Key Points
The Notch ligand Delta-1 reduces membrane bound IL-6R expression, inhibiting IL-6 cis-signaling and the production of myeloid cells.
Combined with a dynamically fed culture system that minimizes IL-6 trans-signaling, Delta-1 produces rapid and sustained HSC engraftment.
Abstract
Increasing evidence supports the importance of cell extrinsic regulation in stem cell fate control. Hematopoietic stem cells (HSC) are responsive to local signals from their niche and to systemic feedback from progenitors and mature cells. The Notch ligand Delta-1 (DL1), a key component of the stem cell niche, regulates human hematopoietic lineage development in a dose-dependent manner and has been used clinically for primitive progenitor expansion. How DL1 acts to regulate HSC fate and whether these actions are related to its lineage skewing effects are poorly understood. Here we demonstrate that, although DL1 activates signal transducer and activator of transcription 3 signaling similarly to the gp130-activating cytokine interleukin-6 (IL-6), it has opposite effects on myeloid cell production. Mechanistically, these different outcomes are attributable to a DL1-mediated reduction in membrane (m)-bound IL-6 receptor (R) expression, converting progenitor cells from being directly IL-6 responsive to requiring both IL-6 and soluble (s) IL-6R for activation. Concomitant reduction of both mIL-6R (by DL1 supplementation) and sIL-6R (using dynamically fed cultures) reduced myeloid cell production and led to enhanced outputs of human HSCs. This work describes a new mode of cytokine action in which DL1 changes cytokine receptor distributions on hematopoietic cells, altering feedback networks and their impact on stem cell fate.
Introduction
Stem cell niche-mediated signaling is complex and context dependent. Notch signaling in hematopoiesis exemplifies this complexity.1,2 Osteoblasts regulate hematopoietic stem cells (HSCs) in the bone marrow niche in a Notch-mediated manner; however, the precise role of Notch pathway activation in HSC cell fate control is less clear.2 In vitro, Notch ligand Delta-1 (DL1) has been shown to enhance short-term severe combined immunodeficient (SCID)-repopulating cells (SRCs), and clinical data indicate that patients transplanted with cells cultured in the presence of DL1 had improved time to neutrophil engraftment after transplantation.3,4
Methods to expand HSC numbers in vitro have traditionally focused on identifying self-renewal factors.4-8 The phenotypic and molecular continuum between HSCs and their closely related but developmentally restricted progeny makes this endeavor challenging. We have previously shown that feedback from mature cells has a significant and cell type–specific impact on HSC expansion.9,10 The role of lineage skewing on local and systemwide feedback may significantly contribute to the regulation of HSC fate in vitro and in vivo. Here, we sought to understand the link between Notch-mediated mature cell lineage skewing and primitive cell fate modulation during human hematopoiesis. By decoupling intrinsic cell fate changes from their microenvironmental consequences using dynamically fed cultures,9 we demonstrate that the myeloid cell lineage skewing observed in the presence of DL1 is predominantly attributable to Notch-mediated regulation of cell type–specific interleukin 6 receptor (IL-6R) expression. This lineage skewing results in an environment supportive for HSC growth, allowing for both rapid and sustained expansion of SRCs.
Methods
Umbilical cord blood cell collection and processing
Umbilical cord blood cell samples were collected from consenting donors according to ethically approved procedures at Mt Sinai Hospital (Toronto, ON, Canada). Mononuclear cells were obtained as described.11 Lineage negative (Lin−) progenitor cells or CD34+ cells were isolated from the mononuclear cell fraction using the EasySep system with the human progenitor cell enrichment kit or human CD34+ enrichment kit (Stemcell Technologies, Vancouver, BC, Canada).
DL1 preparation
DL1 was produced and coated as previously described.12 Both 2.5 μg/mL of DL1 and 5 μg/mL of retronectin (Takara Shuzo, Otsu, Japan) were diluted in chilled phosphate-buffered saline, coated onto nontissue culture–treated plates at 0.16 mL/cm2, and kept overnight at 4°C.
Cell seeding and in vitro culture
CD34+ cells were seeded on DL1-coated plates at an initial density of 1 × 105 cells/mL in serum-free Iscove modified Dulbecco medium (Gibco, Rockville, MD) with 20% bovine serum albumin, insulin, and transferrin serum substitute (Stemcell Technologies) and 1% Glutamax (Gibco). The medium was supplemented with 100 ng/mL Stem Cell Factor (R&D Systems, Minneapolis, MN), 100 ng/mL FMS-like Tryosine Kinase 3 Ligand (R&D Systems), 50 ng/mL Thrombopoietin (R&D Systems), and 1 μg/mL low-density lipoproteins (Calbiochem, La Jolla, CA). Plates were cultured on an orbital shaker to aid in mixing of the system. To mimic the feeding scheme of the previously described fed-batch system,9 cultures were fed every 24 hours with 1 unit volume of fresh media to maintain a dilution rate of D = 1. Cells were transferred to plates with freshly coated ligand every 4 days. Where indicated, IL-6 (R&D) was used at 50 ng/mL, soluble (s)IL-6R (R&D) was used at 1000 ng/mL, Janus kinase inhibitor (JAKi) (Calbiochem) was used at 50 nM, γ-secretase inhibitor L685, 458 (R&D) was used at 1 μM, and SR1 was used at 750 nM. For non–fed-batch conditions, CD34+ cells were cultured at 1 × 105 cells/mL, and a full media exchange was performed every 4 days.
Cell assays
Colony-forming cell (CFC) assay and long-term culture-initiating cell (LTC-IC) assay assays were performed as previously described.11 Surface marker staining was performed with conjugated human antibodies (BD Biosciences, San Jose, CA). All samples were analyzed on a FACSCanto or FACS LSR Fortessa flow cytometer (BD Biosciences). For the sorted cell assay, Lin− cells were sorted for Rhodamine (Rho)loCD34+CD38−CD45RA−CD49f+ with a FACSAria flow cytometer (BD Biosciences), as previously described.13 Forty sorted cells were dispensed per well in a 96-well plate in the previously described media and cultured for 7 days. For the assessment of paracrine signaling, conditioned media were produced by culturing Lin− cells for 10 days in the previously described culture media and then sorting for cells positive for CD14, CD15, CD7, CD41, GlyA, or other (CD34−CD14−CD15−CD7−CD41−GlyA−). Mature cells were seeded at 5 × 105 cells/mL for 7 days and the media were then collected and used to culture the sorted primitive cells as described previously.
Gene expression analysis
Raw gene expression data were obtained from the National Center for Biotechnology Information’s Gene Expression Omnibus through accession numbers GSE42414GSE4241414 and GSE24759.15 The 2 datasets were combined by following a 4-step procedure. First, quantile signals of GSE42414 and GSE24759 datasets were obtained using the normalizeQuantiles() function and the justRMA() function in the open-source limma package (v3.12.3) of BioConductor, respectively. Second, genomewide normalization was performed with respect to a common population between the 2. Third, for each dataset, the average value of probes that target the same gene was calculated. Last, the 2 datasets were combined through Entrez ID. All the analyses were performed in R. Cell populations were labeled as follows: HSC = CD34+CD38−CD90+/−CD49f+CD45RA−, multilymphoid progenitor = Lin−CD34+CD38−CD90−CD45RA+, common myeloid progenitor = CD34+ CD38+CD135+CD45RA−CD7−CD10−, granulocyte/monocyte progenitor = CD34+CD38+CD135+CD45RA+CD7−CD10−, megakaryocyte/erythrocyte progenitor = CD34+CD38+CD135−CD45RA−CD7−CD10−, colony-forming unit (CFU)-macrophage = CD34−CD33+CD13+, monocyte = FSChi SSClo CD14+ CD45dim, CFU-granulocyte = CD34−SSHhiCD45+CD11b−CD16−, neutrophil = FSChiSSChiCD16+CD11b+, CFU-megakaryocyte = CD34+CD41+CD61+CD45−, megakaryocyte = CD34−CD41+CD61+CD45−, CFU-erythroid = CD34−CD71+GlyA−, erythrocyte = CD34−CD71−GlyA+, NK cell/T cell progenitor = CD14−CD19-CD3+CD1d+, CD8 T = CD8+CD62L+/−CD45RA+/−, and CD4 T = CD4+CD62L+/−CD45RA+/−.
pSTAT3 analysis
CD34+ cells cultured for 8-12 days were starved of cytokines and DL1 overnight at low cell densities. Following starvation, cells were stimulated with soluble factors as indicated. For DL1 simulation, starved cells were transferred to plates precoated with DL1 (2.5 μg/mL) to initiate the stimulation period. After the commencement of stimulation, cells were fixed and permeabilized as previously described16 before being stained with phospho-signal transducer and activator of transcription (pSTAT)3 antibody (Cell Signaling Technology, Danvers, MA) and an AF-647 secondary antibody.
Secreted factor analysis
Conditioned media from cells cultured for 12 days in the indicated conditions were collected and frozen. Secreted factor concentrations were sampled in duplicate from thawed conditioned media aliquots using IL-6 and sIL-6R Quantikine ELISA Kits (R&D Systems), according to the manufacturer’s directions.
Limiting dilution transplantation studies
All animal studies were performed according to procedures approved by the appropriate animal ethics boards. Female nonobese diabetic/SCID/IL-2Rgc-null (NSG) mice were sublethally irradiated (250 rad) 24 hours before transplantation. Uncultured CD34+ enriched cells, or cells cultured for 12 days or 16 days, were transplanted via tail vein injection. Cohorts of mice were killed at 3, 9, 16, and 24 weeks after transplantation and bone marrow was collected. Cells were assessed by flow cytometry. Mice were scored positive for human repopulation if at least 0.5% of bone marrow cells were positive for both human CD45 and human HLA-ABC. All limiting dilution analysis and associated statistical analysis was performed with L-calc software (Stemcell Technologies).
Mathematical simulations
The model that we previously developed and described9,11 was used to run all culture simulations with MATLAB 2012 software (Mathworks, Natick, MA). For specific revisions to the model made for this study, see the Blood Web site for supplemental data.
Statistical analysis
Statistical significance was computed using Student t test and analysis of variance. All error bars represent the standard deviation of 3 or more biological replicates. Asterisks or number signs indicate statistical significance between indicated conditions (or if not indicated, between the test condition and control) of P < .05.
Results
DL1-mediated lineage skewing creates a blood progenitor cell–supportive environment
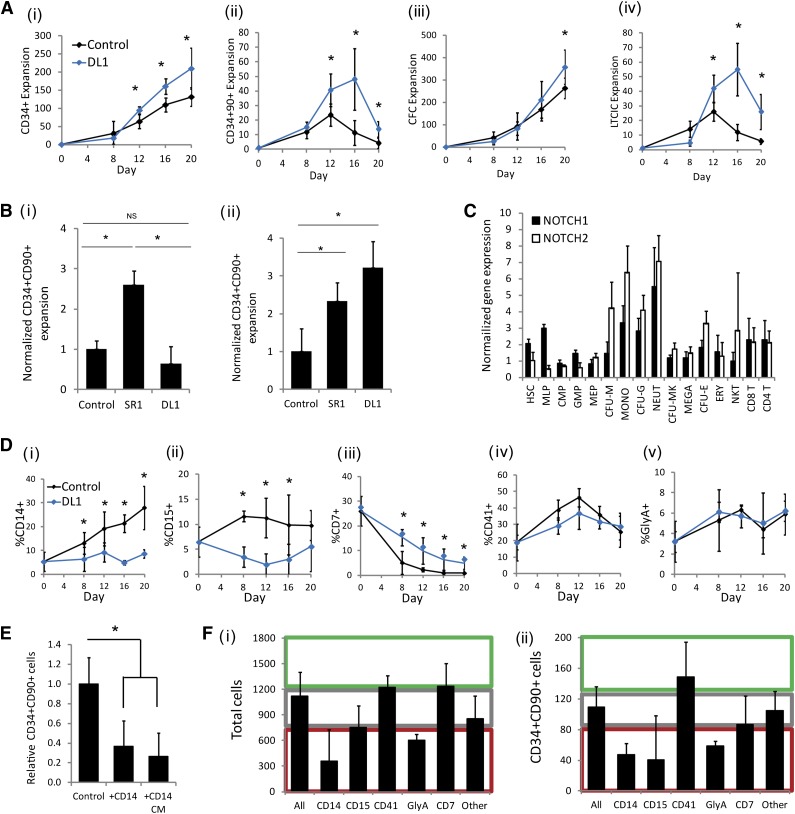
Immobilized DL1 was added to a fed-batch culture platform, which regulates endogenously produced soluble factors.9 The addition of DL1 caused a significant increase in primitive cells, as measured both by phenotype (CD34+ and CD34+CD90+) and by functional in vitro assays (CFC assay and LTC-IC assay) (Figure 1A). The greatest impact of the ligand was seen on the more primitive progenitor populations (CD34+CD90+ and LTC-IC). During the first several days of culture, we observed that DL1 caused a decrease in total cell number because of slower proliferation, as demonstrated by carboxy fluorescein succinimidyl ester staining (supplemental Figure 1). In this heterogeneous culture system, there was not an immediate enhancement in the frequency of progenitor cells as compared with the control; however, as the culture progressed, the anticipated DL1-mediated enhancement of primitive progenitors emerged. We have previously demonstrated that endogenously produced secreted factors can be dominant modulators of HSC fate.9,17 Thus, given the delayed emergence of progenitor expansion with DL1, we next examined whether DL1 was altering the endogenous microenvironment to indirectly regulate primitive cell fate.
Figure 1.
DL1 generates a microenvironment that is progenitor cell expansion supportive. (A) The addition of DL1 to the fed-batch culture system enhanced the expansion of (i) CD34+ cells; (ii) CD34+CD90+ cells; (iii) CFCs; (iv) LTC-ICs. (Bi) Addition of DL1 to HSC-enriched (Lin−RholoCD34+CD38−CD45RA−CD49f+) cells did not lead to a significant increase in the expansion of CD34+CD90+ cells after 7 days of culture. (Bii) In contrast, the addition of DL1 to CD34+ cells for 16 days significantly enhanced CD34+CD90+ expansion as compared with cells cultured without DL1. (C) Microarray analysis indicated that the Notch1 and Notch2 receptors are expressed at varying levels in many hematopoietic populations. (D) DL1 caused lineage skewing in hematopoietic cell culture, most notably a decrease in CD14+ cells and CD15+ cells, and an increase in CD7+ cells. (E) When CD14+ cells or their CM were cultured with Lin−RholoCD34+CD38−CD45RA−CD49f+ cells, there was an inhibitory effect on the proliferation of the CD34+CD90+ population. (F) The CM from the specified mature cell lineages (CD14+, CD15+, CD41+, GlyA+, CD7+, other = CD34−CD14−CD15−CD41−GyA−CD7−) was added to the Lin−RholoCD34+CD38−CD45RA−CD49f+ cells and cultured for 7 days. Different lineages had varying impacts on the expansion of (i) total cells and (ii) CD34+CD90+ cells. Effects were categorized as negative (red region), neutral (gray region), or positive (green region) as compared with CM from unsorted cell populations (all). All error bars indicate standard deviation. n ≥ 3, *P < .05. CFU-E, CFU-erythroid; CFU-M, CFU-macrophage; CFU-G, CFU-granulocyte; CFU-MK, CFU-megakaryocyte; CMP, common myeloid progenitor; ERY, erythrocyte; GMP, granulocyte/monocyte progenitor; MEGA, megakaryocyte; MEP, megakaryocyte/erythrocyte progenitor; MLP, multilymphoid progenitor; MONO, monocyte; NEUT, neutrophil.
To decouple the direct and indirect effects of DL1, we compared the observed impact of DL1 on CD34+ cells (∼0.1% of which are HSCs18) with its impact on purified primitive cells (Lin−RholoCD34+CD38−CD45RA−CD49f+, ∼20% HSCs13). When DL1 was added to the Lin−RholoCD34+CD38−CD45RA−CD49f+ cells, there was no significant increase in primitive cell (CD34+CD90+) expansion beyond what was seen with the cytokine control, and the expansion was lower than that demonstrated with the small molecule SR1, which directly targets primitive cells5 (Figure 1Bi). In contrast, DL1 added to the CD34+ input cell population gave a CD34+CD90+ cell expansion significantly higher than the control population after 16 days of culture (Figure 1Bii). These data suggest that DL1 is not exclusively targeting HSC self-renewal. Instead, we hypothesized that DL1 is acting (at least in part) non–cell-autonomously with respect to HSC fate.
To help determine which lineage populations may be receptive to DL1 and responsible for the indirect enhancement of primitive progenitor cells, we mined gene expression datasets of sorted hematopoietic populations14,15 to assess the expression levels of Notch1 and Notch2, the known receptors of DL1.19 Both of these receptors were expressed at varying levels on many hematopoietic cell populations with particular enrichment on myeloid progenitors (Figure 1Ci), suggesting lineage committed cell populations as targets of DL1. Protein expression analysis of Notch1 and Nocth2 also show variation in their expression among both primitive and lineage-restricted populations (supplemental Figure 2). It has previously been reported that DL1 changes mature lineage output.3,20 We predicted that this lineage skewing may be altering the cell environment and contributing to the effects seen on primitive progenitor expansion. The addition of DL1 to CD34+ cells led to a significant and sustained reduction in the production of CD14+ monocytes and CD15+ granulocytes and an increase in CD7+ lymphocytes, as has been previously reported (Figure 1D).3 The reduction of CD14+ and CD15+ cells seen with the addition of DL1 is particularly interesting because it has been shown that mature myeloid cells have a significant inhibitory effect on the expansion of progenitor populations.10,21
To assess the feedback signaling impact of specific mature cell populations on HSC cell fate, we mixed sorted mature cell populations or their conditioned media with Lin−RholoCD34+CD38−CD45RA−CD49f+ cells. Similar effects on primitive cells were seen with mature cell cocultures or with the corresponding conditioned media (CM), confirming that the effects were soluble factor–mediated (Figure 1E). CM from CD14+, CD15+, and GlyA+ cells caused a significant decrease in expansion of total cells and CD34+CD90+ cells, whereas the effect of CD7+ cells was neutral and CD41+ cells caused a trend toward increased primitive cell expansion (Figure 1F). These results confirmed the differential effects of mature lineage populations on primitive cell expansion and highlighted the inhibitory effect of CD14+ and CD15+ cells. We also confirmed that the expansions of primitive CD34+CD90+ cells were proportionally higher in cultures seeded with purified primitive cells as compared with those seeded with CD34+ cells where myeloid cell accumulation is rapid (supplemental Figure 3), providing further evidence toward the net inhibitory nature of mature myeloid cells. DL1 reduces the CD14+ and CD15+ cell populations that are most inhibitory toward progenitor growth. It is likely that this altered signaling environment contributed to the sustained expansion of the primitive cell populations in the presence of DL1. We next sought to identify the downstream targets through which DL1-mediated signaling was altering myeloid cell production.
DL1 activates JAK-STAT3 signaling but inhibits IL-6 cis-signaling
Given the inhibitory nature of CD14+ and CD15+ cells, we were particularly interested in understanding the mechanism by which DL1 inhibited their production. The production and differentiation of macrophages and granulocytes is known to be largely modulated by gp130 signaling and cytokine-mediated STAT3 activation.22-25 Furthermore, interactions between Notch signaling and STAT3 activation have been reported,26,27 leading us to hypothesize that the DL1-mediated effect on myeloid cell production acted through modulation of the JAK-STAT3 signaling pathway.
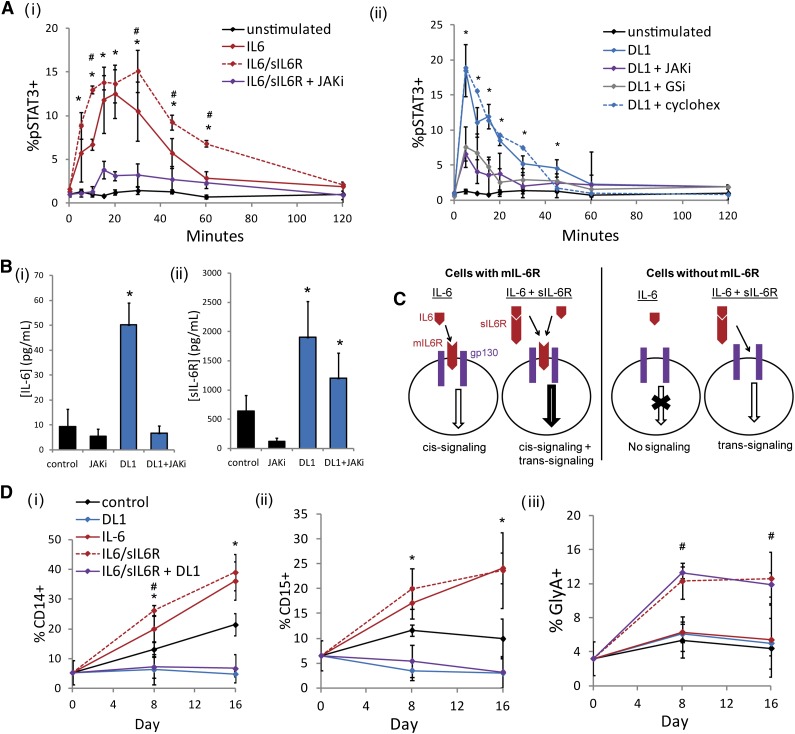
To investigate the impact of DL1 on STAT3, we measured the dynamic activation of STAT3 in response to different stimuli. As expected, stimulation of cultured CD34+ cells with IL-6 or both IL-6 and sIL-6R caused a significant and JAK-dependent upregulation of pSTAT3 (Figure 2Ai). When cultured CD34+ cells were stimulated with DL1, we saw a rapid activation of STAT3 that mimicked that seen with IL-6 and sIL-6R (Figure 2Aii). This activation could be prevented by either JAKi or γ-secretase inhibitor. However, the addition of cycloheximide did not impact the DL1-mediated activation of STAT3, which together with the rapid kinetics of the activation, suggested that it is not a transcriptionally regulated event. The JAK-STAT pathway is known to act in an autoregulatory manner, such that targets of STAT3 upregulate the further production of IL-6 and related ligands.28 Accordingly, we measured the levels of soluble IL-6 family members during culture with or without DL1 and observed that CD34+ cells cultured in the presence of DL1 produced significantly higher concentrations of both IL-6 and sIL-6R (Figure 2B). Notably, DL1-mediated IL-6 production was abrogated by the addition of a JAKi. Conversely, levels of sIL-6R were only somewhat dampened by a JAKi, suggesting that the DL1-mediated increases in sIL-6R were at least partially JAK-STAT3–independent.
Figure 2.
DL1 activates STAT3 similarly to IL-6 but impacts mature cells differently. (Ai) IL-6 stimulation activated STAT3 on cultured CD34+ cells. This effect was enhanced with the combination of IL-6 and sIL-6R and diminished by the addition of a JAKi. *Statistical comparison of IL-6 stimulation to unstimulated condition; #statistical comparison of IL-6/sIL-6R stimulation to IL-6 stimulation. (Aii) DL1 stimulation activated STAT3 on cultured CD34+ cells and was dependent on both JAK and γ-secretase (GSi) but was not impacted by cycloheximide. (B) DL1 upregulated the secretion of (i) IL-6 and (ii) sIL-6R. (C) Schematic of IL-6 cis-signaling and trans-signaling. IL-6 ligand can initiate JAK-STAT signaling in the absence or presence of sIL-6R on cell types that express mIL-6R (cis-signaling). On cells that do not express mIL-6R, IL-6–mediated signaling can occur only in the presence of sIL-6R (trans-signaling). (D) IL-6 increased CD14+ cells and CD15+ cells. The addition of sIL-6R additionally increased GlyA+ cells. The combination of IL-6/sIL-6R and DL1 inhibited the production of (i) CD14+ cells and (ii) CD15+ cells and maintained the production of (iii) GlyA+ cells. *Statistical comparison of IL-6 to control condition; #statistical comparison of IL-6/sIL-6R to IL-6 condition. All error bars indicate standard deviation. n ≥ 3, P < .05.
Our observation that DL1 was modulating IL-6 signaling led us to consider whether DL1 was also changing the population of cells that were responsive to IL-6. IL-6 can initiate JAK-STAT signaling in 1 of 2 ways (Figure 2C). IL-6 cis-signaling results when the IL-6 ligand binds to the membrane (m) IL-6R, which then associates with gp130. In hematopoiesis, mIL-6R and IL-6 cis-signaling are restricted to myeloid lineage cells,29 and we observed that the addition of IL-6 to CD34+ cells produced a significant and JAK-dependent increase in CD14+ and CD15+ cell production (Figure 2D; supplemental Figure 4A). In contrast, combining exogenous IL-6/sIL-6R with DL1 produced the same low levels of CD14+ and CD15+ cells that were generated with DL1 alone, indicating that DL1 is impeding IL-6 cis-signaling. In cells that do not express mIL-6R, IL-6 trans-signaling can occur when IL-6 binds to sIL-6R, and the soluble complex then binds to the ubiquitously expressed gp130 receptor. When both IL-6 and sIL-6R were added to culture, in addition to the increase in CD14+ and CD15+ cells, we also observed a trans-signaling–mediated increase in GlyA+ cells (Figure 2D) consistent with previous reports.30 DL1 in combination with IL-6/sIL-6R maintained GlyA+ levels, suggesting IL-6 trans-signaling is not affected by DL1.
The addition of IL-6 or IL-6/sIL-6R led to a significant increase in total cell expansion but a corresponding decrease in the frequency of CD34+ cells (supplemental Figure 4B). Notably, on the primitive Lin−RholoCD34+CD38−CD45RA−CD49f+ cell population, IL-6 or IL-6/sIL-6R enhanced the expansion of CD34+CD90+ cells (supplemental Figure 4C), but this effect was not evident in the bulk cultures, likely because of the high level of inhibitory factor–secreting mature cells in these cultures. Therefore, despite the similar propensities of DL1 and IL-6/sIL-6R to activate STAT3, these 2 stimuli produce very different effects on both mature cell and primitive cell populations. It was puzzling that both DL1 and IL-6/sIL-6R activated STAT3 but that the effects of the signaling resulted in completely different cell population dynamics. We hypothesized that the reduction of IL-6 cis-signaling was a result of the loss of mIL-6R. To test this hypothesis, we next measured the effect of DL1 on cell population–specific mIL-6R expression.
DL1 converts progenitors from IL-6 cis-signaling to trans-signaling through loss of mIL-6R
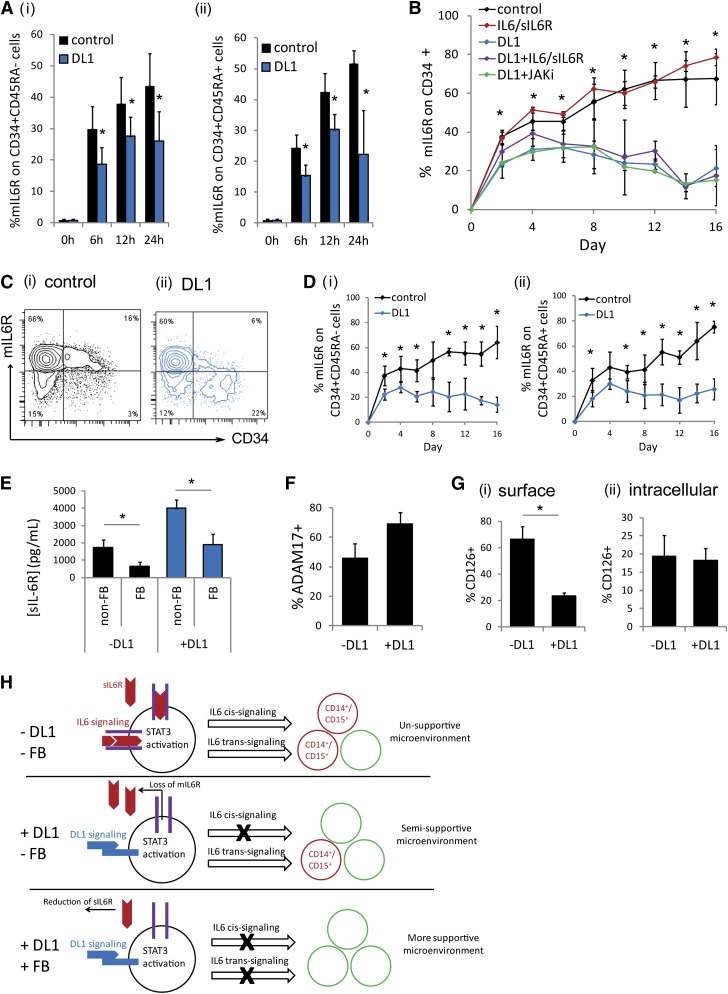
Freshly isolated CD34+ cells do not express mIL-6R; however, upon culture, mIL-6R expression was very quickly (within 6 hours) upregulated (Figure 3A). In the presence of DL1, mIL-6R upregulation was significantly inhibited. This inhibition occurred rapidly, before the first cell division, suggesting that this effect is signaling mediated, as opposed to because of the emergence of a new cell population or changes in cell population composition. The rapid DL1-mediated mIL-6R inhibition was seen on both the more primitive CD34+CD45RA− cell population and the more committed CD34+CD45RA+ cell population (Figure 3Ai-ii). A similar trend was seen on the highly purified CD34+CD38−CD45RA−CD90+ population (supplemental Figure 5A). Throughout culture, CD34+ cells maintained mIL-6R at a significantly lower frequency in the presence of DL1 (Figure 3B-C). This was true on both the CD34+CD45RA− subset and the CD34+CD45RA+ subset (Figure 3D). The addition of either exogenous IL-6/sIL-6R or a JAK inhibitor did not significantly alter the levels of mIL-6R seen with or without DL1 on the CD34+ cells, demonstrating that the inhibition of mIL-6R expression was occurring via a JAK-STAT3–independent mechanism (Figure 3B), consistent with the JAK-STAT3–independent sIL-6R production observed in DL1 culture (Figure 2Bii). Conversely, the DL1-mediated effect was significantly diminished in the presence of the γ-secretase inhibitor (supplemental Figure 5B).
Figure 3.
DL1 causes the loss of mIL-6R, inhibiting IL-6 cis-signaling. (A) DL1 inhibited the rapid upregulation of mIL-6R on (i) CD34+CD45RA− cells and (ii) CD34+CD45RA+ cells on culture of CD34+ cells. (B) DL1 significantly decreased mIL-6R on CD34+ cells throughout culture. The effect was JAK-independent. *Statistical comparison of DL1 to control condition (C) Representative flow cytometry plots at day 16 (i) without DL1 and (ii) with DL1. (D) The impact of DL1 on mIL-6R was similar on (i) CD34+CD45RA− and (ii) CD34+CD45RA+ cells. (E) Fed-batch (FB) conditions reduced sIL-6R production as compared with non–FB conditions. (F) Protein expression of ADAM17 measured by flow cytometry on cells cultured in the absence or presence of DL1 for 10-12 days. (G) Comparison of (i) surface expression of mIL-6R (CD126) and (ii) intracellular expression of mIL-6R on CD34+ cells cultured in the absence or presence of DL1 for 8-12 days. (H) Schematic of proposed mechanism. DL1 activates STAT3 and causes the loss of mIL-6R. The loss of mIL-6R reduces IL-6 cis-signaling and thus reduces the production of CD14+ and CD15+. Instead, only IL-6 trans-signaling can occur, which enhances the production of other cell types, and results in a microenvironment that is more supportive for hematopoietic stem and progenitor cells. The FB conditions further reduce IL-6 trans-signaling. All error bars indicate standard deviation. n ≥ 3, P < .05.
Studies have shown that CD34+mIL-6R+ cells are myeloid-restricted progenitors.20 Therefore, the reduction of mIL-6R on these cells prevents IL-6 cis-signaling–mediated production of mature monocytes and granulocytes. In the absence of mIL-6R, myeloid progenitor cells can still respond to IL-6 via IL-6 trans-signaling in the presence of sIL-6R. However, our use of the fed-batch culture platform, which allowed for the continuous reduction of the impact of endogenously produced soluble signaling factors, allowed us to decouple these aspects by reducing sIL-6R levels (Figure 3E) and thus enhance the impact of DL1-mediated mIL-6R inhibition. It is likely that DL1 causes the ADAM17-mediated shedding of mIL-6R because the metalloprotease ADAM17 is known to facilitate mIL-6R shedding31 and is also responsible for the cleavage of the extracellular domain of Notch receptors to activate Notch signaling.32 Cell-surface ADAM17 is upregulated on cells cultured in the presence of DL1 (Figure 3F). Moreover, intracellular accumulation of mIL-6R is not affected by DL1, providing further evidence that the reduction in mIL-6R is due to receptor shedding and not internalization (Figure 3G).
Taken together, these data indicate that both DL1 and IL-6/sIL-6R activate the JAK-STAT3 pathway. Activation through IL-6/sIL-6R leads primarily to IL-6 cis-signaling, which enhances production of CD14+ and CD15+ cells and results in the generation of a nonsupportive microenvironment with high levels of feedback inhibition. Conversely, DL1 supplementation causes the JAK-STAT3–independent reduction of mIL-6R expression, which inhibits IL-6 cis-signaling. As a result, DL1-mediated STAT3 activation results only in IL-6 trans-signaling, which can be minimized through environmental regulation of sIL-6R with the fed-batch culture system. Together, this results in a decrease in CD14+ and CD15+ cell production that generates a more supportive environment for progenitor cell expansion. A summary model of the system is shown in Figure 3H. Given the degree of environmental control that DL1 generates in combination with the fed-batch platform, we further hypothesized that this system would support the production of both short- and long-term HSCs.
Concurrent modulation of IL-6 cis- and trans-signaling supports the expansion of short- and long-term SRCs
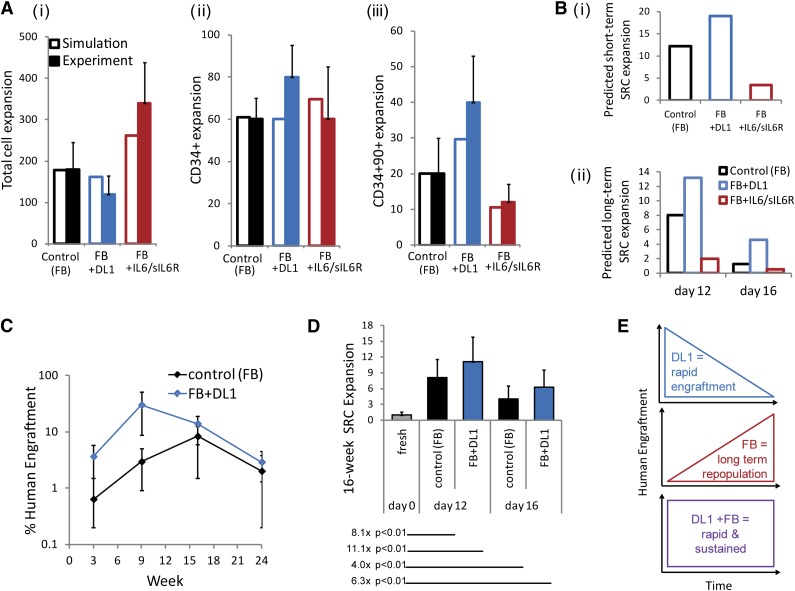
To predict the impact of DL1 addition on HSC fate control, we first used a previously described mathematical model.9,11 The model inputs were revised based on DL1-specific data (details of the mathematical model are described in the supplemental data). Figure 4A shows that the predicted population outputs were comparable to experimental controls, demonstrating the utility of the model in predicting outputs from DL1 or IL-6/sIL-6R conditions. One prediction that emerged from the model simulations was that the combination of the fed-batch system and DL1 would lead to significant expansions of both short- and long-term repopulating HSCs, as measured by SRCs (Figure 4B). The expansion of primitive progenitors and short-term repopulating HSCs by DL1 has been previously demonstrated by quantification of human engraftment at 3 and 9 weeks posttransplantation.4 This study did not see a significant expansion of the longer term repopulating HSCs with DL1. However, the fed-batch system reduces IL-6 trans-signaling and produces a different microenvironmental context onto which DL1-induced Notch signaling is added.
Figure 4.
The impact on short- and long-term repopulating HSCs by feedback-mediated DL1 is predicted and validated. (A) Comparison of day 12 model predictions and experimental expansion outputs from the fed-batch control culture, the DL1 culture, and the IL-6/sIL-6R culture for (i) total cell expansion; (ii) CD34+ cell expansion; and (iii) CD34+CD90+ cell expansion. (Bi) Model predictions of day 12 short-term SRC expansions under each culture condition. (Bii) Model predictions of long-term SRC expansions after 12 and 16 days of culture. (C) Time course of human engraftment after transplantation of day 12–cultured cells into NSG mice. Bone marrow was assessed at 3, 9, 16, and 24 weeks following transplantation with 500 000 cells cultured in the fed-batch control or the fed-batch plus DL1 culture conditions. (D) Quantification of week 16 SRC expansion, based on limiting dilution analysis performed 16 weeks posttransplantation. Cells were cultured for 12 or 16 days in the fed-batch system with or without DL1 before transplantation. Expansions indicated compared cells cultured in vitro with transplantation of the uncultured CD34+ cells. (E) Schematic of the combined effect of DL1 and the fed-batch platform on both rapid and sustained human engraftment.
To experimentally assess the expansion of the SRC populations, we transplanted cells expanded with or without DL1 into NSG mice and quantified human cell repopulation in the bone marrow intermittently for 24 weeks. We confirmed that DL1 showed a trend toward enhancement of human engraftment at 3 and 9 weeks posttransplantation (Figure 4C). We then assessed longer term SRC expansion by limiting dilution analysis at 16 weeks posttransplantation (Figure 4D, supplemental Table 2). The combination of fed-batch and DL1 led to a significant expansion of SRCs as compared with the uncultured CD34+ cells. Thus, the environmental regulation gained with the combination of DL1 and the fed-batch–feeding strategy allowed for both rapid and sustained human engraftment, as illustrated schematically in Figure 4E. Taken together, the findings in this study demonstrate a novel mechanism for signaling rewiring through the conversion of IL-6 cis-signaling to IL-6 trans-signaling, thereby altering the cell populations that are responsive the JAK-STAT pathway. This regulation generates a supportive soluble factor microenvironment that enables enhanced expansion of stem and progenitor cells.
Discussion
Notch signaling plays many important roles in human hematopoiesis, including the in vitro enhancement of primitive cell expansion, which can be achieved through presentation of immobilized DL1 ligand at low concentrations. However, the mechanisms through which this effect is achieved and why it had been limited to short-term SRCs were not fully understood. Here, we demonstrate a non–stem cell autonomous role whereby DL1 acts to alter cell-specific responsiveness to IL-6 signaling, subsequently reducing inhibitory myeloid cell production and unmasking a signaling environment supportive of HSC expansion.
Despite similar STAT3 activation by DL1 and IL-6/sIL-6R, the resulting impact of these 2 stimuli on mature cell populations was very different. We hypothesized that this difference was due to the modulation of IL-6 signaling at the level of mIL-6R and showed that the addition of DL1 causes the loss of mIL-6R from CD34+ myeloid progenitor cells, thus preventing IL-6 cis-signaling. Our data indicate that DL1 signaling is altering mIL-6R receptor expression in individual cells; however, we cannot formally rule out mIL-6R differences resulting from changes in relative numbers of highly enriched stem cell populations.
It is likely that the Notch pathway impacts lineage differentiation via a number of different mechanisms, including the alteration of CEBPA expression on primitive murine myeloid cells33 and the maintenance of T-cell potential in early progenitors.34 This study demonstrates a novel mechanism of myeloid cell inhibition and identifies a new level of control of IL-6-JAK-STAT signaling through the DL1-mediated conversion of cis-signaling to trans-signaling, which allows for different cell populations to respond to IL-6 proliferation signals. Furthermore, with the knowledge gained here, we have been able to demonstrate further enhancement in the generation of progenitors as well as short- and long-term SRCs in a clinically relevant cell culture system.
Supplementary Material
Acknowledgments
The authors thank the donors and the Research Centre for Women’s and Infants’ Health BioBank of Mount Sinai Hospital for the human specimens used in this study, and the members of the P.W.Z. laboratory for their helpful discussion.
This work was funded by the Canadian Stem Cell Network, the Leukemia and Lymphoma Society, the Canadian Institutes of Health Research, The National Heart, Lung, and Blood Institute (grant U01HL100395), an NSERC Graduate Scholarship (E.C.), and an Ontario Stem Cell Initiative Post-doc Fellowship (W.W.). P.W.Z. is the Canada Research Chair in Stem Cell Bioengineering.
Footnotes
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: E.C. undertook the conception and design, collection and assembly of data, data analysis and interpretation, and manuscript writing; W.W. undertook the conception and design and collection of data; T.U. provided technical assistance with in vivo studies; W.Q. provided gene expression analysis; C.D. provided technical advice and reviewed the manuscript; I.D.B. provided study material and technical advice and reviewed manuscript; and P.W.Z. undertook the conception and design, data interpretation, and manuscript writing.
Conflict-of-interest disclosure: The Fred Hutchinson Cancer Research Center holds a patent on “methods for immortalizing cells” that covers the use of Notch ligand for expansion of HSCs. I.D.B. is an inventor on this patent. The remaining authors declare no competing financial interests.
Correspondence: Peter W. Zandstra, Terrence Donnelly Centre for Cellular and Biomolecular Research, Institute for Biomaterials and Biomedical Engineering, 160 College St, Rm 1116, Toronto, ON, Canada M5S 3E1; e-mail: peter.zandstra@utoronto.ca.
References
- 1.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7(9):678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 2.Weber JM, Calvi LM. Notch signaling and the bone marrow hematopoietic stem cell niche. Bone. 2010;46(2):281–285. doi: 10.1016/j.bone.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delaney C, Varnum-Finney B, Aoyama K, Brashem-Stein C, Bernstein ID. Dose-dependent effects of the Notch ligand Delta1 on ex vivo differentiation and in vivo marrow repopulating ability of cord blood cells. Blood. 2005;106(8):2693–2699. doi: 10.1182/blood-2005-03-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delaney C, Heimfeld S, Brashem-Stein C, Voorhies H, Manger RL, Bernstein ID. Notch-mediated expansion of human cord blood progenitor cells capable of rapid myeloid reconstitution. Nat Med. 2010;16(2):232–236. doi: 10.1038/nm.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boitano AE, Wang J, Romeo R, et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. 2010;329(5997):1345–1348. doi: 10.1126/science.1191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amsellem S, Pflumio F, Bardinet D, et al. Ex vivo expansion of human hematopoietic stem cells by direct delivery of the HOXB4 homeoprotein. Nat Med. 2003;9(11):1423–1427. doi: 10.1038/nm953. [DOI] [PubMed] [Google Scholar]
- 7.Peled T, Landau E, Mandel J, et al. Linear polyamine copper chelator tetraethylenepentamine augments long-term ex vivo expansion of cord blood-derived CD34+ cells and increases their engraftment potential in NOD/SCID mice. Exp Hematol. 2004;32(6):547–555. doi: 10.1016/j.exphem.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Zhang CC, Kaba M, Iizuka S, Huynh H, Lodish HF. Angiopoietin-like 5 and IGFBP2 stimulate ex vivo expansion of human cord blood hematopoietic stem cells as assayed by NOD/SCID transplantation. Blood. 2008;111(7):3415–3423. doi: 10.1182/blood-2007-11-122119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Csaszar E, Kirouac DC, Yu M, et al. Rapid expansion of human hematopoietic stem cells by automated control of inhibitory feedback signaling. Cell Stem Cell. 2012;10(2):218–229. doi: 10.1016/j.stem.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Kirouac DC, Ito C, Csaszar E, et al. Dynamic interaction networks in a hierarchically organized tissue. Mol Syst Biol. 2010;6(417):417. doi: 10.1038/msb.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirouac DC, Madlambayan GJ, Yu M, Sykes EA, Ito C, Zandstra PW. Cell-cell interaction networks regulate blood stem and progenitor cell fate. Mol Syst Biol. 2009;5(293):293. doi: 10.1038/msb.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varnum-Finney B, Wu L, Yu M, et al. Immobilization of Notch ligand, Delta-1, is required for induction of notch signaling. J Cell Sci. 2000;113(Pt 23):4313–4318. doi: 10.1242/jcs.113.23.4313. [DOI] [PubMed] [Google Scholar]
- 13.Notta F, Doulatov S, Laurenti E, Poeppl A, Jurisica I, Dick JE. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science. 2011;333(6039):218–221. doi: 10.1126/science.1201219. [DOI] [PubMed] [Google Scholar]
- 14.Laurenti E, Doulatov S, Zandi S, et al. Transcriptional architecture of early human hematopoiesis reveals novel regulators governing lymphoid commitment. Nat Immunol. 2013;14(7):756–763. doi: 10.1038/ni.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Novershtern N, Subramanian A, Lawton LN, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144(2):296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang W, Akbarian V, Audet J. Biochemical measurements on single erythroid progenitor cells shed light on the combinatorial regulation of red blood cell production. Mol Biosyst. 2013;9(2):234–245. doi: 10.1039/c2mb25348h. [DOI] [PubMed] [Google Scholar]
- 17.Madlambayan GJ, Rogers I, Kirouac DC, et al. Dynamic changes in cellular and microenvironmental composition can be controlled to elicit in vitro human hematopoietic stem cell expansion. Exp Hematol. 2005;33(10):1229–1239. doi: 10.1016/j.exphem.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 18.McDermott SP, Eppert K, Lechman ER, Doedens M, Dick JE. Comparison of human cord blood engraftment between immunocompromised mouse strains. Blood. 2010;116(2):193–200. doi: 10.1182/blood-2010-02-271841. [DOI] [PubMed] [Google Scholar]
- 19.Dahlberg A, Delaney C, Bernstein ID. Ex vivo expansion of human hematopoietic stem and progenitor cells. Blood. 2011;117(23):6083–6090. doi: 10.1182/blood-2011-01-283606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamamura K, Ohishi K, Katayama N, et al. Notch ligand Delta-1 differentially modulates the effects of gp130 activation on interleukin-6 receptor alpha-positive and -negative human hematopoietic progenitors. Cancer Sci. 2007;98(10):1597–1603. doi: 10.1111/j.1349-7006.2007.00566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang H, Robinson SN, Lu J, et al. CD3(+) and/or CD14(+) depletion from cord blood mononuclear cells before ex vivo expansion culture improves total nucleated cell and CD34(+) cell yields. Bone Marrow Transplant. 2010;45(6):1000–1007. doi: 10.1038/bmt.2009.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minami M, Inoue M, Wei S, et al. STAT3 activation is a critical step in gp130-mediated terminal differentiation and growth arrest of a myeloid cell line. Proc Natl Acad Sci USA. 1996;93(9):3963–3966. doi: 10.1073/pnas.93.9.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenkins BJ, Grail D, Inglese M, et al. Imbalanced gp130-dependent signaling in macrophages alters macrophage colony-stimulating factor responsiveness via regulation of c-fms expression. Mol Cell Biol. 2004;24(4):1453–1463. doi: 10.1128/MCB.24.4.1453-1463.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Metcalf D. Murine hematopoietic stem cells committed to macrophage/dendritic cell formation: stimulation by Flk2-ligand with enhancement by regulators using the gp130 receptor chain. Proc Natl Acad Sci USA. 1997;94(21):11552–11556. doi: 10.1073/pnas.94.21.11552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H, Nguyen-Jackson H, Panopoulos AD, Li HS, Murray PJ, Watowich SS. STAT3 controls myeloid progenitor growth during emergency granulopoiesis. Blood. 2010;116(14):2462–2471. doi: 10.1182/blood-2009-12-259630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamakura S, Oishi K, Yoshimatsu T, Nakafuku M, Masuyama N, Gotoh Y. Hes binding to STAT3 mediates crosstalk between Notch and JAK-STAT signalling. Nat Cell Biol. 2004;6(6):547–554. doi: 10.1038/ncb1138. [DOI] [PubMed] [Google Scholar]
- 27.Choi B, Chun E, Kim SY, et al. Notch-induced hIL-6 production facilitates the maintenance of self-renewal of hCD34+ cord blood cells through the activation of Jak-PI3K-STAT3 pathway. Am J Pathol. 2012;180(1):351–364. doi: 10.1016/j.ajpath.2011.09.030. [DOI] [PubMed] [Google Scholar]
- 28.Ichiba M, Nakajima K, Yamanaka Y, Kiuchi N, Hirano T. Autoregulation of the Stat3 gene through cooperation with a cAMP-responsive element-binding protein. J Biol Chem. 1998;273(11):6132–6138. doi: 10.1074/jbc.273.11.6132. [DOI] [PubMed] [Google Scholar]
- 29.Tajima S, Tsuji K, Ebihara Y, et al. Analysis of interleukin 6 receptor and gp130 expressions and proliferative capability of human CD34+ cells. J Exp Med. 1996;184(4):1357–1364. doi: 10.1084/jem.184.4.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu Z, Liu W, Liu D, Fan L. The regulatory role of Hyper-IL-6 in the differentiation of myeloid and erythroid progenitors derived from human cord blood. Cell Immunol. 2006;241(1):32–37. doi: 10.1016/j.cellimm.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 31.Garbers C, Jänner N, Chalaris A, et al. Species specificity of ADAM10 and ADAM17 proteins in interleukin-6 (IL-6) trans-signaling and novel role of ADAM10 in inducible IL-6 receptor shedding. J Biol Chem. 2011;286(17):14804–14811. doi: 10.1074/jbc.M111.229393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian L, Wu X, Chi C, Han M, Xu T, Zhuang Y. ADAM10 is essential for proteolytic activation of Notch during thymocyte development. Int Immunol. 2008;20(9):1181–1187. doi: 10.1093/intimm/dxn076. [DOI] [PubMed] [Google Scholar]
- 33.Varnum-Finney B, Halasz LM, Sun M, Gridley T, Radtke F, Bernstein ID. Notch2 governs the rate of generation of mouse long- and short-term repopulating stem cells. J Clin Invest. 2011;121(3):1207–1216. doi: 10.1172/JCI43868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanchez-Dominguez R, Pereira-Mendez S, Gomez A, et al. Notch signals contribute to preserve the multipotentiality of human CD34(+)CD38(-)CD45RA(-)CD90(+) hematopoietic progenitors by maintaining T cell lineage differentiation potential. Exp Hematol. 2012;40(12):983-993.e4. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.