Abstract
Two-component signal transduction systems (TCS), consisting of a sensor histidine protein kinase and its cognate response regulator, are an important mode of environmental sensing in bacteria. Additionally, they have been found to regulate virulence determinants in several pathogens. Bacillus anthracis, the causative agent of anthrax and a bioterrorism agent, harbours 41 pairs of TCS. However, their role in its pathogenicity has remained largely unexplored. Here, we show that WalRK of B. anthracis forms a functional TCS which exhibits some species-specific functions. Biochemical studies showed that domain variants of WalK, the histidine kinase, exhibit classical properties of autophosphorylation and phosphotransfer to its cognate response regulator WalR. Interestingly, these domain variants also show phosphatase activity towards phosphorylated WalR, thereby making WalK a bifunctional histidine kinase/phosphatase. An in silico regulon determination approach, using a consensus binding sequence from Bacillus subtilis, provided a list of 30 genes that could form a putative WalR regulon in B. anthracis. Further, electrophoretic mobility shift assay was used to show direct binding of purified WalR to the upstream regions of three putative regulon candidates, an S-layer protein EA1, a cell division ABC transporter FtsE and a sporulation histidine kinase KinB3. Our work lends insight into the species-specific functions and mode of action of B. anthracis WalRK.
Keywords: Bacillus anthracis, Two-component signal transduction, Histidine kinase, Response regulator, Regulon, YycFG, VicRK
Abbreviations: TCS, two-component signal transduction systems; HK, histidine kinase; RR, response regulator; AcP, acetyl phosphate; EMSA, electrophoretic mobility shift assay; ELISA, enzyme linked immunosorbent assay; HRP, horse radish peroxidase; AP, alkaline phosphatase
Graphical abstract
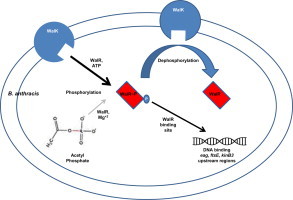
Highlights
-
•
WalRK forms a functional TCS in B. anthracis, expressed throughout the growth phase.
-
•
WalK variants exhibit autophosphorylation and phosphotransfer to WalR.
-
•
WalKc variants also show phosphatase activity towards phosphorylated WalR.
-
•
A potential WalR regulon in B. anthracis was predicted in silico.
-
•
DNA binding ability was demonstrated for WalR.
Introduction
Bacillus anthracis, the causative agent of anthrax, is a notorious pathogen often leading to fatal infections in animals and less frequently, in humans. Early diagnosis of the infection and immediate aggressive antibiotic therapy are the only post exposure measures that may offer optimistic outcome. The spores of B. anthracis can be employed as a weapon of mass destruction in form of a biological warfare agent, which makes it of paramount importance to understand the factors that control its pathogenicity, so as to devise strategies to tackle it effectively. Apart from the classical virulence factors of B. anthracis – a tripartite toxin and an anti-phagocytic capsule [1,2], a number of studies have indicated towards existence of additional virulence determinants such as enzymes, transporters, etc. [3–7] that shape the course of an infection. Moreover, intricate regulatory networks exist (like those of AtxA, AcpA, AcpB, AbrB) that control classical virulence factors indirectly [8–11], thereby facilitating a successful infection.
Two-Component Signal Transduction Systems (TCS) are versatile regulatory networks predominantly found in prokaryotes. These systems are critical components involved in environmental sensing and adaptation of microbes to the dynamic outside environment [12]. In many pathogens, the virulence gene expression and host–pathogen interactions have been observed to be controlled by TCS [13,14]. A typical TCS encodes a sensor Histidine protein Kinase (HK), which is mostly an integral membrane sensor protein, and a Response Regulator (RR). The RR is usually a transcription factor but can also have RNA binding, enzymatic or protein binding functions. The sensor HK and its cognate RR work as a pair specific for a particular signal and often occur in an operon. Signal sensing results in activation of the HK dimer followed by an ATP-dependent autophosphorylation of a conserved histidine residue in the C-terminal catalytic domain of HK. This autophosphorylation of the HK dimer can occur either in trans or in cis [15]. Next, the phosphate group is transferred to a conserved aspartate in the N-terminal receiver domain of the RR (RR∼P). Phosphorylation alters the conformation and hence, the activity of the RR, in turn modifying its function, which is mostly DNA binding [12,16]. The levels of phosphorylation of RRs can be further controlled by phosphatase activity of the HK, RR (autodephosphorylation) or specific aspartyl phosphatases [17,18]. Often, it is the HK that assumes the bifunctional role of kinase and phosphatase towards its cognate RR to regulate the levels of phosphorylation of RR [17]. Phosphatase activity assumes importance so as to limit cross-talk from other non-cognate kinases and small molecule phosphoryl donors, in addition to regulating the longevity of response. TCS have also been discovered in an expanded version in eukaryotes like yeast, fungi, and higher plants in the form of multistep phosphorelays, which involve multiple phosphotransfers between His- and Asp-containing proteins. Increased number of proteins in a phosphorelay provides scope for greater and more complex regulation [16].
TCS control important elements of adaptation in bacteria and virulence in many pathogens. However, they have largely remained unexplored in B. anthracis, with the exception of two [19,20]. HKs that function in sporulation have been identified in B. anthracis but these do not exist as a TCS pair [21]. The genome of B. anthracis strain Sterne is reported to house 52 HKs and 51 RRs. Out of these, 41 occur as pairs and can be speculated to form a functional TCS [22]. Five of the HKs and 1 RR from this list have been found to be truncated in B. anthracis, but whether the truncations have made the proteins non-functional has not been checked [22]. Nevertheless, this number is considerably higher in comparison to a related non-pathogenic bacteria Bacillus subtilis (29 TCS pairs) [22] or other related pathogens: Staphylococcus aureus (17 TCS pairs) and Staphylococcus pneumoniae (13 TCS pairs) [23]. Genomes of organisms that thrive in fluctuating environments have been seen to encode higher number of TCS genes [24]. This further highlights the importance of signal transduction through TCS in B. anthracis, a human pathogen.
WalRK (earlier referred to as YycFG/VicRK) is a TCS that was originally identified in B. subtilis [25] and has been implicated in the virulence of several low GC %, Gram positive pathogens like S. aureus, [14,26] S. pneumoniae [27–29] and Streptococcus mutans [30] where it has been shown to regulate virulence associated genes. It has also attracted considerable interest as a probable antimicrobial target against low GC % Gram positive pathogens since it has been found to be essential for the viability in most of the organisms studied [25,26,28,30,31].
In the present study, we carried out the functional characterization of WalRK, a TCS from B. anthracis. WalRK was found to be expressed throughout the growth period. The domain variants of HK- WalKc, WalKc(PH) and WalKc(H)- exhibited proficient autophosphorylation and phosphotransfer to its cognate RR WalR. In addition to this, WalK variants could also show phosphatase activity displaying some species-specific functions. An in silico DNA motif search in the intergenic regions of B. anthracis was used to identify genes that could be potentially regulated by WalR. WalR binding to the upstream regions of three putative regulon genes was shown in vitro.
Results
Cloning, expression and purification of WalR, WalKc, WalKc(PH) and WalKc(H)
WalR is the full length RR, while WalKc (consisting of HAMP, PAS, DHp and CA domains) is the complete cytoplasmic portion of the HK WalK. WalKc(PH) (consisting of PAS, DHp and CA domains) and WalKc(H) (consisting of DHp and CA domains) are two domain deletion variants of WalKc (Fig. 1). All the ORFs were cloned in the expression vector pET28a+ that adds a hexa-histidine tag to their C-terminus. The proteins were overexpressed and purified using Ni+2–NTA affinity chromatography. The histidine-tagged purified proteins were analyzed using SDS–PAGE. The expressed recombinant proteins ran at molecular weight of ∼30 kDa for WalR (migrating slightly higher than its expected size of ∼28 kDa), ∼48 kDa for WalKc, ∼44 kDa for WalKc(PH) and ∼28 kDa for WalKc(H) (Fig. S1).
Fig. 1.
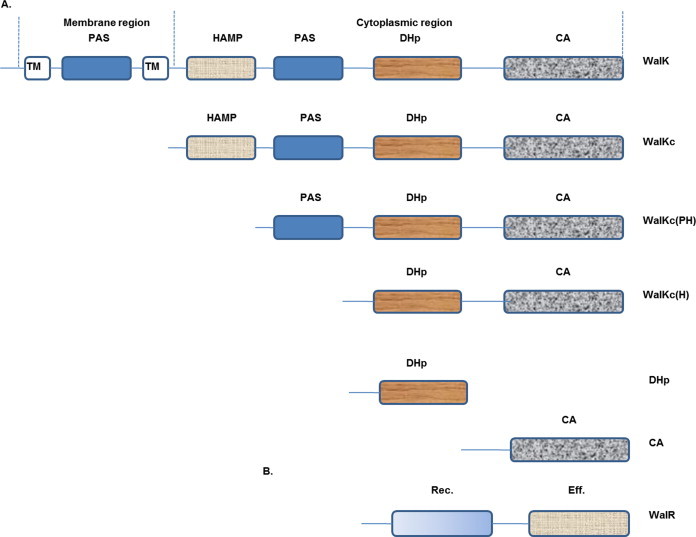
Domain organization of WalK & WalR of B. anthracis (SMART database). (A) WalK, histidine kinase, has two transmembrane domains (TM), a HAMP (a domain found in Histidine kinases, Adenylyl cyclases, Methyl binding proteins, Phosphatases) domain, two PAS (a domain common to Per-Arnt-Sim proteins) domains, a DHp (Dimerization and Histidine phosphotransfer, housing conserved histidine) domain and a CA (Catalytic and ATP-binding) domain. (B) WalR, response regulator, has a receiver (housing conserved aspartate) domain and an effector (DNA binding) domain.
In vivo expression of walR and walK
Quantitative real time PCR: qRT-PCR was done to analyze the growth stage specific expression of transcript levels of walR and walK in B. anthracis. Four growth stages were chosen according to the growth curve of B. anthracis Sterne strain-early exponential (O.D.600nm ∼0.3), mid exponential (O.D.600nm ∼0.6), late exponential (O.D.600nm ∼0.9) and onset of stationary phase (O.D.600nm ∼1.2). The mRNAs for both the genes could be detected throughout the growth period, indicating that the genes are transcribed throughout the phases (Fig. 2A). After the transcript levels, expression at the protein level was also checked using immunoblotting.
Fig. 2.
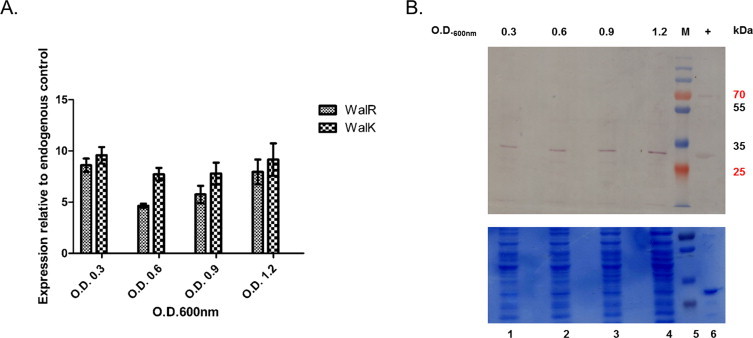
In vivo expression of walR and walK in B. anthracis at different growth stages – O.D.600nm ∼0.3 (early exponential phase), 0.6 (mid exponential phase), 0.9 (late exponential phase), 1.2 (onset of stationary phase). (A) qRT-PCR for walR and walK transcripts from B. anthracis total RNA. Transcripts for both the genes could be detected at all the tested growth stages. Normalized Ct values are plotted against different growth stages, using GraphPad Prism 5 software. Mean with SEM values, from two independent experiments carried out in triplicate are shown. (B) Immunoblotting for endogenous WalR protein expression in B. anthracis. WalR could be detected in B. anthracis lysates from all the tested growth stages using anti-WalR antisera. A single band could be observed at the same position as recombinant WalR monomer (+). Equal protein was loaded for all the samples as shown in the coomassie stained SDS–PAGE gel. No band could be detected in any of the B. anthracis fractions with anti-WalKc antisera. M – Molecular weight standards in kDa.
Immunoblotting: The endogenous protein expression was also validated by immunoblotting of the soluble/cytoplasmic and cell wall plus membrane subcellular fractions of B. anthracis. WalR was found to be expressed throughout the growth period, as detected in the soluble fractions of B. anthracis at different growth stages. A single specific band at the expected size was observed for WalR (∼30 kDa) and the specificity of the band was confirmed using recombinant WalR as the control (Fig. 2B). Both the recombinant and native WalR migrated to nearly the same distance because there was only 1 kDa difference in their molecular weights. In case of WalK, no specific band could be detected on probing with anti-WalKc antisera (data not shown). This could be due to very low amount of WalK in the respective fractions.
The endogenous protein expression and localization for WalR and WalK was also checked using confocal immunofluorescence microscopy. WalR could be localized to the cytoplasm of B. anthracis but WalK could not be detected once again (data not shown). Though the expression of walR and walK could be observed throughout the exponential and stationary stages, this does not necessarily indicate its constitutive activation. It is the phosphorylation of any HK and RR that governs its activity. A true picture of the activity of any TCS can be drawn by an assessment of the transcript levels of the gene candidates that it can specifically regulate.
Autophosphorylation of WalK variants and phosphotransfer to WalR
The primary catalytic function of a HK is autophosphorylation followed by phosphotransfer to its cognate RR. Biochemical studies were done to test both these activities in WalKc and its variants. When WalKc was incubated with labeled ATP mixture, autophosphorylation could be detected as early as 5 min post ATP addition and activity increased with time, reaching a maximum in 60 min (Lane 2 & 5, Fig. 3A). When WalKc and WalR were incubated together, autophosphorylation of WalKc and phosphotranfer to WalR could be observed after addition of labeled ATP mixture. Phosphotransfer also could be detected within 5 min of incubation that increased with time of incubation and reached maximum level in 60 min (Lane 1 & 4, Fig. 4A), indicating WalKc and WalR form a functional HK-RR pair in vitro.
Fig. 3.
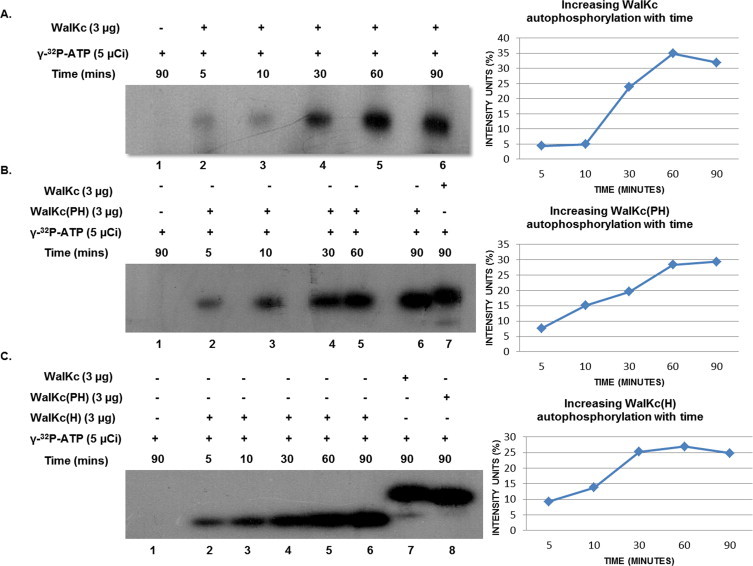
Autophosphorylation of WalK and its variants, with its intensity analysis (ImageJ software). Autophosphorylation was observed within 5 min of γ-32P-ATP addition and increased with time for each variant as shown. (A) Autophosphorylation of WalKc. (B) Autophosphorylation of WalKc(PH) with WalKc as a positive control. (C) Autophosphorylation of WalKc(H) with WalKc and WalKc(PH) as the positive controls. The reaction mixtures were analyzed by a 10% SDS–PAGE, followed by autoradiography. For each autoradiogram, intensity units (%) for phosphorylation are plotted against time in minutes.
Fig. 4.
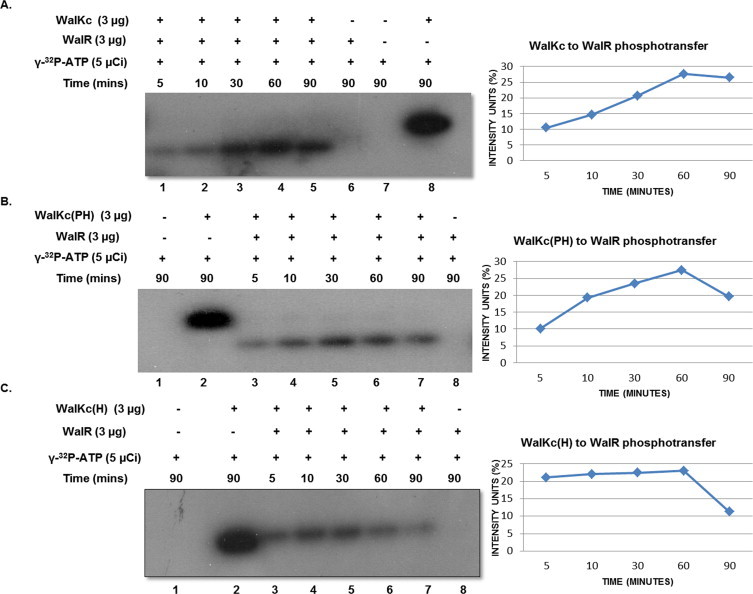
Phosphotransfer from WalK variants to WalR, with its intensity analysis (ImageJ software). Phosphotransfer was observed within 5 min of γ-32P-ATP addition and increased with time as shown. (A) WalKc to WalR phosphotransfer. (B) WalKc(PH) to WalR phosphotransfer. (C) WalKc(H) to WalR phosphotransfer. The reaction mixtures were analyzed by a 10% SDS–PAGE, followed by autoradiography. For each autoradiogram, intensity units (%) for phosphotransfer are plotted against time in minutes.
WalKc, after purification, partly degraded into a lower molecular weight protein which tested positive in immunoblotting and autophosphorylation. This issue was resolved by removing the HAMP linker from WalKc resulting in WalKc(PH). WalKc(PH) harboring PAS, DHp and CA domains, and WalKc(H) comprising of DHp and CA domains were also tested for autophosphorylation and phosphotransfer to WalR. Both, WalKc(PH) and WalKc(H), appeared to be more efficient than WalKc in autophosphorylation. The detectable levels of phosphorylation within the first 10 min of incubation with labeled ATP mixture were higher for WalKc(PH) and WalKc(H) than for WalKc (Lane 3, Fig. 3A–C). Phosphotransfer could also be observed in both the cases indicating that both, WalKc(PH) and WalKc(H), are functionally active and competent for phosphotransfer to WalR (Fig. 4).
Phosphorylation of WalR by small molecule phosphoryl donors
Low molecular weight phosphoryl donors like acetyl phosphate, carbamoyl phosphate and phosphoramidate have been shown to phosphorylate some RRs [32,33]. We tested whether acetyl phosphate and carbamoyl phosphate could phosphorylate WalR in vitro. When WalR was incubated with acetyl phosphate, it was observed that only 20–25% of WalR was phosphorylated under the conditions tested (Fig. 5A). The phosphorylated form was stable at 37 °C for 60 min. Acetyl phosphate mediated phosphorylation has not been checked for B. subtilis WalR (WalRBsu), but a similar observation was made with a related RR- PhoPBsu [34] which has 83% similarity (62% identity) with B. anthracis WalR (WalRBan) in the receiver domain.
Fig. 5.
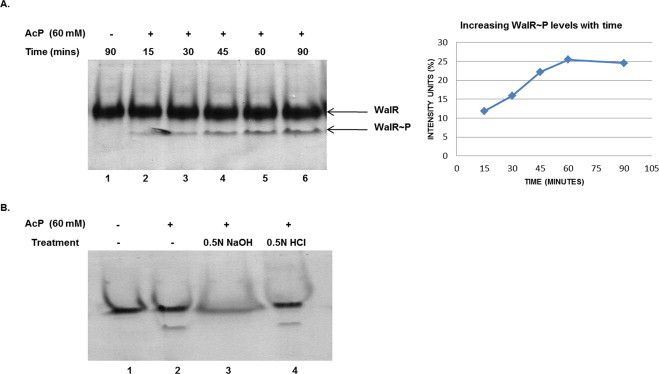
Phosphorylation of WalR by acetyl phosphate (AcP). (A) WalR could be phosphorylated with AcP which increased with time, reaching a maximum of 20–25% only. The phosphorylation of WalR was analyzed by 12% polyacrylamide native gel electrophoresis followed by coomassie staining and intensity analysis. Intensity units (%) of WalR phosphorylation are plotted against time in minutes. B. Acid/base treatment of WalR∼P. The stability of WalR∼P to acid/base treatment was tested in the presence of 0.5 N HCl and 0.5 N NaOH. The products were analyzed by 12% polyacrylamide native gel electrophoresis and coomassie staining. The phosphorylated form was base labile and acid stable as shown, which is characteristic of phospho-aspartate bonds in response regulator.
The stability of WalR∼P was tested under acidic (0.5 N HCl) and basic (0.5 N NaOH) conditions to analyze the properties of the residue that has undergone phosphorylation in WalR. The acetyl phosphate mediated phosphorylation of WalR was acid stable, base labile (Fig. 5B) and required Mg+2 ions, displaying characteristics of a phospho-aspartate bond in RRs as generated by the cognate kinase [34]. Carbamoyl phosphate mediated phosphorylation of WalR was also found to be inefficient, reaching 25–30% only in 60 min (data not shown), suggesting small molecule phosphoryl donors may not act as efficient substrates for WalR phosphotransfer activity.
WalKc variants mediated dephosphorylation of WalR∼P
The domain organization for WalK is shown in Fig. 1. WalK HK possesses a DHp domain which belongs to HisKA subfamily [35]. The HisKA family, which is the largest subfamily of bacterial DHp domains, bears an E/DxxT/N motif which has been implicated in phosphatase activity [36,37]. The T/N residue of this motif (hydroxyl/amide group) is critical for coordinating a nucleophilic water molecule for an inline attack on RR∼P resulting in hydrolysis of the phospho-aspartate bond [36,37]. A similar motif was found in the DHp domain of WalK just next to phosphorylatable histidine (Fig. 6). This observation led us to test WalKc variants for their phosphatase activity.
Fig. 6.

B. anthracis WalK DHp domain (SMART database) with associated E/DxxT/N motif (Underlined). Residues highlighted in red form helix 1 of the DHp domain, which houses the conserved histidine (*) and interacts with the response regulator. Residues of helix 1 are marked on the basis of homology of WalKBan DHp with EnvZE.coli DHp domain which is also a HisKA domain (PDB database). The residues are numbered according to WalKBan amino acid sequence.
The WalR∼P obtained after phosphorylation of WalR by acetyl phosphate was used to assay the phosphatase activity of WalKc(PH) and WalKc(H). Both WalKc(PH) and WalKc(H) could cause loss of phosphate from WalR∼P (Lane 3, 4, 9–12, Fig. 7A). However, WalKc(H) appeared to be more robust than WalKc(PH) in phosphatase activity as is indicated by the residual WalR∼P levels after 10 min of incubation (Lane 3 & 5, Fig. 7B and C). This indicated that it is the DHp and/or CA domains which are the essential element for phosphatase activity. DHp domains alone have been seen to exhibit phosphatase activity in HKs [38]. To test the same, DHp domain of WalK was cloned. However, it could not be expressed under the same conditions which were tested for other WalK variants in the study. No other condition was tested for DHp expression. Hence, WalK possesses both the kinase and phosphatase activity, thereby, adding it to the growing list of bifunctional HKs of TCS.
Fig. 7.
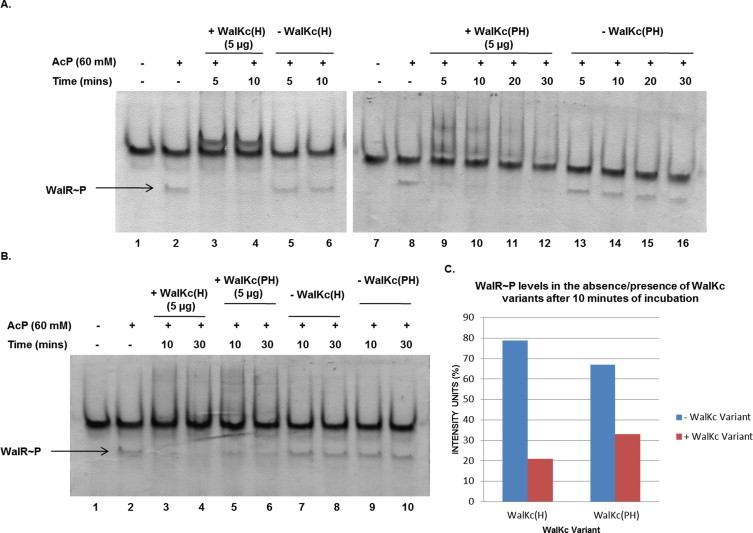
Phosphatase activity of WalKc variants. WalR was phosphorylated with AcP for 60 min, following which the reaction was stopped with 40 mM EDTA. WalKc(H)/WalKc(PH) (5 μg each) was then added to assay for phosphatase activity at different time points. The reaction mixtures were analyzed on a 12% polyacrylamide native gel followed by coomassie staining and intensity analysis. (A) Both WalKc(H) and WalKc(PH) could mediate dephosphorylation of WalR∼P. (B) Comparative analysis of phosphatase activity of WalKc(H) and WalKc(PH). (C) WalR∼P levels remaining after 10 min of incubation with WalKc(H)/WalKc(PH) (5 μg each) in comparison with control. Intensity units (%) are plotted for both the variants.
Identification of putative WalR regulon in B. anthracis genome
The WalR RR of B. anthracis shares 79% identity with its ortholog in B. subtilis in the primary amino acid sequence. Their C-terminal DNA binding domains exhibit a significant 93% amino acid identity. Such a high level of identity indicates that the two domains may have a similar structure and recognize similar DNA sequence. Therefore, the consensus WalR DNA binding sequence was expected to be similar to that obtained from B. subtilis [39]. Using the methodology described, a total of 33 prospective WalR binding sites were found, on both the strands, corresponding to upstream regions of 30 genes (Table 1) that could form a potential WalR regulon. Domain annotation and broad functional prediction was carried out for the candidate genes and those with significant predicted functions were shortlisted for further DNA binding studies. Of the 30 hits, only five were annotated for their function in the Uniprot database (eag, ftsE, kinB3, phoX and hppD). The eag gene codes for an S-layer protein Extractable Antigen 1 (EA1), ftsE codes for a cell division ABC transporter and kinB3 codes for a sporulation histidine kinase (Table 2). Structure function annotation of the deduced genes having WalR binding site in the upstream region using the conserved domain database and COG database, is given in Table 1. None of the classical virulence factors of B. anthracis appeared to be regulated by WalR, in contrast to what has been observed in related pathogens S. aureus and S. pneumoniae [14,26,29]. By COG categorization, 9 genes from the total of 30 were predicted to be implicated in transport and metabolism, while 2 were hydrolases and 1 was a member of Major Facilitator Superfamily. This reflects that WalRK may control a diverse set of functions in B. anthracis, but direct regulation of virulence is not indicated.
Table 1.
Putative WalR regulon in B. anthracis.
| S.No. | Gene (Protein) | Protein name | Function prediction (COG database) | Conserved domain/ superfamily (CD search) |
|---|---|---|---|---|
| 1. | BAS0842 (YP_027118.1) | EA1 | S-layer component | S-layer homology domain (pfam00395 (PSSMID 201204)) |
| 2. | BAS5034 (YP_031273.1) | Cell division ABC transporter, ATP-binding protein FtsE | Cell division and chromosome partitioning | P-loop containing nucleoside triphosphate hydrolases ([Superfamily] cl09099 (PSSMID 213440)) |
| 3. | kinB3 BAS3916 (YP_030166.1) | Sporulation kinase B | Signal transduction mechanisms | Histidine kinase A (dimerization/phosphoacceptor) domain; histidine kinase-like ATPases (cd00082 (PSSMID 119399)) |
| 4. | phoX BAS4174 (YP_030423.1) | Phosphate ABC transporter, phosphate-binding protein | Inorganic ion transport and metabolism | LysR-type transcriptional regulators (LTTRs) ([Superfamily] cl11398 (PSSMID 209302)) |
| 5. | hppD BAS0226 (YP_026511.1) | 4-Hydroxyphenylpyruvate dioxygenase | Amino acid transport and metabolism | TIGR01263 (PSSMID 162275) |
| 6. | BAS0506 (YP_026786.1) | UPF0295 protein | – | – |
| 7. | BAS0379 (YP_026659.1) | Membrane protein, putative | – | Major Facilitator Superfamily ([Superfamily] cl11420 (PSSMID 213446)) |
| 8. | BAS3327 (YP_029584.1) | Lipoprotein, putative | Defense mechanisms | VanW like protein (pfam04294 (PSSMID 146762)) |
| 9. | BAS4032 (YP_030282.1) | Lipase/acylhydrolase, putative | Amino acid transport and metabolism | SGNH_hydrolase, or GDSL_hydrolase ([Superfamily] cl01053 (PSSMID 207299)) |
| 10. | BAS0534 (YP_026812.1) | Glycerol uptake operon antiterminator regulatory protein, putative | Transcription | Glycerol-3-phosphate responsive antiterminator ([Superfamily] cl00852 (PSSMID 120175)) |
| 11. | BAS0748 (YP_027024.1) | Na/Pi-cotransporter family protein | Inorganic ion transport and metabolism | Na+/phosphate symporter (pfam02690 (PSSMID 202351)) |
| 12. | BAS4373 (YP_030620.1) | Putative uncharacterized protein | – | Protein of unknown function (DUF1696) ([Superfamily] cl06850 (PSSMID 208562))/ YvbH-like oligomerisationregion (pfam11724 (PSSMID 152160)) |
| 13. | BAS3917 (YP_030167.1) | Putative uncharacterized protein | – | – |
| 14. | BAS1831 (YP_028095.1) | Lipoprotein, putative | – | – |
| 15. | BAS3475 (YP_029730.1) | Phosphonate ABC transporter, phosphonate-binding protein, putative | Inorganic ion transport and metabolism | [Superfamily] cl15306 (PSSMID 199164) |
| 16. | BAS1813 (YP_028077.1) | Alpha/beta hydrolase family protein | – | pfam12697 (PSSMID 205026) |
| 17. | BAS0990 (YP_027263.1) | Lipoprotein, putative | Amino acid transport and metabolism | – |
| 18. | BAS4372 (YP_030619.1) | Putative uncharacterized protein | – | – |
| 19. | BAS0651 (YP_026928.1) | Putative uncharacterized protein | – | 3D domain ([Superfamily] cl01439 (PSSMID 207412)) |
| 20. | BAS3956 (YP_030206.1) | Putative uncharacterized protein | – | – |
| 21. | BAS0680 (YP_026957.1) | Ferrous iron transport protein A, putative | Inorganic ion transport and metabolism | FeoAdomain ([Superfamily] cl00838 (PSSMID 207215)) |
| 22. | BAS2439 (YP_028699.1) | Putative uncharacterized protein | – | – |
| 23. | BAS3476 (YP_029731.1) | 2′,3′-cyclic-nucleotide 2′-Phosphodiesterase, N-terminus | Nucleotide transport and metabolism | N-terminal metallophosphatase domain (cd07410 (PSSMID 163653)) |
| 24. | BAS3328 (YP_029585.1) | Putative uncharacterized protein | – | – |
| 25. | BAS0650 (YP_026927.1) | Methyl-accepting chemotaxis protein | Cell motility and signal transduction | Methyl-accepting chemotaxis protein (MCP), signaling domain (cd11386 (PSSMID 206779)) |
| 26. | BAS1954 (YP_028217.1) | Putative uncharacterized protein | – | May be ornothinecarbamoyl transferase |
| 27. | BAS2440 (YP_028700.1) | Putative uncharacterized protein | – | Two different domains of unknown functions |
| 28. | BAS1814 (YP_028078.1) | ABC transporter, ATP-binding protein, N-terminus | Defense mechanisms | ABC-type multidrug transport system, ATPase and permease components ([Superfamily] cl00549 (PSSMID 207103)) |
| 29. | BAS4031 (YP_030281.1) | 2′,3′-cyclic-nucleotide 2′-phosphodiesterase | Nucleotide transport and metabolism | N-terminal metallophosphatase domain (cd07410 (PSSMID 163653)/ 5′-nucleotidase (pfam02872 (PSSMID 202440)) |
| 30. | BAS0681 (YP_026958.1) | Phosphate ABC transporter, phosphate-binding protein, putative | Inorganic ion transport and metabolism | substrate binding domain of LysR-type transcriptional regulators ([Superfamily] cl11398 (PSSMID 209302)) |
Table 2.
WalR binding site in the upstream regions of putative regulon genes.a
| Gene | Sequenceb | Function |
|---|---|---|
| eag (BAS0842)c | S-layer protein | |
| ftsE (BAS5034)c | TGTAACATAACTGTAAC…………N212…………ATG | Cell division ABC transporter |
| hppD (BAS0226)d | TGTTACATTAATGTTAC…………N676…………ATG | 4-Hydroxyphenyl pyruvatedioxygenase |
| kinB3 (BAS3916)c | Sporulation kinase B3 | |
| phoX(BAS4174) | TTTACAAAACTTTTACA…………N85……………ATG | Phosphate ABC transporter |
All the binding sites were present in the intergenic regions of B. anthracis chromosome.
Arrows and underlining indicate the WalR binding site with orientation on either strand.
DNA binding experimentally shown.
Two promoters predicted within 700 bp upstream intergenic region from ORF start (PromBase and Bprom).
DNA binding ability of WalR
RRs mostly function as transcription factors. WalR belongs to the OmpR family of RRs on the basis of its C-terminal domain sequence which contains a winged helix-turn-helix DNA binding motif. To test if WalR could bind to DNA, increasing amounts of purified WalR was incubated with probes from eag, ftsE and kinB3 upstream regions, which contained the WalR binding site, in the presence of an excess of non-specific competitor DNA poly (dI-dC). The incubation of WalR with eag, ftsE and kinB3 probes resulted in formation of higher molecular weight DNA-protein complexes while WalR did not cause a comparable shift with a control probe lacking the binding site (Fig. 8). This shows that WalR can bind to the upstream regions of eag, ftsE and kinB3 in vitro in a sequence dependent manner, indicating that they can be a probable part of WalR regulon.
Fig. 8.
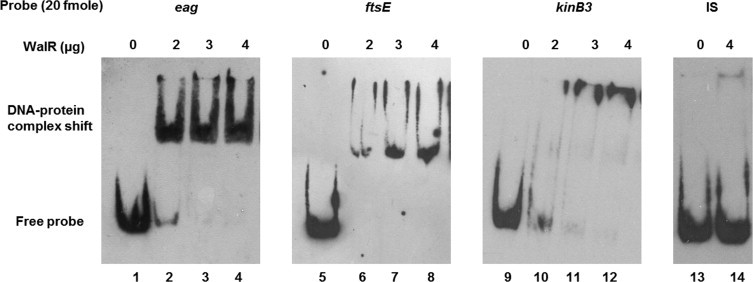
DNA binding ability of WalR. EMSA was used to check for WalR binding to DNA by incubating increasing amounts of purified WalR (2, 3 and 4 μg) with eag, ftsE and kinB3 probes containing the WalR binding site sequence. IS probe, lacking the WalR binding site sequence, was the negative control. Binding reactions were performed at room temperature and the reaction products were run on 8% polyacrylamide native gels in 0.5X TBE at 4 °C. Bands were detected after transfer to a positively charged nylon membrane using a chemiluminescent nucleic acid detection module. WalR could bind to eag, ftsE and kinB3 probes, but not IS probe as shown.
Discussion
In the current study, HK WalK and RR WalR of B. anthracis were shown to form a functional TCS in vitro, exhibiting the classical properties of autophosphorylation and phosphotransfer. WalK variants were also found to display phosphatase activity towards WalR∼P. Growth stage specific qRT-PCR indicated that both the genes were expressed throughout the growth period. WalR could be detected in B. anthracis soluble/cytoplasmic fractions throughout the growth phases through immunoblotting, but WalK could not be detected in any of the fractions. The in silico regulon determination for WalR in B. anthracis comprised of 30 genes, out of which only 5 were annotated for their function. DNA binding ability of WalR to three of the regulon candidates, eag, ftsE and kinB3, could be demonstrated in vitro.
Interestingly, WalRK TCS illustrates some species-specific distinctions in its function. The phosphorylation achieved for B. anthracis WalR (WalRBan) with acetyl phosphate in vitro was suboptimal, whereas WalR from S. pneumoniae (WalRSpn) could undergo ∼95% phosphorylation [29]. The WalKc variants displayed robust phosphatase activity towards WalR∼P, indicating WalKBan to be a bifunctional HK, similar to WalK from another human pathogen S. pneumoniae [40]. The phosphatase activity in HKs has been attributed to DHp domain in most of the bifunctional HKs [41–43], but in S. pneumoniae, a role of PAS domain has also been indicated [40]. In contrast, absence of PAS domain did not diminish the phosphatase activity of WalKBan. Moreover, the WalR∼P levels in phosphotransfer assays of WalKc(PH) and WalKc(H) also did not point towards any difference in their phosphatase activity. WalKBan and WalKSpn differ in HK domain architecture and hence, their domains may serve different roles suitable for their respective physiology.
The in silico WalR regulon analysis gave a list of 30 possible candidates. One of the genes, eag, codes for an S-layer protein, EA1. S-layers are paracrystalline surface structures which cover the surface of a bacterium and are often found in pathogenic organisms. In B. anthracis, two S-layer proteins cover the surface in a growth phase dependent manner-Sap in the exponential phase and EA1 in the stationary phase [44], though in vivo EA1 is the dominant S-layer component [45]. EA1 has been recognized as a major cell associated antigen of B. anthracis and elicits a strong humoral response [45]. It has been shown to have murein hydrolase activity as well [46]. The WalR binding sequence was found upstream of the −35 region of EA1 sigma-H promoter [44], suggesting a positive regulation. It is intriguing to envisage what role WalR, which is primarily activated during the exponential phase as seen in related organisms [25,47], may play in the eag regulation.
Another candidate from the regulon search was ftsE, which codes for a cell division ABC transporter. In B. anthracis, FtsE forms the ATP binding moiety of an ABC transporter complex FtsEX and both the genes occur in an operon. FtsEX has been shown to regulate sporulation and more recently, peptidoglycan hydrolase activity in B. subtilis [48,49]. Interestingly, FtsX has been revealed to mediate killing of B. anthracis vegetative cells in a CXCL10 chemokine dependent manner [50]. Related pathogen S. aureus also interacts with host chemokines and this interaction triggers the release of one of its virulence factor [51]. Such interactions, as recognized before [50], may represent another mode of host-pathogen interactions which influence the fate of an infection. WalR binding site was found upstream of the −35 box of a bioinformatically predicted promoter for ftsE, indicative of a possible positive regulation.
The kinB3 gene also had a WalR binding site in its upstream region. KinB3 is one the nine sporulation HKs of B. anthracis [21]. Sporulation occurs in B. anthracis upon sensing of adverse conditions. These spores are the infective particles of B. anthracis, and can survive harsh environments. The kinB3 in B. anthracis Ames strain, BA4223, was found to be the most active gene for complementation of sporulation deficient B. subtilis. Also, its promoter was found to be the second most active promoter among all the sporulation HKs of B. anthracis Ames strain under conditions favorable for sporulation. However, its deletion did not affect sporulation significantly [21], which could be due to redundancy of its function with other genes. The high activity of kinB3 promoter suggests kinB3 to be playing an important role in vivo. Again, the position of WalR binding site relative to a bioinformatically predicted promoter hints towards a positive regulation, suggesting WalRK TCS to be linked to sporulation in B. anthracis.
The positive results in binding studies with all three of the regulon candidates tested indicate that WalR may additionally bind to the upstream regions of the other gene candidates also. Structure function annotation of the rest of the 27 genes showed that 9 are involved in ion, nucleotide and amino acid transport and metabolism, 2 are hydrolases and 1 each is a member of Major Facilitator Superfamily (MFS), a VanW like protein implicated in vancomycin resistance and a chemotaxis protein. However, for some of the hits, no conserved domain or function could be deduced from the databases (Table 1). Though none of the direct virulence factors emerged in the in silico analysis, negating a direct role of WalRK TCS in B. anthracis virulence, the possibility of modulation of sporulation by WalR appears exciting.
Overall, our study establishes that B. anthracis has a functional WalRK, encoding a bifunctional histidine kinase/phosphatase and a cognate response regulator, which displays some species-specific functions. It may control a diverse set of genes with functions ranging from structure, cell division, sporulation, transport and metabolism. Further studies are required, under conditions mimicking the host environment, to elucidate the role it may play in the viability and pathogenicity of B. anthracis.
Materials and methods
Materials
Escherichia coli strains DH5α and BL21(λDE3) were used as cloning and expression hosts, respectively. An avirulent strain of B. anthracis Sterne 34F2 (pXO1+ pXO2−) was used in this study. E. coli strains were grown in Luria Bertani medium and B. anthracis Sterne strain in Brain Heart Infusion (BHI) medium, supplemented with antibiotics ampicillin (100 μg/ml) and kanamycin (50 μg/ml), wherever required.
pET28a+ from Novagen (Madison, WI, USA) was used as the expression vector for heterologous gene expression. DNA polymerases and restriction enzymes from New England Biolabs Inc. (Ipswich, MA, USA) were used. Ni+2–NTA agarose resin was from Qiagen (Hilden, Germany). Nitrocellulose membrane was from MDI membrane technologies (Ambala Cantt., India). Isopropyl β-D-thiogalactopyranoside (IPTG), bovine serum albumin (BSA) and antibiotics were from USB chemicals (Affymetrix, Ohio, USA). Anti-histidine murine monoclonal, alkaline phosphatase conjugated, horseradish peroxidase conjugated and FITC conjugated antibodies were from Sigma–Aldrich (St. Louis, MO, USA). Bradford reagent used for protein estimation was from Bio-rad (CA, USA). [γ-32P]-ATP was obtained from BRIT (Hyderabad, India). Oligonucleotides were synthesized from Sigma (St. Louis, MO, USA).
Cloning, expression and purification of WalR, WalKc, WalKc(PH), WalKc(H) and DHp of B. anthracis
The ORFs corresponding to walR comprising of receiver and effector domains; walKc comprising of HAMP, PAS, DHp and CA domains; walKc(PH) comprising of PAS, DHp and CA domains; walKc(H) comprising of DHp and CA domains and DHp domain; were amplified from the B. anthracis genomic DNA using the indicated primers (Table S1). After restriction digestion, the amplicons were ligated to NcoI–XhoI digested pET28a+ expression vector to obtain pETwalR, pETwalKc, pETwalKc(PH), pETwalKc(H) and pETDHp constructs. The sequence of the cloned genes was verified through automated dideoxy DNA sequencing. pETwalR was transformed into competent BL21(λDE3) cells and protein expression was induced at O.D.600nm ∼0.4, with the addition of 1 mM IPTG for 8 h at 37 °C. For pETwalKc, pETwalKc(PH), pETwalKc(H) and pETDHp, the protein expression was induced in BL21(λDE3) cells at O.D.600nm ∼0.6 with 0.1 mM IPTG at 18 °C for 16 h. The identity of all the recombinant proteins was checked by immunoblotting. MALDI-TOF was also used for recombinant protein identification. The coomassie stained band at the requisite position was excised and digested with sequencing grade trypisin (Promega, Madison, USA) followed by MALDI-TOF/MS analysis at Central Instrumentation Facility, School of Life Sciences, Jawaharlal Nehru University, New Delhi. MASCOT search engine (http://www.matrixscience.com; Matrix Science, London, UK) was used for protein identification using Peptide Mass Fingerprinting. Purification of the proteins was done from the soluble fraction using Ni+2–NTA affinity chromatography as described by Qiagen. WalKc was also purified from the inclusion bodies to raise protein specific polyclonal antibodies in mice. The protein concentration was estimated using Bradford reagent, with BSA as the standard.
Raising of polyclonal antibodies against WalR and WalKc
Swiss-albino mice (4–6 weeks old) were immunized subcutaneously with 20 μg of recombinant WalR and WalKc proteins emulsified in complete Freund's adjuvant. Subsequent booster doses were given on the 14th and 28th day with proteins emulsified in incomplete Freund's adjuvant. The mice were bled after the 14th day of each immunization and sera were collected. Enzyme-linked immunosorbant assay (ELISA) was done for determining antibody titers for each protein and ascertaining dilutions that may be used for immunoblotting. The regulations of Institutional Animal Ethics Committee (IAEC) were followed in all mice experiments.
Immunoblotting
The cell lysates of E. coli expressing WalR or WalKc (or WalKc(PH) or WalKc(H)) were run on a 10–12% SDS–PAGE and electroblotted onto nitrocellulose membrane followed by immunodetection. The membrane was blocked for 1 h at RT with 2% BSA solution in PBS, then incubated with primary antibody- anti-His antibody (1:10,000), anti-WalR (1:7000) or anti-WalKc (1:50,000)- as the case may be, for 1 h at RT followed by incubation with alkaline phosphatase conjugated anti-mouse IgG secondary antibody (1:10,000) for 1 h at RT. All antibody incubations were followed by three washes with PBST (PBS + 0.05% Tween 20) for 5 min each. The immunoreactive bands were visualized using BCIP–NBT substrate solution.
In vivo expression of walR and walK in B. anthracis
-
(a).
Quantitative real time PCR (qRT-PCR): Transcript abundance for walR and walK was determined by qRT-PCR using SYBR Green chemistry (Applied Biosystems). Total RNA was isolated from 10 ml of B. anthracis cells (O.D.600nm ∼0.3, 0.6, 0.9, 1.2 corresponding to early exponential, mid-exponential, late-exponential and onset of stationary phase, respectively) grown in BHI medium using trizol method followed by RNase free-DNase (Qiagen) treatment. A growth curve of B. anthracis was used to designate the different stages of growth. Genomic DNA contamination was checked by PCR using gene specific primers. 1 μg of RNA was used to synthesize cDNA using the high capacity cDNA reverse transcription kit (Applied Biosystems). Gene-specific primers for qRT-PCR were designed using the Primer Express software, version 2.0. The reaction was performed in a 10 μl volume containing 5 μl of 2X SYBR Green master mix, 40 nM of primers, and 1 μl of 10-fold diluted reverse transcription products (5 ng cDNA per 10 μl reaction mix). The PCRs were run in ABI 7500 using the following program: 50 °C for 2 min, 95 °C for 10 min, and 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Following PCR amplification, thermal dissociation curves were analyzed for each reaction to detect non specific amplification products. The dissociation program was 95 °C for 15 s, 60 °C for 15 s followed by 20 min of slow ramp from 60 to 95 °C. For each gene, cDNA dilution curves were generated to calculate the individual real time PCR efficiencies that were determined by measuring the values of the slopes obtained. Three technical replicates of each reaction were run. A constitutively expressed gene, dna gyrase, was used as an endogenous control for data normalization. Ct values for walR and walK were plotted after subtraction of the endogenous control (−ΔCt). Normalized Ct values, mean with SEM, are plotted against different growth stages, using GraphPad Prism 5 software.
-
(b).
Immunoblotting: B. anthracis cells were grown in BHI medium (O.D.600nm ∼0.3, 0.6, 0.9, 1.2) and subjected to sonication for 15 min at 30% amplitude (2 mm microtip, 750W Sonic Vibra Cell Sonicator) followed by centrifugation where the supernatant was the soluble/cytoplasmic fraction, while the pellet was treated as the cell wall plus membrane fraction. The fractions were checked for the presence of WalR and WalK, by subjecting them to 10% SDS–PAGE followed by immunoblotting with protein specific antisera as described above. Equal protein was loaded for each fraction, with recombinant protein as the positive control.
Autophosphorylation and phosphotransfer assay
Phosphorylation assays were performed as described before [26]. Briefly, for WalKc, WalKc(PH) and WalKc(H) autophosphorylation, 3 μg of protein was incubated in a final volume of 20 μl phosphorylation buffer (100 mM Tris HCl [pH 8], 200 mM KCl, 4 mM MgCl2, 20 mM DTT, 0.1 mM EDTA, 3.5% glycerol, 2.5 μM ATP, 5 μCi of [γ-32P]-ATP) for 5, 10, 30, 60 and 90 min at 37 °C. Reaction was started by addition of radiolabelled ATP mixture and stopped by addition of 4 μl SDS loading buffer. For phosphotransfer reactions, 3 μg of both proteins were mixed together in a final volume of 30 μl and the reaction was initiated by addition of radiolabelled ATP mixture. The reaction mixtures were analyzed by a 10% SDS–PAGE, followed by autoradiography. Intensity analysis was performed using ImageJ 1.45S software [52].
Phosphorylation of WalR by small molecule phosphoryl donors
WalR (20 μg) was incubated with 60 mM of acetyl phosphate or carbamoyl phosphate (Sigma) in 50 μl of phosphorylation buffer (50 mM Tris HCl [pH 7.5], 50 mM KCl, 20 mM MgCl2, 20 mM DTT) for 15, 30, 45, 60 and 90 min at 37 °C. The reaction was stopped by addition of 40 mM EDTA (final concentration) which halts any further phosphorylation. The stability of the phosphorylated form to acid/base treatment was tested in the presence of 0.5 N HCl and 0.5 N NaOH at room temperature for 20 min. The phosphorylation status of WalR was analyzed by 12% polyacrylamide native gel electrophoresis and coomassie staining. Intensity analysis was done using ImageJ 1.45S software.
WalKc variants mediated dephosphorylation of WalR∼P
WalR (20 μg in 100 μl reaction mix) was phosphorylated using acetyl phosphate for 60 min at 37 °C as described above and the reaction was terminated with 40 mM EDTA so that no more phosphorylation occurs (acetyl phosphate mediated phosphorylation is Mg+2 dependent). The reaction mix was then divided into two 50 μl aliquots (10 μg WalR each). The phosphatase activity of WalKc(PH) and WalKc(H) was tested by incubating WalR∼P, from one of the 50 μl aliquots, with 5 μg of each protein separately at 37 °C for 5, 10, 20 and 30 min. The second 50 μl aliquot was used as the control with only buffer incubation. The reaction mixtures were analyzed on a 12% polyacrylamide native gel followed by coomassie staining and ImageJ intensity analysis.
Identification of putative WalR regulon in B. anthracis genome
A consensus WalR binding sequence, consisting of two hexanucleotide direct repeats separated by five nucleotides [5′-TGT(A/T)A(A/T/C)-N5-TGT(A/T)A(A/T/C)-3′], known from B. subtilis [39] was used to perform an in silico motif search in the intergenic regions of complete B. anthracis Sterne chromosome using the DNA pattern search tool available at http://rsat.ulb.ac.be/genome-scale-dna-pattern_form.cgi [53]. The final list of genes obtained was checked for annotations using Uniprot database and domain annotation was performed using CD search [54]. Functional prediction was carried out using the COG database [55]. Prokaryotic promoter identification was done using PromBase [56] and Bprom program (Softberry), if experimental data was not available.
Electrophoretic mobility shift assay (EMSA)
For EMSA, the upstream regions of the candidate genes that emerged from the bioinformatic analysis were used. The upstream regions containing WalR binding site from three putative WalR regulon genes, namely, eag (52 bp), ftsE (54 bp) and kinB3 (73 bp) along with adjoining sequences was synthesized from Sigma (St. Louis, MO, USA) (Table S2) as sense and anti-sense oligonucleotides. The oligonucleotides were 3′-end labeled with biotin using biotin 3′-end DNA labeling kit from Pierce Biotechnology (Rockford, IL, USA) as per the manufacturer's instructions. The labeled oligonucleotides were then annealed to form a double stranded DNA probe for EMSA. Binding reactions were performed at room temperature in a final volume of 20 μl binding buffer (10 mM Tris HCl [pH 7.5], 50 mM NaCl, 10% glycerol, 5 mM EDTA, 20 mM DTT, 0.5 μg poly (dI-dC)) for 20 min with varying amounts of WalR (2, 3 and 4 μg) and 20 fmole of the labeled probe. Reaction mixtures were electrophoresed on 8% polyacrylamide native gels (29:1 acrylamide:bisacrylamide ratio) in 0.5X TBE at 4 °C. Bands were detected after transfer to a positively charged nylon membrane (MDI membrane technologies) using Chemiluminescent Nucleic Acid Detection Module (Pierce Biotechnology) as per the manufacturer's instructions. An oligo having an internal gene sequence of walR (IS, 53 bp) was labeled and used as described above, for a negative control.
Acknowledgements
We acknowledge the financial support from CSIR, Government of India. Alisha Dhiman is a recipient of Senior Research Fellowship from CSIR, Government of India. The funding agency has no role in study design or data analysis and interpretation. This work was in part supported by DST-PURSE grant. AIRF, JNU is acknowledged for providing the LSCM facility and Mr. Ashok Kumar Sahu is acknowledged for his technical help in the same. We thank Dr. Samer Singh for critical review of the manuscript.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Contributor Information
Alisha Dhiman, Email: alisha.dhiman1986@gmail.com.
Sonika Bhatnagar, Email: ecc999@gmail.com.
Parul Kulshreshtha, Email: parulkuls26@gmail.com.
Rakesh Bhatnagar, Email: rakeshbhatnagar@jnu.ac.in.
Supplementary material
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.fob.2013.12.005.
Appendix. Supplementary materials
(A) Overexpression of WalR, WalKc, WalKc(PH) and WalKc(H) recombinant proteins. (B) Purified WalR (+β-mercaptoethanol with WalR monomer, −β-mercaptoethanol with WalR monomer and dimer), WalKc, WalKc(PH) and WalKc(H). M – Molecular weight standards in kDa, UI – Uninduced, IN – Induced.
Primers used in the study.
Oligos used in EMSA.
References
- 1.Mock M, Fouet A. Anthrax. Annu. Rev. Microbiol. 2001;55:647–671. doi: 10.1146/annurev.micro.55.1.647. 55/1/647 [pii] [DOI] [PubMed] [Google Scholar]
- 2.Collier RJ, Young JA. Anthrax toxin. Annu. Rev. Cell Dev. Biol. 2003;19:45–70. doi: 10.1146/annurev.cellbio.19.111301.140655. [DOI] [PubMed] [Google Scholar]
- 3.Chitlaru T, Gat O, Gozlan Y, Ariel N, Shafferman A. Differential proteomic analysis of the Bacillus anthracis secretome: distinct plasmid and chromosome CO2-dependent cross talk mechanisms modulate extracellular proteolytic activities. J. Bacteriol. 2006;188:3551–3571. doi: 10.1128/JB.188.10.3551-3571.2006. 188/10/3551 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gat O, Mendelson I, Chitlaru T, Ariel N, Altboum Z, Levy H, Weiss S, Grosfeld H, Cohen S, Shafferman A. The solute-binding component of a putative Mn(II) ABC transporter (MntA) is a novel Bacillus anthracis virulence determinant. Mol. Microbiol. 2005;58:533–551. doi: 10.1111/j.1365-2958.2005.04848.x. MMI4848 [pii] [DOI] [PubMed] [Google Scholar]
- 5.Wilson AC, Hoch JA, Perego M. Two small c-type cytochromes affect virulence gene expression in Bacillus anthracis. Mol. Microbiol. 2009;72:109–123. doi: 10.1111/j.1365-2958.2009.06627.x. MMI6627 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chitlaru T, Zaide G, Ehrlich S, Inbar I, Cohen O, Shafferman A. HtrA is a major virulence determinant of Bacillus anthracis. Mol. Microbiol. 2011;81:1542–1559. doi: 10.1111/j.1365-2958.2011.07790.x. [DOI] [PubMed] [Google Scholar]
- 7.Dixon SD, Janes BK, Bourgis A, Carlson PE, Jr., Hanna PC. Multiple ABC transporters are involved in the acquisition of petrobactin in Bacillus anthracis. Mol. Microbiol. 2012;84:370–382. doi: 10.1111/j.1365-2958.2012.08028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fouet A, Mock M. Regulatory networks for virulence and persistence of Bacillus anthracis. Curr. Opin. Microbiol. 2006;9:160–166. doi: 10.1016/j.mib.2006.02.009. S1369–5274(06)00029–4 [pii] [DOI] [PubMed] [Google Scholar]
- 9.Dai Z, Sirard JC, Mock M, Koehler TM. The atxA gene product activates transcription of the anthrax toxin genes and is essential for virulence. Mol. Microbiol. 1995;16:1171–1181. doi: 10.1111/j.1365-2958.1995.tb02340.x. [DOI] [PubMed] [Google Scholar]
- 10.Drysdale M, Bourgogne A, Hilsenbeck SG, Koehler TM. atxA controls Bacillus anthracis capsule synthesis via acpA and a newly discovered regulator, acpB. J. Bacteriol. 2004;186:307–315. doi: 10.1128/JB.186.2.307-315.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saile E, Koehler TM. Control of anthrax toxin gene expression by the transition state regulator abrB. J. Bacteriol. 2002;184:370–380. doi: 10.1128/JB.184.2.370-380.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu. Rev. Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. 69/1/183 [pii] [DOI] [PubMed] [Google Scholar]
- 13.Beier D, Gross R. Regulation of bacterial virulence by two-component systems. Curr. Opin. Microbiol. 2006;9:143–152. doi: 10.1016/j.mib.2006.01.005. S1369–5274(06)00020–8 [pii] [DOI] [PubMed] [Google Scholar]
- 14.Delaune A, Dubrac S, Blanchet C, Poupel O, Mader U, Hiron A, Leduc A, Fitting C, Nicolas P, Cavaillon JM. The WalKR system controls major staphylococcal virulence genes and is involved in triggering the host inflammatory response. Infect. Immun. 2012;80:3438–3453. doi: 10.1128/IAI.00195-12. IAI.00195–12 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashenberg O, Keating AE, Laub MT. Helix bundle loops determine whether histidine kinases autophosphorylate in cis or in trans. J. Mol. Biol. 2013;425:1198–1209. doi: 10.1016/j.jmb.2013.01.011. S0022–2836(13)00014–4 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao R, Stock AM. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 2009;63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silversmith RE. Auxiliary phosphatases in two-component signal transduction. Curr. Opin. Microbiol. 2010;13:177–183. doi: 10.1016/j.mib.2010.01.004. S1369–5274(10)00009–3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huynh TN, Stewart V. Negative control in two-component signal transduction by transmitter phosphatase activity. Mol. Microbiol. 2011;82:275–286. doi: 10.1111/j.1365-2958.2011.07829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vetter SM, Schlievert PM. The two-component system Bacillus respiratory response A and B (BrrA-BrrB) is a virulence factor regulator in Bacillus anthracis. Biochemistry. 2007;46:7343–7352. doi: 10.1021/bi700184s. [DOI] [PubMed] [Google Scholar]
- 20.Stauff DL, Skaar EP. Bacillus anthracis HssRS signalling to HrtAB regulates haem resistance during infection. Mol. Microbiol. 2009;72:763–778. doi: 10.1111/j.1365-2958.2009.06684.x. MMI6684 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brunsing RL, La Clair C, Tang S, Chiang C, Hancock LE, Perego M, Hoch JA. Characterization of sporulation histidine kinases of Bacillus anthracis. J. Bacteriol. 2005;187:6972–6981. doi: 10.1128/JB.187.20.6972-6981.2005. 187/20/6972 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Been M, Francke C, Moezelaar R, Abee T, Siezen RJ. Comparative analysis of two-component signal transduction systems of Bacillus cereus, Bacillus thuringiensis and Bacillus anthracis. Microbiology. 2006;152:3035–3048. doi: 10.1099/mic.0.29137-0. 152/10/3035 [pii] [DOI] [PubMed] [Google Scholar]
- 23.Barakat M, Ortet P, Whitworth DE. P2CS: a database of prokaryotic two-component systems. Nucleic Acids Res. 2011;39:D771–776. doi: 10.1093/nar/gkq1023. gkq1023 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capra EJ, Laub MT. Evolution of two-component signal transduction systems. Annu. Rev. Microbiol. 2012;66:325–347. doi: 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fabret C, Hoch JA. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J. Bacteriol. 1998;180:6375–6383. doi: 10.1128/jb.180.23.6375-6383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dubrac S, Msadek T. Identification of genes controlled by the essential YycG/YycF two-component system of Staphylococcus aureus. J. Bacteriol. 2004;186:1175–1181. doi: 10.1128/JB.186.4.1175-1181.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner C, Saizieu Ad A, Schonfeld HJ, Kamber M, Lange R, Thompson CJ, Page MG. Genetic analysis and functional characterization of the Streptococcus pneumoniae vic operon. Infect. Immun. 2002;70:6121–6128. doi: 10.1128/IAI.70.11.6121-6128.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng WL, Robertson GT, Kazmierczak KM, Zhao J, Gilmour R, Winkler ME. Constitutive expression of PcsB suppresses the requirement for the essential VicR (YycF) response regulator in Streptococcus pneumoniae R6. Mol. Microbiol. 2003;50:1647–1663. doi: 10.1046/j.1365-2958.2003.03806.x. doi: 3806 [pii] [DOI] [PubMed] [Google Scholar]
- 29.Ng WL, Tsui HC, Winkler ME. Regulation of the pspA virulence factor and essential pcsB murein biosynthetic genes by the phosphorylated VicR (YycF) response regulator in Streptococcus pneumoniae. J. Bacteriol. 2005;187:7444–7459. doi: 10.1128/JB.187.21.7444-7459.2005. 187/21/7444 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Senadheera MD, Guggenheim B, Spatafora GA, Huang YC, Choi J, Hung DC, Treglown JS, Goodman SD, Ellen RP, Cvitkovitch DG. A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J. Bacteriol. 2005;187:4064–4076. doi: 10.1128/JB.187.12.4064-4076.2005. 187/12/4064 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gotoh Y, Eguchi Y, Watanabe T, Okamoto S, Doi A, Utsumi R. Two-component signal transduction as potential drug targets in pathogenic bacteria. Curr. Opin. Microbiol. 2010;13:232–239. doi: 10.1016/j.mib.2010.01.008. S1369–5274(10)00013–5 [pii] [DOI] [PubMed] [Google Scholar]
- 32.Lukat GS, McCleary WR, Stock AM, Stock JB. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc. Natl. Acad. Sci. U.S.A. 1992;89:718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCleary WR, Stock JB. Acetyl phosphate and the activation of two-component response regulators. J. Biol. Chem. 1994;269:31567–31572. [PubMed] [Google Scholar]
- 34.Liu W, Hulett FM. Bacillus subtilis PhoP binds to the phoB tandem promoter exclusively within the phosphate starvation-inducible promoter. J. Bacteriol. 1997;179:6302–6310. doi: 10.1128/jb.179.20.6302-6310.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. gkr1065 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao R, Collins EJ, Bourret RB, Silversmith RE. Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ. Nat. Struct. Biol. 2002;9:570–575. doi: 10.1038/nsb816. nsb816 [pii] [DOI] [PubMed] [Google Scholar]
- 37.Wolanin PM, Webre DJ, Stock JB. Mechanism of phosphatase activity in the chemotaxis response regulator CheY. Biochemistry. 2003;42:14075–14082. doi: 10.1021/bi034883t. [DOI] [PubMed] [Google Scholar]
- 38.Zhu Y, Qin L, Yoshida T, Inouye M. Phosphatase activity of histidine kinase EnvZ without kinase catalytic domain. Proc. Natl. Acad. Sci. U.S.A. 2000;97:7808–7813. doi: 10.1073/pnas.97.14.7808. doi: 97/14/7808 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Howell A, Dubrac S, Andersen KK, Noone D, Fert J, Msadek T, Devine K. Genes controlled by the essential YycG/YycF two-component system of Bacillus subtilis revealed through a novel hybrid regulator approach. Mol. Microbiol. 2003;49:1639–1655. doi: 10.1046/j.1365-2958.2003.03661.x. doi: 3661 [pii] [DOI] [PubMed] [Google Scholar]
- 40.Gutu AD, Wayne KJ, Sham LT, Winkler ME. Kinetic characterization of the WalRKSpn (VicRK) two-component system of Streptococcus pneumoniae: dependence of WalKSpn (VicK) phosphatase activity on its PAS domain. J. Bacteriol. 2010;192:2346–2358. doi: 10.1128/JB.01690-09. JB.01690–09 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russo FD, Silhavy TJ. EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J. Mol. Biol. 1991;222:567–580. doi: 10.1016/0022-2836(91)90497-t. doi: 0022–2836(91)90497-T [pii] [DOI] [PubMed] [Google Scholar]
- 42.Willett JW, Kirby JR. Genetic and biochemical dissection of a HisKA domain identifies residues required exclusively for kinase and phosphatase activities. PLoS Genet. 2012;8:e1003084. doi: 10.1371/journal.pgen.1003084. PGENETICS-D-12–00740 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huynh TN, Noriega CE, Stewart V. Missense substitutions reflecting regulatory control of transmitter phosphatase activity in two-component signalling. Mol. Microbiol. 2013;88:459–472. doi: 10.1111/mmi.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mignot T, Mesnage S, Couture-Tosi E, Mock M, Fouet A. Developmental switch of S-layer protein synthesis in Bacillus anthracis. Mol. Microbiol. 2002;43:1615–1627. doi: 10.1046/j.1365-2958.2002.02852.x. doi: 2852 [pii] [DOI] [PubMed] [Google Scholar]
- 45.Mesnage S, Tosi-Couture E, Mock M, Gounon P, Fouet A. Molecular characterization of the Bacillus anthracis main S-layer component: evidence that it is the major cell-associated antigen. Mol. Microbiol. 1997;23:1147–1155. doi: 10.1046/j.1365-2958.1997.2941659.x. [DOI] [PubMed] [Google Scholar]
- 46.Ahn JS, Chandramohan L, Liou LE, Bayles KW. Characterization of CidR-mediated regulation in Bacillus anthracis reveals a previously undetected role of S-layer proteins as murein hydrolases. Mol. Microbiol. 2006;62:1158–1169. doi: 10.1111/j.1365-2958.2006.05433.x. MMI5433 [pii] [DOI] [PubMed] [Google Scholar]
- 47.Howell A, Dubrac S, Noone D, Varughese KI, Devine K. Interactions between the YycFG and PhoPR two-component systems in Bacillus subtilis: the PhoR kinase phosphorylates the non-cognate YycF response regulator upon phosphate limitation. Mol. Microbiol. 2006;59:1199–1215. doi: 10.1111/j.1365-2958.2005.05017.x. MMI5017 [pii] [DOI] [PubMed] [Google Scholar]
- 48.Garti-Levi S, Hazan R, Kain J, Fujita M, Ben-Yehuda S. The FtsEX ABC transporter directs cellular differentiation in Bacillus subtilis. Mol. Microbiol. 2008;69:1018–1028. doi: 10.1111/j.1365-2958.2008.06340.x. MMI6340 [pii] [DOI] [PubMed] [Google Scholar]
- 49.Meisner J, Montero Llopis P, Sham LT, Garner E, Bernhardt TG, Rudner DZ. FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis. Mol. Microbiol. 2013 doi: 10.1111/mmi.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crawford MA, Lowe DE, Fisher DJ, Stibitz S, Plaut RD, Beaber JW, Zemansky J, Mehrad B, Glomski IJ, Strieter RM. Identification of the bacterial protein FtsX as a unique target of chemokine-mediated antimicrobial activity against Bacillus anthracis. Proc. Natl. Acad. Sci. U.S.A. 2011;108:17159–17164. doi: 10.1073/pnas.1108495108. 1108495108 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yung SC, Parenti D, Murphy PM. Host chemokines bind to Staphylococcus aureus and stimulate protein A release. J. Biol. Chem. 2011;286:5069–5077. doi: 10.1074/jbc.M110.195180. M110.195180 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Helden J. Regulatory sequence analysis tools. Nucleic Acids Res. 2003;31:3593–3596. doi: 10.1093/nar/gkg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–331. doi: 10.1093/nar/gkh454. 32/suppl_2/W327 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. doi: gkd013 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rangannan V, Bansal M. PromBase: a web resource for various genomic features and predicted promoters in prokaryotic genomes. BMC Res. Notes. 2011;4:257. doi: 10.1186/1756-0500-4-257. 1756–0500–4–257 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Overexpression of WalR, WalKc, WalKc(PH) and WalKc(H) recombinant proteins. (B) Purified WalR (+β-mercaptoethanol with WalR monomer, −β-mercaptoethanol with WalR monomer and dimer), WalKc, WalKc(PH) and WalKc(H). M – Molecular weight standards in kDa, UI – Uninduced, IN – Induced.
Primers used in the study.
Oligos used in EMSA.


