aegypti as a model to study the transmission of F. tularensis subsp. holarctica in Sweden is open to question since the species does not occur naturally in Sweden. Nevertheless, to our knowledge, there is no suitable laboratory mosquito model available for any of the species native to Sweden. Aedes sp. or other mosquito species may be more or less competent vectors for transmission of F. tularensis subsp. holarctica to susceptible hosts. In order to confirm that a transmission route for F. tularensis subsp. holarctica from the environment (water, soil or host) via a mosquito vector is relevant for transmission of the disease, vector competence of relevant locally occurring mosquito species needs to be demonstrated.
The frequency of F. tularensis-positive mosquitoes after feeding from infected mice was higher than the frequency of infection of mosquitoes exposed to F. tularensis subsp. holarctica as larvae (approximately 58 and 25 %, respectively). This indicates the possibility of mosquito-mediated transmission of F. tularensis subsp. holarctica directly between susceptible hosts during an ongoing outbreak. It is possible that the mosquitoes play a dual role as vectors during outbreaks of tularaemia, both as a link between a reservoir and susceptible host and as an amplifier of an ongoing outbreak by direct transmission of the bacterium between susceptible hosts.
Sampling of haemolymph from pupae and adult mosquitoes exposed to F. tularensis subsp. holarctica as larvae showed a high frequency of F. tularensis subsp. holarctica-positive samples (29 and 19 %, respectively), suggesting that mosquitoes used in the model do maintain F. tularensis subsp. holarctica, internally. Based on our real-time PCR assay, we did not, however, find any evidence that the bacterium replicates in the mosquito A. aegypti. The real-time PCR assay used in this study can detect low numbers (in hundreds) of F. tularensis in mosquitoes, but the range of linearity was established between 103 and 106 bacteria per mosquito. However, it cannot be excluded that the PCR assay detected DNA from non-viable bacteria.
The bacterial counts of F. tularensis subsp. holarctica reached approximately 1010 bacteria/ml in mouse blood during infection. Consequently, a deceased animal can contaminate the local environment (soil and water) with an extremely high concentration of F. tularensis subsp. holarctica. Even though the retention time for the bacterium varies depending upon a variety of factors such as temperature, salinity, direct exposure to sunlight and other physical factors that generally affect the survival of microbes, it has been documented that surface waters and mud in stream bottoms and ponds can be contaminated with F. tularensis. The bacterium has been shown to survive at infective levels in those substrates for many months [10]. Other studies have shown that although F. tularensis subsp. holarctica persists locally in the environment for prolonged periods (in years), under specific conditions, the bacterium is reported to enter a non-infective, viable but non-culturable (VBNC) state [9, 13, 34, 35]. Hot spots of recently introduced infective bacteria may act as temporary sources for tularaemia outbreaks via dispersal of the bacterium as aerosols (contaminated dust) or contaminated water. However, the role of such hotspots in the perpetuation of F. tularensis subsp. holarctica is unclear. Previously published data suggest that F. tularensis subsp. holarctica persists in association with water and protozoa and thus could be transported up through the microbial food web to mosquito larvae [11]. Moreover, insect antimicrobial peptides have been shown partially to inhibit the growth of F. novicida in a fly model involving Drosophila melanogaster [36]. The increasing body of whole-genome data from Francisella species and subspecies indicate that there is almost no exchange of genetic material between F. tularensis subspecies [37]. This could be due to them occupying separate environmental niches or having different life cycles and suggests that the bacteria replicate in environments where there is little opportunity to interact with other bacteria. In addition, an epidemiological investigation combined with high-resolution genotyping of F. tularensis subsp. holarctica isolates obtained from patients in the Örebro region indicated that genetic subpopulations of the bacteria were present throughout the summer season and also persisted over several years with little genetic variation [38]. Taken together, current data indicate that between outbreaks, the causative agent of tularaemia, F. tularensis subsp. holarctica, persists in the environment in a dormant state with little or no replication.
In conclusion, we have shown that F. tularensis subspecies holarctica is found in several genera and species of wild-caught mosquitoes in Sweden. Subsequent laboratory studies of A. aegypti larvae exposed to fully virulent F. tularensis subsp. holarctica demonstrated transstadial maintenance of the bacteria during metamorphosis into adult mosquitoes. The possibility of mosquito transmission of the bacteria to blood vials during artificial feeding was confirmed as well as transmission of the bacteria from infected mice to mosquitoes. Under our experimental conditions, we did not demonstrate transmission from mosquitoes to mice that confirm the viability of the bacteria. We found no evidence for the replication of F. tularensis subsp. holarctica in mosquitoes, but the results from the molecular analysis indicate that the bacterium does reside inside mosquitoes.
To further clarify the transmission mechanism, future studies should investigate the potential of mechanical transmission of the bacterium via analysis of mouthparts after feeding on infected host and possible transmission via interrupted feeding. A very prolonged incubation of the susceptible hosts (i.e. longer than the 21 days used in this study) may also ensure that potential VBNC Francisella cells resuscitate. Further, F. tularensis infection models show a great variation in ID50 depending on the site of infection, an examination of the dermal site where vectors probe might be suitable to indicate local infection.
The ability of F. tularensis subsp. holarctica to be transstadially maintained through the mosquito life cycle, as demonstrated in this study, may have implications for the mode of transmission of the microorganism. We suggest that mosquitoes take up the bacteria during their larval stage and, once they become adults, transmit the disease, at very low frequencies, to a susceptible host where a massive burst of replication occurs. These susceptible hosts act as local amplifiers of F. tularensis subsp. holarctica and facilitate the spread of disease via vectors (including mosquitoes) and, after death, through carcasses contaminating the local environment. This plausible scenario involves the dynamics of at least two complex cycles: aquatic microbial dynamics that, via mosquito larvae, may lead to infection of mammals and terrestrial dynamics with potential arthropod vectors and mammal population densities that may favour the spread of the disease. Both cycles would exhibit common source–sink dynamics, with outbreaks during a limited time (i.e. months) resulting in massive growth of F. tularensis subsp. holarctica in susceptible hosts and thus passive contamination of the environment with F. tularensis subsp. holarctica, followed by bacterial long-term persistence and a slow decline over time. The need for the intertwining of these two complex cycles in space and time, as well as the specific biotic and abiotic factors governing the spread of disease, may explain the inherent unpredictability of tularaemia outbreaks and illustrates the need for further studies to elucidate the enigmatic life cycle of F. tularensis.
Acknowledgments
We thank Per Bülow for assistance with the sampling and packaging of samples from Örebro; Amandine Collado and Derek Nimmo at Oxitec for their assistance with mosquito delivery and rearing protocol; Anna-Lena Forslund and Karin Wallgren for their excellent technical assistance with the animal infection experiments; and Petter Lindgren and Göran Bucht for their valuable comments. This project was supported by grants from the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (formas no. 209-2006-1311) and the Swedish Ministry of Defence (no. A404213)
References
- 1.Huber B, Escudero R, Busse H, Seibold E, Scholz H, Anda P, Kämpfer P, Splettstoesser W (2009) Description of Francisella hispaniensis sp. nov., isolated from human blood, reclassification of Francisella novicida (Larson et al. 1955) Olsufiev et al. 1959 as Francisella tularensis subsp. novicida comb. nov., and emended description of the genus Francisella. Int J Syst Evol Microbiol 60:1887–1896 [DOI] [PubMed]
- 2.Sjöstedt AB, Brenner DJ, Krieg NR, Staley JT, Garrity GM (2005) Family XVII. Francisellaceae, genus I. Francisella. In: Anonymous (ed.) Bergey's manual of systematic bacteriology. Springer, New York, pp. 200–210
- 3.Johansson A, Celli J, Conlan W, Elkins KL, Forsman M, Keim PS, Larsson P, Manoil C, Nano FE, Petersen JM, Sjöstedt A. Objections to the transfer of Francisella novicida to the subspecies rank of Francisella tularensis. Int J Syst Evol Microbiol. 2010;60:1717–1718. doi: 10.1099/ijs.0.022830-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Busse HJ, Huber B, Anda P, Escudero R, Scholz HC, Seibold E, Splettstoesser WD, Kämpfer P. Objections to the transfer of Francisella novicida to the subspecies rank of Francisella tularensis—response to Johansson et al. Int J Syst Evol Microbiol. 2010;60:1718–1720. doi: 10.1099/00207713-60-8-1718. [DOI] [PubMed] [Google Scholar]
- 5.Keim P, Johansson A, Wagner DM. Molecular epidemiology, evolution, and ecology of Francisella. Ann N Y Acad Sci. 2007;1105:30–66. doi: 10.1196/annals.1409.011. [DOI] [PubMed] [Google Scholar]
- 6.Telford SR, Goethert HK. Toward an understanding of the perpetuation of the agent of tularemia. Front Microbiol. 2010;1:150. doi: 10.3389/fmicb.2010.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petersen J, Mead P, Schriefer M. Francisella tularensis: an arthropod-borne pathogen. Vet Res. 2009;40:7. doi: 10.1051/vetres:2008045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hopla CE. The ecology of tularemia. Adv Vet Sci Comp Med. 1974;18:25–53. [PubMed] [Google Scholar]
- 9.Broman T, Thelaus J, Andersson AC, Backman S, Wikstrom P, Larsson E, Granberg M, Karlsson L, Back E, Eliasson H, Mattsson R, Sjostedt A, Forsman M. Molecular detection of persistent Francisella tularensis subspecies holarctica in natural waters. Int J Microbiol. 2011;2011:851946. doi: 10.1155/2011/851946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parker R, Steinhaus E, Kohls G, Jellison W. Contamination of natural waters and mud with Pasteurella tularensis and tularemia in beavers and muskrats in the northwestern United States. Bull Natl Inst Health. 1951;193:1–161. [PubMed] [Google Scholar]
- 11.Abd H, Johansson T, Golovliov I, Sandstrom G, Forsman M. Survival and growth of Francisella tularensis in Acanthamoeba castellanii. Appl Environ Microbiol. 2003;69:600–606. doi: 10.1128/AEM.69.1.600-606.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berdal BP, Mehl R, Meidell NK, Lorentzen-Styr AM, Scheel O. Field investigations of tularemia in Norway. FEMS Immunol Med Microbiol. 1996;13:191–195. doi: 10.1111/j.1574-695X.1996.tb00235.x. [DOI] [PubMed] [Google Scholar]
- 13.Thelaus J, Andersson A, Mathisen P, Forslund A, Noppa L, Forsman M. Influence of nutrient status and grazing pressure on the fate of Francisella tularensis in lake water. FEMS Microbiol Ecol. 2009;67:69–80. doi: 10.1111/j.1574-6941.2008.00612.x. [DOI] [PubMed] [Google Scholar]
- 14.El-Etr S, Margolis J, Monack D, Robison R, Cohen M, Moore E, Rasley A. Francisella tularensis type A strains cause the rapid encystment of Acanthamoeba castellanii and survive in amoebal cysts for three weeks postinfection. Appl Environ Microbiol. 2009;75:7488–7500. doi: 10.1128/AEM.01829-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eliasson H, Bäck E. Tularaemia in an emergent area in Sweden: an analysis of 234 cases in five years. Scand J Infect Dis. 2007;39:880–889. doi: 10.1080/00365540701402970. [DOI] [PubMed] [Google Scholar]
- 16.Rydén P, Björk R, Schäfer ML, Lundström JO, Petersén B, Lindblom A, Forsman M, Sjöstedt A, Johansson A. Outbreaks of tularemia in a boreal forest region depends on mosquito prevalence. J Infect Dis. 2012;205:297–304. doi: 10.1093/infdis/jir732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olin G. Occurrence and mode of transmission of tularemia in Sweden. Acta Microbiol Scand. 1942;19:220–247. doi: 10.1111/j.1699-0463.1942.tb03345.x. [DOI] [Google Scholar]
- 18.Lundstrom JO, Andersson AC, Backman S, Schafer ML, Forsman M, Thelaus J. Transstadial transmission of Francisella tularensis holarctica in mosquitoes, Sweden. Emerg Infect Dis. 2011;17:794–799. doi: 10.3201/eid1705.100426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schäfer M, Lundström J, Pfeffer M, Lundkvist E, Landin J. Biological diversity versus risk for mosquito nuisance and disease transmission in constructed wetlands in southern Sweden. Med Vet Entomol. 2004;18:256–267. doi: 10.1111/j.0269-283X.2004.00504.x. [DOI] [PubMed] [Google Scholar]
- 20.Becker N, Zgomba M, Petric D, Dahl C, Boase C, Lane J, Kaiser A. Mosquitoes and their control. New York: Springer; 2003. [Google Scholar]
- 21.Broman T, Thelaus J, Andersson AC, Bäckman S, Wikström P, Larsson E, Granberg M, Karlsson L, Bäck E, Eliasson H, Mattsson R, Sjöstedt A, Forsman M (2011) Molecular detection of persistent Francisella tularensis subspecies holarctica in natural waters. Int J Microbiol 2011. doi: 10.1155/2011/851946 [DOI] [PMC free article] [PubMed]
- 22.Petersen J, Carlson J, Yockey B, Pillai S, Kuske C, Garbalena G, Pottumarthy S, Chalcraft L. Direct isolation of Francisella spp. from environmental samples. Lett Appl Microbiol. 2009;48:663–667. doi: 10.1111/j.1472-765X.2009.02589.x. [DOI] [PubMed] [Google Scholar]
- 23.Gaspar A, Tresselt H, Ward M. New solid medium for enhanced growth of Pasteurella tularensis. J Bacteriol. 1961;82:564–569. doi: 10.1128/jb.82.4.564-569.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biggerstaff B. Add-in to compute prevalence estimates from pooled samples. Fort Collins: Centers for Disease Control and Prevention; 2009. [Google Scholar]
- 25.Svensson K, Sjödin A, Byström M, Granberg M, Brittnacher MJ, Rohmer L, Jacobs MA, Sims-Day EH, Levy R, Zhou Y, Hayden HS, Lim R, Chang J, Guenthener D, Kang A, Haugen E, Gillett W, Kaul R, Forsman M, Larsson P, Johansson A. Genome sequence of Francisella tularensis subspecies holarctica strain FSC200, isolated from a child with tularemia. J Bacteriol. 2012;194:6965–6966. doi: 10.1128/JB.01040-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandström G, Tärnvik A, Wolf-Watz H, Löfgren S. Antigen from Francisella tularensis: nonidentity between determinants participating in cell-mediated and humoral reactions. Infect Immun. 1984;45:101–106. doi: 10.1128/iai.45.1.101-106.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pavlov VM, Mokrievich AN, Volkovoy K. Cryptic plasmid pFNL10 from Francisella novicida-like F6168: the base of plasmid vectors for Francisella tularensis. FEMS Immunol Med Microbiol. 1996;13:253–256. doi: 10.1111/j.1574-695X.1996.tb00247.x. [DOI] [PubMed] [Google Scholar]
- 28.Hill L, Davis J, Hapgood G, Whelan P, Smith G, Ritchie S, Cooper R, van den Hurk A. Rapid identification of Aedes albopictus, Aedes scutellaris, and Aedes aegypti life stages using real-time polymerase chain reaction assays. Am J Trop Med Hyg. 2008;79:866–875. [PubMed] [Google Scholar]
- 29.Simşek H, Taner M, Karadenizli A, Ertek M, Vahaboğlu H. Identification of Francisella tularensis by both culture and real-time TaqMan PCR methods from environmental water specimens in outbreak areas where tularemia cases were not previously reported. Eur J Clin Microbiol Infect Dis. 2012;31:2353–2357. doi: 10.1007/s10096-012-1576-z. [DOI] [PubMed] [Google Scholar]
- 30.Barns SM, Grow CC, Okinaka RT, Keim P, Kuske CR. Detection of diverse new Francisella-like bacteria in environmental samples. Appl Environ Microbiol. 2005;71:5494–5500. doi: 10.1128/AEM.71.9.5494-5500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duodu S, Larsson P, Sjödin A, Forsman M, Colquhoun DJ. The distribution of Francisella-like bacteria associated with coastal waters in Norway. Microb Ecol. 2012;64:370–377. doi: 10.1007/s00248-012-0023-0. [DOI] [PubMed] [Google Scholar]
- 32.Berrada Z, Sr T. Diversity of Francisella species in environmental samples from Martha's Vineyard, Massachusetts. Microb Ecol. 2010;59:277–283. doi: 10.1007/s00248-009-9568-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Triebenbach AN, Vogl SJ, Lotspeich-Cole L, Sikes DS, Happ GM, Hueffer K. Detection of Francisella tularensis in Alaskan mosquitoes (Diptera: Culicidae) and assessment of a laboratory model for transmission. J Med Entomol. 2010;47:639–648. doi: 10.1093/jmedent/47.4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forsman M, Henningson EW, Larsson E, Johansson T, Sandström G. Francisella tularensis does not manifest virulence in viable but non-culturable state. FEMS Microbiol Ecol. 2000;31:217–224. doi: 10.1111/j.1574-6941.2000.tb00686.x. [DOI] [PubMed] [Google Scholar]
- 35.Mironchuk I, Mazepa AV. Viability and virulence of Francisella tularensis subsp. holarctica in water ecosystems (experimental study) Zh Mikrobiol Epidemiol Immunobiol. 2002;2:9–13. [PubMed] [Google Scholar]
- 36.Vonkavaara M, Pavel ST, Hölzl K, Nordfelth R, Sjöstedt A, Stöven S. Francisella is sensitive to insect antimicrobial peptides. J Innate Immun. 2012;5:50–59. doi: 10.1159/000342468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sjödin A, Svensson K, Ohrman C, Ahlinder J, Lindgren P, Duodo S, Hnath J, Burans JP, Johansson A, Colquhoun DJ, Larsson P, Forsman M. Genome characterisation of the genus Francisella reveals insight into similar evolutionary paths in pathogens of mammals and fish. BMC Genomics. 2012;13:268. doi: 10.1186/1471-2164-13-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Svensson K, Bäck E, Eliasson H, Berglund L, Granberg M, Karlsson L, Larsson P, Forsman M, Johansson A. Landscape epidemiology of tularemia outbreaks in Sweden. Emerg Infect Dis. 2009;15:1937–1947. doi: 10.3201/eid1512.090487. [DOI] [PMC free article] [PubMed] [Google Scholar]