Abstract
Stable and soluble proteins are ideal candidates for functional and structural studies. Unfortunately, some proteins or enzymes can be difficult to isolate, being sometimes poorly expressed in heterologous systems, insoluble and/or unstable. Numerous methods have been developed to address these issues, from the screening of various expression systems to the modification of the target protein itself. Here we use a hydrophobic, aggregation-prone, phosphate-binding protein (HPBP) as a case study. We describe a simple and fast method that selectively uses ancestral mutations to generate a soluble, stable and functional variant of the target protein, here named sHPBP. This variant is highly expressed in Escherichia coli, is easily purified and its structure was solved at much higher resolution than its wild-type progenitor (1.3 versus 1.9 Å, respectively).
Keywords: Ancestral librairies, Protein engineering, Protein solubilization, Hydrophobic proteins, Phosphate-binding proteins, DING proteins
Highlights
-
•
Selective use of ancestral mutations can efficiently solubilize hydrophobic proteins.
-
•
The variant protein solubilized by ancestral mutations is fully functional.
-
•
The soluble variant is an excellent model for functional and structural studies.
Introduction
Stable, soluble and functional proteins comprise ideal models for functional and structural studies. However, when overexpressed in heterologous systems such as in Escherichia coli, natural proteins from various sources can sometimes be insoluble, unstable or poorly expressed [1]. These difficulties considerably hamper studies of certain proteins, and have yielded a considerable bias in protein functional and structural analysis toward soluble and expressible proteins [2]. Several strategies have been developed to skirt these limitations. Classical methods involving expression in heterologous systems usually screen the host nature, the culture conditions, and media composition [1,3–5]. Codon optimization, protein fusion or the co-expression with chaperones [6] may also represent useful strategies to successfully express proteins [1,5,6]. Nevertheless, these trials may remain inefficient in some cases; particularly for numerous mammalian proteins [1]. Thus, methodologies tuning the protein target itself emerged, with the aim of producing soluble and expressible models for further studies [7,8]. In that respect, site-directed mutagenesis could be used to substitute surface residues and therefore generate more soluble proteins. However, this technique is limited by our ability to precisely identify problematic surface residues [9]. Conversely, directed evolution allows to extensively mutate the target protein and to select for more soluble variants [10]. The explored sequence space is however huge, and the method thus requires a high throughput screening method [11]. An alternative method, called DNA shuffling, uses several genes sharing high sequence identity with the target protein and shuffles them all. The screening of the resulting gene library for solubility may yield soluble and expressible variants, which can subsequently be subjected to functional and structural studies [7].
In regard to protein stabilization or solubilization, phylogenetic-based protein engineering may represent a powerful method. Indeed, consensus libraries, which are composed of mutations that bring the sequence of the target gene closer to the family consensus sequence, can efficiently stabilize proteins [12]. Ancestral mutations also have proven ability to yield soluble and stabilized protein variants [13,14]. Moreover, the use of ancestral mutations libraries can yield interesting protein variants with altered enzymatic activity and/or stability [11,15–18]. Ancestral mutations may therefore be turned into an easy and fast method to solubilize/stabilize contemporary proteins.
In this study, we focus on ancestral mutations, and their ability to solubilize a protein target. We therefore used the human phosphate binding protein (HPBP) as a case study. HPBP belongs to DING proteins family, a clade of the phosphate binding protein (PBP) superfamily [19]. HPBP is a hydrophobic, possibly an apolipoprotein, crystallized from supposedly pure human paraoxonase (PON1) preparations [20–22]. HPBP possesses a venus-flytrap topology identical to the high affinity phosphate-binding protein (PBP or PstS) carriers of the ABC transporter systems, and a similar phosphate-binding ability [22]. Interestingly, and as for other related DING proteins [23–25], HPBP has been shown to inhibit HIV-1 replication by targeting the transcriptional step [26].
Nevertheless, functional and structural studies on HPBP are considerably hampered by its high hydrophobicity [22], its propensity to aggregate, and the failure to express it heterologously in soluble form (Chabriere, unpublished results). The existing purification procedure, starting from human plasma samples, is complex and laborious, and yields to little amounts of pure HPBP [20]. We hereby describe a simple and fast method to generate an ancestral-mutations based, fully functional, soluble variant of HPBP.
Materials and methods
Phylogenetic analysis and ancestral resurrection
The sequences of phosphate-binding proteins (including DING and PstS proteins) were collected from the National Center of Biotechnology Information (NCBI) using protein alignment BLAST (blastp) [27,28] with default parameters versus the non-redundant protein sequence database (nr). Only complete protein sequences were selected, and redundancy was subsequently removed (maximum 95% of sequence identity) with Cd-hit [29]. The sequence alignment was performed with clustalW 2.0 software [30] and manually improved (Fig. 1-1). The substitution matrix corresponding to the sequence alignment was determined using the Prottest software [31]. The alignment was subsequently submitted to PhyML software [32] with the JTT substitution matrix with 100 iterations. The prediction of the putative ancestral sequences at each nodes was performed using FastML [33]. We have chosen the putative ancestor of HPBP and a related, bacterial, soluble homologue, PfluDING from Pseudomonas fluorescens (Node 10) (Fig. 1-2) [34,35]. The ancestral mutation library contains the substitutions of the putative ancestral sequence, as compared to HPBP sequence. By applying four simple filters:
-
(1)
include substitutions of surface apolar residue into polar residue,
-
(2)
include substitution from Gly to X, with the exception of Gly residue involved in the start/end of secondary structure,
-
(3)
include core mutations,
-
(4)
include mutations that change a surface hydrophobic residue into a less hydrophobic one, we have reduced the numbers of substitutions from 93 to 22 (Fig. 1-3).
Fig. 1.
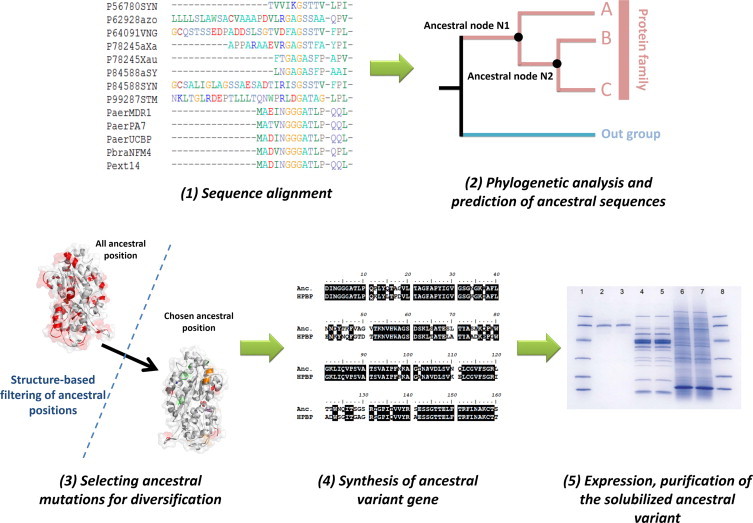
Selective use of ancestral mutations for protein solubilization. (1) Orthologous and paralogous protein sequences of the target protein has been identified and aligned. (2) A phylogenetic tree was constructed using this alignment, and the most probable ancestral sequence at each node were inferred. The substitutions between the most probable ancestral sequence at a given node and the contemporary target protein sequence (i.e wt HPBP) delineate the ancestral library. (3) Using a structural model and simple criteria (see methods), ancestral positions were filtered. (4) The gene coding for the ancestral variant was synthetized and (5) the protein was expressed, purified and crystallized.
Gene synthesis and cloning of HPBP and sHPBP
The genes encoding for HPBP and sHPBP were optimized for E. coli expression and synthesized by service providers (Genecust, Luxembourg, and GeneArt, Life Technologies, France, respectively) (Fig. 1-4). The genes were subsequently subcloned into pET22b (+) (Novagen) using NcoI and XhoI as cloning sites.
Production and purification of HPBP and sHPBP
Productions of HPBP and sHPBP were performed using E. coli BL21(DE3)-pGro7/GroEL cells (TaKaRa) in 6 l of ZYP medium [1] (100 μg ml−1 ampicillin, 34 μg ml−1 chloramphenicol). The cultures were grown at 37 °C to reach OD600nm = 0.6 and then induced by starting the consumption of lactose in ZYP medium coupled to temperature transition to 17 °C during 16 h. Cells were harvested by centrifugation (4500g, 4 °C, 15 min) and pellets were suspended in 400 ml of lysis buffer (20 mM TRIS, pH 8, 100 mM NaCl, Lysozyme 0.25 mg ml−1, DNAse I 10 μg ml−1, PMSF 0.1 mM, MgSO4 20 mM and 8 tablets of anti-protease EDTA-free (Roche)) and stored at −80 °C for 2 h. Cells were then thawed at 37 °C for 15 min and disrupted by 3 steps of 30 s of sonication (QSonica sonicator; amplitude 40). Debris was removed by centrifugation (12,500g, 4°C, 30 min). Supernatant was loaded on a Nickel affinity column (HisTrap 5 ml, FFCrude from GE Healthcare) at a flow rate of 5 ml min−1. Proteins gripped to the column were eluted by imidazol, using an elution buffer (20 mM TRIS, pH 8, 100 mM NaCl and 250 mM imidazole). Then, a size exclusion chromatography step (Superdex 75 16/60, GE Healthcare) was performed using buffer 20 mM TRIS, pH 8 and 100 mM NaCl. Protein production and purity were checked by 15% SDS–PAGE analysis (Fig. 1-5) and mass spectrometry analysis (Plateforme Timone, Marseille, France).
Crystallization of sHPBP
sHPBP was concentrated to 4 mg ml−1 using a centrifugation device (Vivaspin 500, MWCO 3 kDa, Sartorius stedim, Germany). Crystallization trials were performed at 298 K using the same condition as the homologue PfluDING (i.e. LiSO4 1 M, 20–30% PEG 8000 and Sodium Acetate 2 M at pH 4.5–5.5, [34,36,37]). Since only thin crystal plates were obtained, commercial screens conditions were tested (i.e. Stura and MDL, Molecular Dimension, England) using a Mosquito instrument (TTP Labtech, England) with the sitting-drop vapor diffusion method setup in a 96-well plate. Drops were monitored using a Discovery V8 binocular microscope and an AxioCam ERc5S camera (Zeiss, Germany). Crystals were obtained in the MDL screen, in a condition containing 0.2 M Sodium Acetate, 0.1 M Sodium Cacodylate pH 6.5 and 25% PEG 8000, and using a 2:1 (protein:reservoir) ratio (200 nl:100 nl). In order to obtain bigger crystals, this condition was optimized using the hanging drop method. The final condition (0.2 M Sodium Acetate, 0.1 M Sodium Cacodylate pH 6.5 and 25% PEG 8000), and using a 2:1 (protein:precipitant) ratio (500 nl:250 nl), led to the apparition of three-dimensional crystals (around 75–100 μm). Reproducible crystals appeared after 3 days at 298 K.
Data collection and structure resolution of sHPBP
The crystal was transferred few seconds in a drop (1 μl) containing a cryo-protectant solution made out of the mother liquor plus 10% (v/v) of glycerol. After mounting on a CryoLoop (Hampton research), crystal was flash-frozen in liquid nitrogen. X-ray diffraction intensities were collected on the ID23-1 beamline at the ESRF (Grenoble, France) using a wavelength of 0.97655 Å and a Pilatus 6M detector with 0.15 s exposures. Diffraction data were collected from 1027 images; each frames consisted of 0.15° step oscillations, over a range of 154.05° (Table 1). The molecular replacement using the HPBP structure as model (PDB: 2V3Q) was performed with Phaser [38]. The solution was then used for refinement performed using REFMAC5 and phenix [39,40], the model was improved using Coot [41]. The model and structure factor were deposited under the Protein Data Bank code PDB: 4M1V (Table 1).
Table 1.
Data collection and refinement statistics of sHPBP structure.
| Data collection | |
|---|---|
| Dataset | Native |
| PDB ID | 4M1V |
| Beamline | ID23-1 |
| Wavelength (Å) | 0.97655 |
| Detector | PILATUS 6M |
| Oscillation (°) | 0.15 |
| Number of frames | 1027 |
| Resolution (Å) (last bin) | 1.3 (1.4–1.3) |
| Space group | C2 |
| Unit-cell parameters (Å) | a = 125.04, b = 71.99, c = 38.98, β = 103.12 |
| No. of observed reflections (last bin) | 268471 (52411) |
| No. of unique reflections (last bin) | 81625 (16226) |
| Completeness (%)(last bin) | 98.8 (99.3) |
| Rmeas (%) (last bin) | 4.1 (27.5) |
| CC (1/2) (last bin) | 99.9 (95.3) |
| I/σ(I) (last bin) | 21.36 (4.65) |
| Redundancy (last bin) | 3.28 (3.23) |
| Refinement statistics | |
| Rfree/Rwork | 13.96/10.47 |
| No. of total model atoms | 3541 |
| Ramachandran favored (%) | 98.4 |
| Ramachandran outliers (%) | 0 |
| Generously allowed rotamers (%) | 1.6 |
| Rmsd from ideal | |
| Bond lengths (Å) | 0.027 |
| Bond angles (°) | 2.466 |
HIV inhibition by HPBP and sHPBP
Inhibition assays were performed in HeLa cells, as previously described [23]. Briefly, HeLa cells were maintained in DMEM + 10 % FBS with antibiotics (100 U/ml of Penicillin and 100 mg/ml of Streptomycin). Cells were pre-incubated with proteins (0.25 μg/ml) for 48 h, then transfected using Lipofectamine 2000 transfection reagent with 0.5 μg of HIV-LTR-luciferase reporter plasmid. Each transfection was done in triplicate.
Results
Ancestral mutations as a tool for protein solubilization
A phylogenetic tree of the PBPs, including DING proteins was generated (Figs. 1 and 2A), and the most probable ancestral sequence at Node 10 was reconstructed (Fig. 2A, see methods). The sequence of this putative ancestor exhibits 93 substitutions with the contemporary HPBP sequence (Fig. 2B). Among them, 22 substitutions were rationally selected (Fig. 2B and C). The selection was made using very simple criteria, such as (i) include substitutions of surface apolar/hydrophobic residues into polar/less hydrophobic ones, (ii) include core mutations, and (iii) include substitutions of glycine to X, when the glycine residue is not involved in the start/end of a secondary structure (see methods). The selected substitutions are in fact mainly located at the protein surface (Fig. 2D), and are predicted to solubilize the target protein, as shown by the hydrophobic profile comparison of sHPBP and HPBP (Fig. 3).
Fig. 2.
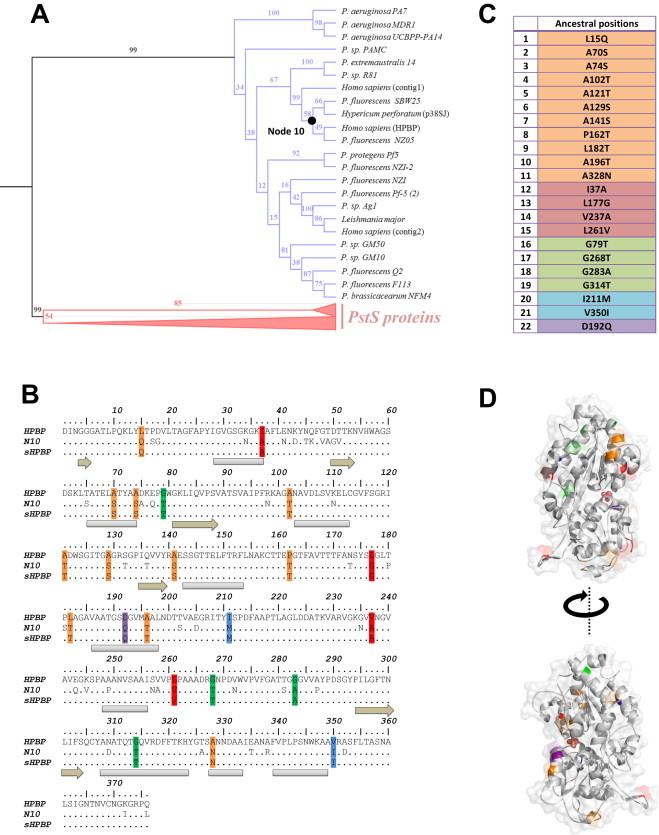
Phylogeny of PBPs and selected ancestral mutations. (A) Phylogenetic tree of PBPs, including PstS proteins (in pink) and DING proteins (in blue). Bootstrap values are shown for each node. (B) Sequence alignment of the target protein sequence (wt HPBP), the most probable ancestor sequence (Node 10) and the ancestral variant sequence containing selected substitutions (sHPBP). (C) Ancestral substitutions that were retained after structure-based filtering: substitutions changing apolar residues into polar residues (orange), hydrophobic residues into less hydrophobic ones (magenta), core substitutions (cyan) and D192Q substitution (purple) are listed. Secondary structures are represented in gray and light brown, for α-helix and β-sheet, respectively. (D) Retained ancestral substitutions locations on sHPBP structure are colored as in (C). The protein surface is represented as transparent gray and the bound phosphate anion is shown as red spheres. (For interpretation of the references to colour in this figure legend, the reader is reffered to the web version of this article.)
Fig. 3.
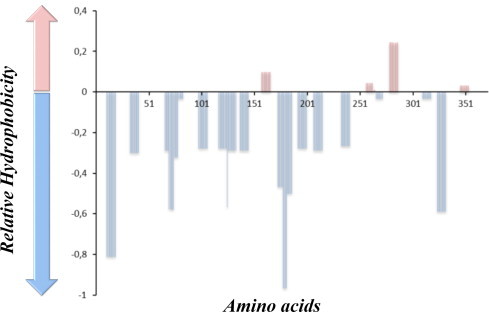
Hydropathy profile discrepancy between sHPBP and HPBP. The hydropathy profiles of both proteins were determined using the software “ProtScale” in Expasy website (algorithm Kyte and Doolitle, Window size 5). The graph represents, in y-axis, the difference of the hydrophobic potential between sHPBP and HPBP along the protein sequence (in x-axis). The light blue bars are for the sequence regions where sHPBP is predicted to be the more hydrophilic than HPBP, whereas regions where sHPBP is predicted to be more hydrophobic are shown as pink bars. (For interpretation of the references to colour in this figure legend, the reader is reffered to the web version of this article.)
The gene coding for sHPBP, the ancestral variant that includes 22 substitutions, was synthetized, and both sHPBP and wt HPBP were heterologously expressed in E. coli strain BL21(DE3)-pGro7/EL, using the same protocol. Protein expression profiles show that wt HPBP is expressed in E. coli, albeit solely in the insoluble fraction (Fig. 4, left part). Conversely, sHPBP is largely present in the soluble fraction (Fig. 4, middle part). Noteworthy, both proteins share more than 94% of sequence identity but possess opposite solubility profiles in E. coli. The expression of sHPBP is significant, as illustrated by the purification yield: about 12 mg of pure protein per liter of culture (Fig. 4, right part). Additionally, whereas HPBP is aggregation-prone, sHPBP is soluble, including in the absence of detergent, and present a melting temperature (Tm) of 47.2 ± 0.13 °C.
Fig. 4.
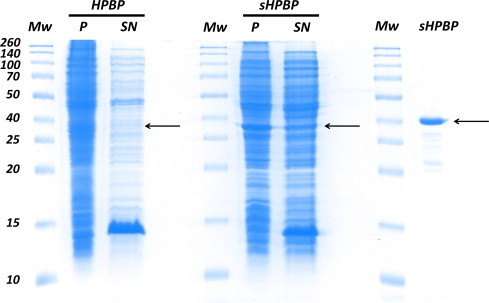
Solubility of wt HPBP and sHPBP when overexpressed in E. coli. The left part, middle part and left part of this Coomassie-stained SDS–PAGE gel correspond to the expression of HPBP, the expression of sHPBP, and the purified sHPBP, respectively. The lanes “P” and “SN” are for pellet and supernatant, respectively. Molecular weight markers (lane “Mw”) are indicated in kiloDalton (kDa; Spectra Multicolor broad range protein ladder). Arrows indicate the expected position of HPBP and sHPBP.
sHPBP and wt HPBP share nearly identical structures
sHPBP yielded high quality crystals and its structure was solved at 1.3 Å resolution (Table 1), a resolution that is higher than the previous structure of HPBP (1.9 Å) [42]. As expected by the high sequence identity between both proteins (94% identity), the structure of sHPBP is extremely similar to that of HPBP. The structures of both proteins exhibit identical topology, where the two globular domains are linked together by a flexible hinge. Each domain is composed of a central core β-sheets which is flanked by α-helix and present at their interface the phosphate binding cleft. Both structures superimpose extremely well, with a RMSD on all carbon α positions of 0.2 Å (Fig. 5A), including in the phosphate binding cleft region (Fig. 5B). Nonetheless, small differences relate to slightly different conformations of the 4 protuberant surface loops, some being possibly affected by ancestral mutations. The crystal packings of HPBP and sHPBP being different, we cannot exclude that packings affected the conformations of these surface loops. Interestingly, the calculation of the surface electrostatic potential of both structures reveals significantly different patterns, consistent with a superior solubility of sHPBP, since the apolar (or hydrophobic) surface patches present in HPBP structure are smaller in the sHPBP structure (Fig. 5C).
Fig. 5.
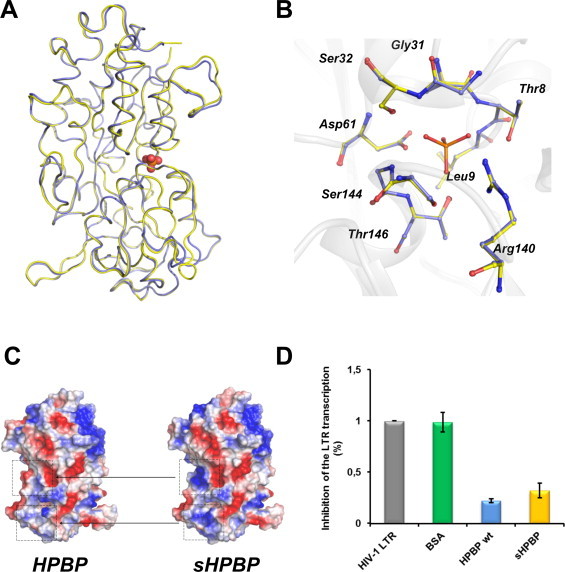
Structural and functional comparison of sHPBP and wt HPBP. (A) Structural superposition of sHPBP (yellow) and wt HPBP (blue). Phosphate is represented as red spheres. (B) Superposition of the phosphate binding cleft of both proteins (sHPBP (yellow) and HPBP (blue)). (C) Electrostatic potential comparison of both proteins. Positive and negative charges are respectively represented in red and blue. White regions are neutrally charged or hydrophobic. (D) Inhibition of the HIV-1 transcription, measured using a LTR-luciferase reporter system in HeLa cells. The constitutive luminescence of the system is represented in HIV-1 LTR (gray). The inhibition properties of the different samples (0.25 μg/ml) are shown: Bovine Serum Albumin (green), wt HPBP (blue) and sHPBP (yellow). (For interpretation of the references to colour in this figure legend, the reader is reffered to the web version of this article.)
The soluble variant sHPBP is functional
Besides the phosphate binding ability, HPBP has been shown to exhibit anti-HIV-1 properties [26]. The anti-HIV properties of HPBP and sHPBP have thus been compared using a LTR-reporter system in HeLa cells. Each assay was performed by pre-incubation of proteins at the estimated IC50 value of HPBP (i.e. 5 nM), previously determined using derived-immune system cells (Peripheral blood lymphocytes and primary macrophages) [26]. At this concentration and on HeLa cells, HPBP has a strong inhibitory effect on the HIV transcription (78%). We here show that sHPBP inhibits the HIV transcription with a very similar efficiency (68%) (Fig. 5D). This strongly suggests that sHPBP shares similar inhibition properties (e.g. IC50, CC50) and mechanisms than wt HPBP.
Discussion
Ancestral variants such as sHPBP comprise excellent model for functional studies of poorly soluble proteins
HPBP is a protein with an increasing interest because of its HIV-1 inhibition properties. However, its study has long been hampered because of its intrinsic hydrophobic character, and the resulting difficulty to obtain and store purified protein. We used ancestral mutations to generate a soluble variant of HPBP. This variant, sHPBP, can be expressed and purified from a convenient host, E. coli, with a significant purification yield (∼12 mg/l of culture), whereas this classical approach failed with wt HPBP. The enhanced expression level observed for sHPBP is concomitant to its higher solubility, compared to the wt protein. This dramatic change in behavior may be mainly ascribed to the decrease of the protein's surface hydrophobicity. sHPBP is therefore a more convenient protein to work with, and a critical consequence of this fact resides in the obtaining of sHPBP structure at 1.3 Å resolution; whereas HPBP structure was solved at 1.9 Å. Indeed, a better control on the purification process and the availability of high amounts of purified protein are key factors for optimization of crystallization conditions.
The structure of sHPBP reveals that it is extremely similar to that of wt HPBP. Most importantly, the phosphate binding cleft of both proteins is nearly identical. Consequently to this high similarity, sHPBP shares a similar HIV-1 inhibition capacity with wt HPBP. sHPBP therefore represents an excellent model for future structural and functional studies, with the aim of deciphering the biological function(s) of HPBP.
A fast, easy and efficient method based on ancestral mutations can produce soluble variants
We describe here the construction of a soluble variant of the hydrophobic, aggregation-prone, HPBP using ancestral mutations. The employed methodology is fast, simple and yielded a variant that is both soluble and active. Ancestral mutations and libraries indeed comprise an efficient tool for focusing substitutions to positions that can readily promote changes in protein stability, solubility or even substrate specificity [11,13,43]. A primordial property of ancestral mutations resides in the possibility of incorporating a very large number of these mutations, while maintaining the produced variants viable [11,43]. Therefore, by properly choosing ancestral mutations (e.g. active site substitutions), one can efficiently alter the substrate specificity of enzymes [11].
Here, we chose ancestral node 10 that relates the hydrophobic HPBP to a bacterial homologue dubbed PfluDING, reported to be a soluble and stable protein [34]. Then, among the substitutions between the putative ancestors and the HPBP sequence, we have mainly retained surface residues that may have a stabilizing and/or a solubilizing effect by applying 4 simple criteria (see Results). The resulting sequence was subsequently synthetized and yielded a soluble variant, sHPBP. The requirements for this method are limited to a good sequence sampling of the protein family, as well as a structure model for proper selection of the inferred ancestral mutations. This method can therefore be applied in numerous cases.
We believe that the selective use of ancestral mutations described here complements the existing approaches for producing more stable proteins, such as directed evolution, family shuffling, consensus libraries and others. However, its unique feature and its simplicity make it attractive to use with challenging protein targets, with the aim of producing, stable, soluble variants with increased propensity to crystallize.
Acknowledgments
This research was supported by a grant to EC from “Agence Nationale pour la Recherche sur le sida et les hépatites virales” (Grant No. 12264). GG and DG are granted by AP-HM (Marseille, France). JH is a PhD supported by the Delegation General pour l’Armement. We particularly thank Magali Richez for its helpful technical support.
Contributor Information
Eric Chabriere, Email: eric.chabriere@univ-amu.fr.
Mikael Elias, Email: mikael.elias@gmx.fr.
Appendix. Supplementary materials
Supplementary materials for Ancestral mutations as a tool for solubilizing proteins: the case of a hydrophobic phosphate-binding protein.
References
- 1.Graslund S, Nordlund P, Weigelt J, Hallberg BM, Bray J, Gileadi O, Knapp S, Oppermann U, Arrowsmith C, Hui R, Ming J, dhe-Paganon S, Park HW, Savchenko A, Yee A, Edwards A, Vincentelli R, Cambillau C, Kim R, Kim SH, Rao Z, Shi Y, Terwilliger TC, Kim CY, Hung LW, Waldo GS, Peleg Y, Albeck S, Unger T, Dym O, Prilusky J, Sussman JL, Stevens RC, Lesley SA, Wilson IA, Joachimiak A, Collart F, Dementieva I, Donnelly MI, Eschenfeldt WH, Kim Y, Stols L, Wu R, Zhou M, Burley SK, Emtage JS, Sauder JM, Thompson D, Bain K, Luz J, Gheyi T, Zhang F, Atwell S, Almo SC, Bonanno JB, Fiser A, Swaminathan S, Studier FW, Chance MR, Sali A, Acton TB, Xiao R, Zhao L, Ma LC, Hunt JF, Tong L, Cunningham K, Inouye M, Anderson S, Janjua H, Shastry R, Ho CK, Wang D, Wang H, Jiang M, Montelione GT, Stuart DI, Owens RJ, Daenke S, Schutz A, Heinemann U, Yokoyama S, Bussow K, Gunsalus KC. Protein production and purification. Nat. Methods. 2008;5:135–146. doi: 10.1038/nmeth.f.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peng, K, Obradovic, Z, Vucetic, S. (2003). Exploring bias in the Protein Data Bank using contrast classifiers. in: Paper Presented at the Pacific Symposium on Biocomputing 2004: Hawaii, USA, pp. 6–10, January 2004. [DOI] [PubMed]
- 3.Sorensen HP, Mortensen KK. Soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microb. Cell Fact. 2005;4:1. doi: 10.1186/1475-2859-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sorensen HP, Mortensen KK. Advanced genetic strategies for recombinant protein expression in Escherichia coli. J. Biotechnol. 2005;115:113–128. doi: 10.1016/j.jbiotec.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Vincentelli R, Bignon C, Gruez A, Canaan S, Sulzenbacher G, Tegoni M, Campanacci V, Cambillau C. Medium-scale structural genomics: strategies for protein expression and crystallization. Acc. Chem. Res. 2003;36:165–172. doi: 10.1021/ar010130s. [DOI] [PubMed] [Google Scholar]
- 6.Tokuriki N, Tawfik DS. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature. 2009;459:668–673. doi: 10.1038/nature08009. [DOI] [PubMed] [Google Scholar]
- 7.Harel M, Aharoni A, Gaidukov L, Brumshtein B, Khersonsky O, Meged R, Dvir H, Ravelli RB, McCarthy A, Toker L, Silman I, Sussman JL, Tawfik DS. Structure and evolution of the serum paraoxonase family of detoxifying and anti-atherosclerotic enzymes. Nat. Struct. Mol. Biol. 2004;11:412–419. doi: 10.1038/nsmb767. [DOI] [PubMed] [Google Scholar]
- 8.Aharoni A, Gaidukov L, Yagur S, Toker L, Silman I, Tawfik DS. Directed evolution of mammalian paraoxonases PON1 and PON3 for bacterial expression and catalytic specialization. Proc. Natl. Acad. Sci. U.S.A. 2004;101:482–487. doi: 10.1073/pnas.2536901100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyazaki K, Arnold FH. Exploring nonnatural evolutionary pathways by saturation mutagenesis: rapid improvement of protein function. J. Mol. Evol. 1999;49:716–720. doi: 10.1007/pl00006593. [DOI] [PubMed] [Google Scholar]
- 10.Waldo GS. Genetic screens and directed evolution for protein solubility. Curr. Opin. Chem. Biol. 2003;7:33–38. doi: 10.1016/s1367-5931(02)00017-0. [DOI] [PubMed] [Google Scholar]
- 11.Alcolombri U, Elias M, Tawfik DS. Directed evolution of sulfotransferases and paraoxonases by ancestral libraries. J. Mol. Biol. 2011;411:837–853. doi: 10.1016/j.jmb.2011.06.037. [DOI] [PubMed] [Google Scholar]
- 12.Jochens H, Aerts D, Bornscheuer UT. Thermostabilization of an esterase by alignment-guided focussed directed evolution. Protein Eng. Des. Sel. 2010;23:903–909. doi: 10.1093/protein/gzq071. [DOI] [PubMed] [Google Scholar]
- 13.Gaucher EA, Govindarajan S, Ganesh OK. Palaeotemperature trend for Precambrian life inferred from resurrected proteins. Nature. 2008;451:704–707. doi: 10.1038/nature06510. [DOI] [PubMed] [Google Scholar]
- 14.Risso VA, Gavira JA, Mejia-Carmona DF, Gaucher EA, Sanchez-Ruiz JM. Hyperstability and substrate promiscuity in laboratory resurrections of Precambrian beta-lactamases. J. Am. Chem. Soc. 2013;135:2899–2902. doi: 10.1021/ja311630a. [DOI] [PubMed] [Google Scholar]
- 15.Afriat-Jurnou L, Jackson CJ, Tawfik DS. Reconstructing a missing link in the evolution of a recently diverged phosphotriesterase by active-site loop remodeling. Biochemistry. 2012 doi: 10.1021/bi300694t. [DOI] [PubMed] [Google Scholar]
- 16.Khersonsky O, Rosenblat M, Toker L, Yacobson S, Hugenmatter A, Silman I, Sussman JL, Aviram M, Tawfik DS. Directed evolution of serum paraoxonase PON3 by family shuffling and ancestor/consensus mutagenesis, and its biochemical characterization. Biochemistry. 2009;48:6644–6654. doi: 10.1021/bi900583y. [DOI] [PubMed] [Google Scholar]
- 17.Bershtein S, Goldin K, Tawfik DS. Intense neutral drifts yield robust and evolvable consensus proteins. J. Mol. Biol. 2008;379:1029–1044. doi: 10.1016/j.jmb.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 18.Jochens H, Aerts D, Bornscheuer UT. Thermostabilization of an esterase by alignment-guided focussed directed evolution. Protein Eng. Des. Sel. 2010;23:903–909. doi: 10.1093/protein/gzq071. [DOI] [PubMed] [Google Scholar]
- 19.Berna A, Scott K, Chabriere E, Bernier F. The DING family of proteins: ubiquitous in eukaryotes, but where are the genes? Bioessays. 2009;31:570–580. doi: 10.1002/bies.200800174. [DOI] [PubMed] [Google Scholar]
- 20.Renault F, Chabriere E, Andrieu JP, Dublet B, Masson P, Rochu D. Tandem purification of two HDL-associated partner proteins in human plasma, paraoxonase (PON1) and phosphate binding protein (HPBP) using hydroxyapatite chromatography. J. Chromatogr. 2006;836:15–21. doi: 10.1016/j.jchromb.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 21.Rochu D, Chabriere E, Elias M, Renault F, Clery-Barraud C, Masson P. The Paraoxonases: Their Role in Disease Development and Xenobiotic Metabolism. Springer; Netherlands: 2008. Stabilisation of active form of natural human PON1 requires HPBP; pp. 171–183. [Google Scholar]
- 22.Morales R, Berna A, Carpentier P, Contreras-Martel C, Renault F, Nicodeme M, Chesne-Seck ML, Bernier F, Dupuy J, Schaeffer C, Diemer H, Van-Dorsselaer A, Fontecilla-Camps JC, Masson P, Rochu D, Chabriere E. Serendipitous discovery and X-ray structure of a human phosphate binding apolipoprotein. Structure. 2006;14:601–609. doi: 10.1016/j.str.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 23.Darbinian, N, Gomberg, R, Mullen, L, Garcia, S, White, MK, Khalili, K, Amini, S. Suppression of HIV-1 transcriptional elongation by a DING phosphatase J. Cell. Biochem. 112, 225–232. [DOI] [PMC free article] [PubMed]
- 24.Sachdeva Rakhee, Darbinian N, Khalili Kamel, Amini Shohreh, Gonzalez Daniel, Djeghader Ahmed, Chabriére Eric, Suh Andrew, Scott Ken, Simm Malgorzata. DING proteins from phylogenetically different species share high degree of sequence and structure homology and block transcription of HIV-1 LTR promoter. PloS One. 2013 doi: 10.1371/journal.pone.0069623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lesner A, Shilpi R, Ivanova A, Gawinowicz MA, Lesniak J, Nikolov D, Simm M. Identification of X-DING-CD4, a new member of human DING protein family that is secreted by HIV-1 resistant CD4(+) T cells and has anti-viral activity. Biochem. Biophys. Res. Commun. 2009;389:284–289. doi: 10.1016/j.bbrc.2009.08.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cherrier T, Elias M, Jeudy A, Gotthard G, Le Douce V, Hallay H, Masson P, Janossy A, Candolfi E, Rohr O, Chabriere E, Schwartz C. uman-phosphate-binding-protein inhibits HIV-1 gene transcription and replication. Virol. J. 2011;8:352. doi: 10.1186/1743-422X-8-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 28.Mount DW. Using the Basic Local Alignment Search Tool (BLAST) CSH Prot. 2007 doi: 10.1101/pdb.top17. 2007, pdb top17. [DOI] [PubMed] [Google Scholar]
- 29.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 30.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 31.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 33.Ashkenazy H, Penn O, Doron-Faigenboim A, Cohen O, Cannarozzi G, Zomer O, Pupko T. FastML: a web server for probabilistic reconstruction of ancestral sequences. Nucleic acids research. 2012;40:W580–W584. doi: 10.1093/nar/gks498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moniot S, Elias M, Kim D, Scott K, Chabriere E. Crystallization, diffraction data collection and preliminary crystallographic analysis of DING protein from Pseudomonas fluorescens. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2007;63:590–592. doi: 10.1107/S1744309107028102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahn S, Moniot S, Elias M, Chabriere E, Kim D, Scott K. Structure–function relationships in a bacterial DING protein. FEBS Lett. 2007;581:3455–3460. doi: 10.1016/j.febslet.2007.06.050. [DOI] [PubMed] [Google Scholar]
- 36.Liebschner D, Elias M, Moniot S, Fournier B, Scott K, Jelsch C, Guillot B, Lecomte C, Chabriere E. Elucidation of the phosphate binding mode of DING proteins revealed by subangstrom X-ray crystallography. J. Am. Chem. Soc. 2009;131:7879–7886. doi: 10.1021/ja901900y. [DOI] [PubMed] [Google Scholar]
- 37.Elias M, Wellner A, Goldin-Azulay K, Chabriere E, Vorholt JA, Erb TJ, Tawfik DS. The molecular basis of phosphate discrimination in arsenate-rich environments. Nature. 2012;491:134–137. doi: 10.1038/nature11517. [DOI] [PubMed] [Google Scholar]
- 38.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 41.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diemer H, Elias M, Renault F, Rochu D, Contreras-Martel C, Schaeffer C, Van Dorsselaer A, Chabriere E. Tandem use of X-ray crystallography and mass spectrometry to obtain ab initio the complete and exact amino acids sequence of HPBP, a human 38-kDa apolipoprotein. Proteins. 2008;71:1708–1720. doi: 10.1002/prot.21866. [DOI] [PubMed] [Google Scholar]
- 43.Gaucher EA, Thomson JM, Burgan MF, Benner SA. Inferring the palaeoenvironment of ancient bacteria on the basis of resurrected proteins. Nature. 2003;425:285–288. doi: 10.1038/nature01977. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials for Ancestral mutations as a tool for solubilizing proteins: the case of a hydrophobic phosphate-binding protein.


