Abstract
In Agrobacterium tumefaciens, horizontal transfer and vegetative replication of oncogenic Ti plasmids involve a cell-to-cell communication process called quorum-sensing (QS). The determinants of the QS-system belong to the LuxR/LuxI class. The LuxI-like protein TraI synthesizes N-acyl-homoserine lactone molecules which act as diffusible QS-signals. Beyond a threshold concentration, these molecules bind and activate the LuxR-like transcriptional regulator TraR, thereby initiating the QS-regulatory pathway. For the last 20 years, A. tumefaciens has stood as a prominent model in the understanding of the LuxR/LuxI type of QS systems. A number of studies also unveiled features which are unique to A. tumefaciens QS, some of them being directly related to the phytopathogenic lifestyle of the bacteria. In this review, we will present the current knowledge of QS in A. tumefaciens at both the genetic and molecular levels. We will also describe how interactions with plant host modulate the QS pathway of A. tumefaciens, and discuss what could be the advantages for the agrobacteria to use such a tightly regulated QS-system to disseminate the Ti plasmids.
Keywords: quorum-sensing, opines, conjugation, genetic, plant host, quorum-quenching, gene expression regulation
INTRODUCTION
In its canonical definition, quorum-sensing (QS) refers to a process through which a bacterial population is able to monitor its cell density and accordingly to mount coordinate responses (Fuqua et al., 1994). This phenomenon relies on the synthesis, diffusion, and perception of small signal molecules (autoinducers) that allow bacteria to communicate with each other and to regulate gene expression. In the last 40 years, a number of studies have established that QS is widespread in the bacterial kingdom although the nature of the signal molecules and/or signaling networks as well as the functions regulated by QS may vary considerably depending on the species (Miller and Bassler, 2001; Frederix and Downie, 2011; Stevens et al., 2012; Pereira et al., 2013).
In Proteobacteria, the typical QS model is epitomized by the LuxI/LuxR bioluminescence system of Vibrio fischeri that was described as early as 1970 (Nealson et al., 1970; Eberhard, 1972). In summary, LuxI catalyzes the synthesis of an N-acyl-homoserine lactone, namely the 3-oxo-hexanoyl-homoserine lactone (3OC6HSL), that acts as an autoinducer and accumulates in a cell density-dependent manner. At a threshold concentration, the 3OC6HSL molecules bind to their ligands, the transcriptional factor LuxR, and the newly formed LuxR dimers induce the expression of the lux operon which includes the genes responsible for bioluminescence but also luxI. This last autoregulatory action results in an exponential increase of the production of autoinducers and accounts for the characteristic pattern of QS-dependent bioluminescence in V. fischeri populations which rapidly shift at the quorum concentration from an “off” state to an “on” state.
Interestingly many homologs of LuxI and LuxR proteins have been found in other bacterial species such as Pseudomonas aeruginosa, Pectobacterium atrosepticum, and Agrobacterium tumefaciens (Fuqua et al., 1994, 1996). The first milestone in the study of A. tumefaciens QS was the functional characterization of the TraR protein, the LuxR homolog (Piper et al., 1993; Zhang et al., 1993). This seminal finding opened a new area of research in horizontal transfer of virulence Ti plasmids in A. tumefaciens that made this phytopathogenic species a leading model for the investigation of LuxI/LuxR QS systems. In this review, we will recap the most striking results obtained in deciphering the genetic network as well as the molecular basis of A. tumefaciens QS. We will also present how this QS system, consistent with the phytopathogenic lifestyle of A. tumefaciens, is integrated into an exquisite regulatory process, including various opine-induced regulons and lactonase activities. Finally we will discuss the biological/evolutionary relevance of this complex network in terms of dissemination of Ti plasmid genes in the plant tumor environment.
OVERVIEW OF A. tumefaciens QS
A LuxI/LuxR TYPE QS INTEGRATING AN ANTAGONIST COMPONENT
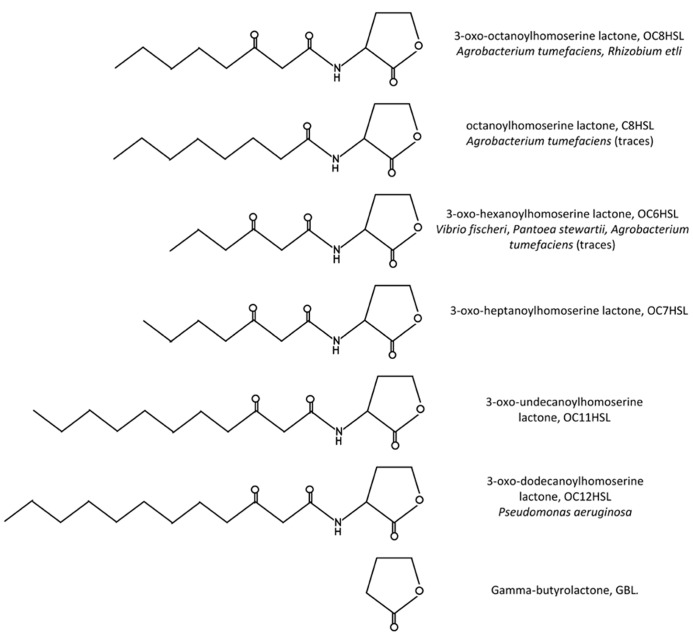
The first insight of a QS system in A. tumefaciens was gained with the functional characterization of a traR gene, homologous to V. fischeri luxR, the product of which acted as a transcriptional activator in the presence of a co-inducer. Actually two versions of the traR gene were found almost concomitantly in nopaline- and octopine-type Ti plasmids (Piper et al., 1993; Fuqua and Winans, 1994). These genes displayed high homology between them but were located in dissimilar regions of the two Ti plasmids, the expression of each of these regions being controlled by specific opines. Along with these discoveries, the chemical structure of the co-inducer required for TraR activity was determined by spectrometry analysis as 3-oxo-octanoyl-homoserine lactone (OC8HSL, see structure in Figure 1; Zhang et al., 1993). Soon afterward the gene traI, for which very closely related sequences also exist in nopaline- and octopine-type Ti plasmids, was shown to be responsible for OC8HSL synthesis (Hwang et al., 1994).
FIGURE 1.
Structures of the QS signals mentioned in this review. In parallel are indicated the full name of the molecule, its abbreviation as well as some bacterial species known to produce it.
Like other LuxI/LuxR type QS systems, A. tumefaciens QS comprises another component that negatively modulates the activity of TraR and OC8HSL and this component is the Ti plasmid-encoded protein TraM which can suppress TraR transcriptional activity. Versions of the traM gene were identified in both nopaline- and octopine-type Ti-plasmids (Fuqua et al., 1995; Hwang et al., 1995). The octopine-type Ti plasmid A6 even possesses a second functional traM gene borne on a chromosome, surely as a result of gene duplication (Wang et al., 2006a). For long it has been thought that TraM proteins were not related to any other proteins found in the databases, but recent characterization of the Pseudomonas aeruginosa QslA protein contradicted this view (Seet and Zhang, 2011), suggesting that TraM-type functions might be relatively common in bacteria.
At a mechanistic level, yeast two-hybrid assays revealed that TraM and TraR could directly interact. From these data it was deduced that the association between the two proteins was responsible for the inhibition of TraR-mediated responses by preventing proper TraR binding to DNA (Hwang et al., 1999). Two subsequent findings strengthened the negative regulatory functions exerted by TraM on QS. First it was established that this protein could block TraR activity even after the transcription factor has bound to DNA (Luo et al., 2000) and second TraM was demonstrated to promote TraR proteolysis (Costa et al., 2012).
The implications of TraM action for the dynamics of the QS system will be discussed in the following section.
QS-REGULATED GENES ARE INVOLVED IN FEEDBACK CONTROL AND Ti PLASMID DISSEMINATION
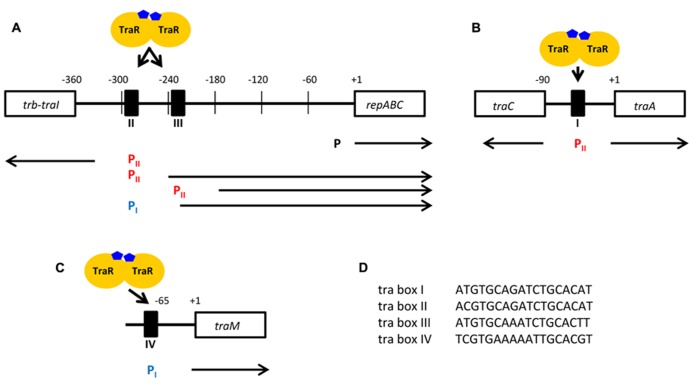
Chronologically the first TraR-regulated, hence QS-regulated, genes were the OC8HSL synthesis traI gene and the tra genes involved in conjugation of the Ti plasmid (Piper et al., 1993; Fuqua and Winans, 1994; Hwang et al., 1994). Next, were the regulatory gene traM (Fuqua et al., 1995; Hwang et al., 1995) and finally the rep genes required for vegetative replication of the Ti plasmid (Li and Farrand, 2000). Concomitantly, four 18 bp-inverted repeat operator sequences (called tra box I, II, III, and IV), the disruption of which abolished the TraR transactivation, were found in the promoter regions of the QS-regulated genes. These promoters were assigned to two distinct classes (class I-type and class II-type) according to the position of the tra boxes relatively to the transcription initiation site. In promoters of class I-type, the tra box is located approximately 65 nucleotides upstream of the transcription start site and in promoters of class II-type, the tra box is located about 45 nucleotides upstream of the transcription start site, partially overlapping with the -35 element of the promoter (Figure 2; Fuqua and Winans, 1996a). The traR gene has also been reported as being self-regulated though no tra box was detected in its promoter region (Fuqua and Winans, 1996b).
FIGURE 2.
Promoter architecture of the TraR-regulated genes in A. tumefaciens. (A–C) Representation of the regions upstream of the traI-trbBCDEJKLFGHI and repABC operons (A), traCDGyci and traAFBH operons (B) traM gene (C). The tra boxes (I, II, III, and IV) are indicated by black boxes. Under each tra box are presented the associated promoters, the activations of which are dependent on the binding of TraR. The promoters of class I-type are in blue while those of class II-type are in red. The fourth identified promoter controlling the expression of repABC in a TraR-independent way is also displayed (P). The arrows indicate the direction of transcription. (D) The nucleotide sequences of the four tra boxes. (Adapted from Fuqua and Winans, 1996a; Pappas and Winans, 2003a,b; White and Winans, 2005).
In line with the above studies, an extensive survey of QS-regulated genes has been recently carried out both in nopaline- and octopine-type Ti plasmids, using gene arrays and a TraR-overexpressing system (Cho and Winans, 2007). The results globally confirmed the previous data. Only genes located in the Ti plasmids were affected. In nopaline-type Ti plasmid, 31 genes were up-regulated in response to TraR overexpression and 25 in octopine-type Ti plasmid. Among the up-regulated genes common to the two plasmids, were the tra, rep, and traM genes. Moreover the operon structures, the presence of tra boxes in the promoter regions and the overall regulation of the expression of these genes were well conserved within the two plasmids.
Table 1 summarizes the identities and functions of the A. tumefaciens QS-regulated genes which are detailed in the following. The traCDGyci and traAFBH operons are divergently transcribed from a single class II-type promoter activated by a tra box I. These genes code for a DNA transfer and replication machinery involved in the conjugative processing of the Ti plasmid (Farrand et al., 1996; Cook et al., 1997; Cho and Winans, 2007). The proteins TraA, TraC, and TraD are notably thought to form a relaxosome at the oriT of the Ti plasmid which can also repress the expressions of both traCDGyci and traAFBH operons (Cho and Winans, 2007). The promoter of traI-trbBCDEJKLFGHI operon belongs to the class II-type of QS-regulated promoter but is characterized by the presence of a tra box II. The trb genes encode a mating pair formation system for the transfer of the Ti plasmid which is related to type IV secretion systems (Li et al., 1998). Among the proteins encoded by these genes, TrbJ and TrbK also act synergistically to implement an entry exclusion mechanism which ensures that conjugation events cannot occur between donor and recipient A. tumefaciens cells harboring similar Ti plasmids (Cho et al., 2009). In agreement with the gene functions, TraR-mediated up-regulation of the three traCDGyci, traAFBH and traI-trbCDEJKLFGHI operons results in induction of Ti plasmid conjugation. On the other hand the control of traI expression by TraR leads to a positive feedback loop which amplifies, through increase in OC8HSL production, the QS responses of A. tumefaciens (Fuqua and Winans, 1994; Hwang et al., 1994). As an illustration of this effect, exogenous supply of OC8HSL to A. tumefaciens cells accelerated the TraR-mediated induction of Ti plasmid conjugation (Fuqua and Winans, 1996a).
Table 1.
List of QS-regulated genes in nopaline- and octopine-type Ti plasmids (adapted from Cho and Winans, 2007).
| Gene name | Function | atu code |
|---|---|---|
| traC | Conjugal transfer protein | atu6126 |
| traD | Conjugal transfer protein | atu6125 |
| traG | Conjugal transfer protein | atu6124 |
| yci | Nuclease | atu6122 |
| traA | Conjugal transfer protein | atu6127 |
| traF | Conjugal transfer protein | atu6128 |
| traB | Conjugal transfer protein | atu6129 |
| traH | Conjugal transfer protein | atu6130 |
| traI | Acyl-homoserine-lactone synthase | atu6042 |
| trbB | Conjugal transfer protein | atu6041 |
| trbC | Conjugal transfer protein | atu6040 |
| trbD | Conjugal transfer protein | atu6039 |
| trbE | Conjugal transfer protein | atu6038 |
| trbJ | Conjugal transfer protein | atu6037 |
| trbK | Entry-exclusion protein | atu6036 |
| trbL | Conjugal transfer protein | atu6035 |
| trbF | Conjugal transfer protein | atu6034 |
| trbG | Conjugal transfer protein | atu6033 |
| trbH | Conjugal transfer protein | atu6032 |
| trbI | Conjugal transfer protein | atu6031 |
| traM | Transcriptional anti-activator | atu6131 |
| repA | Plasmid-partitioning protein | atu6043 |
| repB | Plasmid-partitioning protein | atu6044 |
| repC | Replication initiation protein | atu6045 |
Curiously the traM gene coding for the TraR antiactivator appears also to be up-regulated by TraR (Hwang et al., 1995). It was proposed that this regulatory mechanism allows the cells to produce TraM proteins at levels sufficient to inhibit the available TraR under conditions of basal-level expression. Later on, when the expression of traR is induced, the resulting increased levels of TraR protein would overcome the available TraM, thence triggering the QS response. This model actually highlights the importance of relative TraR and TraM protein levels in QS regulation and suggests that TraM significantly contributes to the quorum-dependent dimension of the system by delaying the moment when TraR is able to transactivate target genes (Su et al., 2008). Consistently, a traM defective strain was shown to be QS active in a cell density-independent manner (Piper and Farrand, 2000). Furthermore, a mathematical approach claimed that TraM was necessary for the existence of the A. tumefaciens QS “off” state (Goryachev et al., 2005). Another implication of the traM regulation by TraR is that the rate of TraR production must at one point exceed that of TraM production, otherwise QS activation would continuously be inhibited. Evidence that TraM is specifically transcribed from a mildly activated promoter with a tra box IV (White and Winans, 2005) is in line with this requirement. Alternatively an interesting but yet unexplored possibility to explain the induction of traM expression by TraR would be that this mechanism provides the cells with a mean to limit or shut off the QS process when this one is too strongly activated and becomes for instance too demanding energetically. This down-regulation loop is indeed common in other LuxI/LuxR systems (Gelencser et al., 2012). Either way a more critical examination of TraM regulation is still needed to fully clarify its role in QS. Additionally it has been shown that acetosyringone, a phenolic compound released by wounded plant cells, could also induce expression of traM, suggesting that during first steps of tumorigenesis TraM could efficiently inhibit QS activity (Cho and Winans, 2005).
The A. tumefaciens Ti-plasmids use an original system of replication and partitioning encoded in a single locus named repABC. While RepC is essential for replicative DNA synthesis, RepA and RepB are thought to be involved in stable partitioning of plasmids into daughter cells (Pinto et al., 2012). Initially the expression of the operon repABC was shown to be strongly stimulated by TraR in bacterial backgrounds with both nopaline- and octopine-type plasmids. This stimulation was also correlated with induction of vegetative replication, i.e., with a drastic increase in number of Ti plasmid copies per cell (Li and Farrand, 2000; Pappas and Winans, 2003a). However, in the array experiment mentioned previously (Cho and Winans, 2007), repABC up-regulation by TraR was barely detectable. The authors argued that this result was probably due to the very weak basal expression of the operon and that it did not question the role of QS in controlling the number of Ti plasmid copies because under their experimental conditions the number of Ti plasmids per cell was still higher than one. Another interpretation of this result might be that increased Ti plasmid copies culminate in a negative feedback control possibly bringing back the expression of the repABC genes to their basal levels, thereby avoiding continuous and anarchic replication of the replicon. The promoter architecture of repABC may support this hypothesis as three different TraR-dependent (repAP1, 2, and 3) and one TraR-independent (repAP4) promoters control the expression of the operon (Pappas and Winans, 2003b). Promoter repAP4 is thought to mediate the Ti plasmid replication associated with cell division but it is also autorepressed by RepA and RepB. Moreover repAP4 is located downstream of repAP1, 2, and 3. It is therefore conceivable that autorepression of repAP4 might impair activation of TraR-dependent promoters. Additionally expression of repABC can be induced by the virulence proteins VirA and VirG, further suggesting that the regulation of this operon is complex and might be sensitive to different physiological states (Cho and Winans, 2005; Pappas, 2008).
MECHANISTIC INSIGHTS INTO A. tumefaciens QS
A central aspect of the LuxI/LuxR type QS systems resides in the way autoinducers, transcriptional factors and gene promoters interact with each other. A better understanding of these mechanisms is therefore crucial to evaluate the specificity of the system. Given the large variety of acyl-homoserine lactone derivatives which can serve as QS signals, it may also represent a privileged opportunity to get insight into possible crosstalk between different bacterial QS or to develop strategies of quorum-quenching. By combining biochemical and structural approaches with analysis of mutant strains and in vivo expression assays, the investigations on A. tumefaciens QS undoubtedly assemble one of the most elaborate sets of data in this domain.
TraI AND OC8HSL SYNTHESIS
To identify the substrates of OC8HSL synthesis, the enzymatic activity of a purified A. tumefaciens TraI protein was tested in the presence of different molecules (More et al., 1996). It was thus determined that 3-oxo-octanoyl-acyl carrier protein (OC8-ACP) was the fatty acid donor and S-adenosylmethionine (SAM) the homoserine lactone precursor involved in OC8HSL synthesis. Mechanistically the synthesis reaction is proposed to occur in a “bi-ter” (two substrates, three products) way. The donation of the 3-oxo-octanoyl branch to the amine of SAM leads to the releases of first apo-ACP, then OC8HSL and finally methylthioadenosine (Parsek et al., 1999). All enzymes of the LuxI family are expected to share similar mechanisms of reaction, though variations in the acyl chain length and oxidation state at C3 of their acyl-ACP substrates exist. High-resolution crystal structures were obtained for two TraI orthologs: EsaI of Pantoea stewartii that synthesizes 3OC6HSLs and LasI of Pseudomonas aeruginosa that synthesizes 3-oxo-dodecanoyl-homoserine lactones (Watson et al., 2001; Gould et al., 2004). Analyses of these structures revealed that conserved residues in the N-terminal part of the protein were essential for SAM-binding and that selectivity of the acyl-ACP substrate was dependent on a V-shaped cleft passing through the enzyme. Other results also suggested that selectivity of LuxI-like proteins could be affected by availability of different acyl-ACP substrates. Noticeably, besides OC8HSL, A. tumefaciens produces traces of OC6HSL and octanoyl-homoserine lactone (C8HSL; Zhu et al., 1998).
OC8HSL SPECIFICALLY INTERACTS WITH TraR
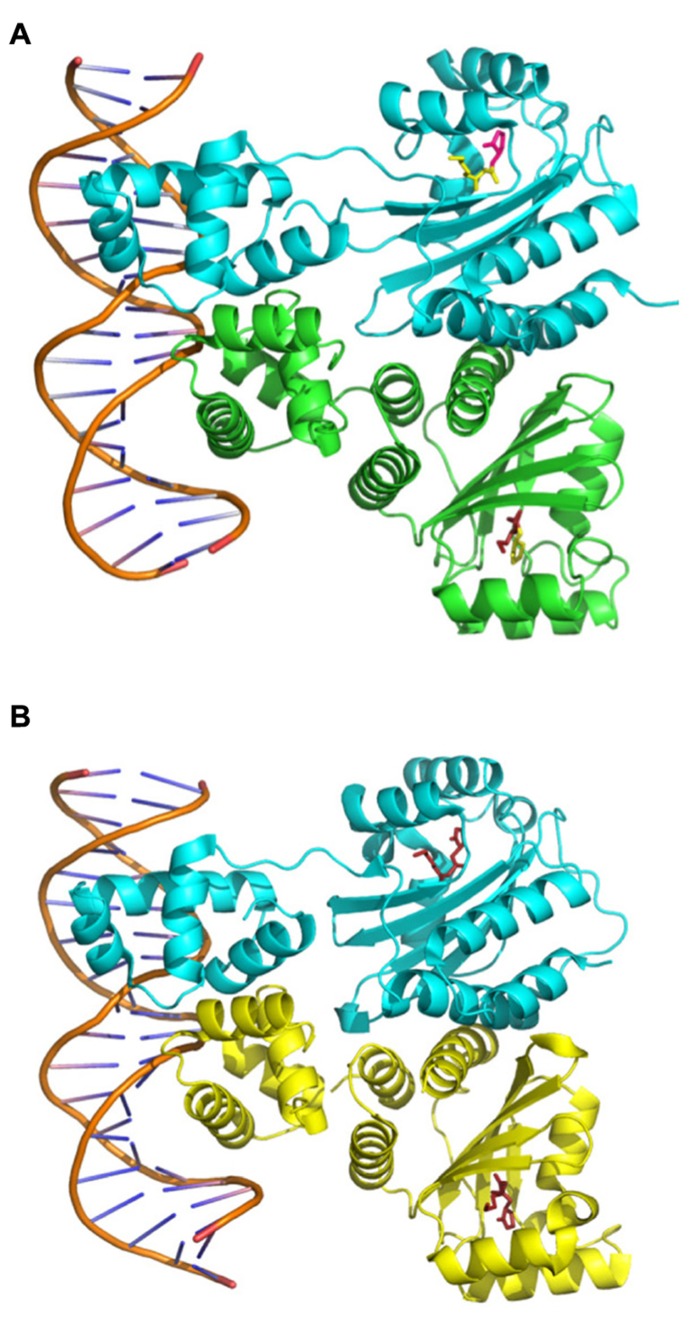
The first evidence of the interaction between TraR and OC8HSL was obtained through purified active TraR complexes which co-eluted with OC8HSLs in a ratio 1:1 (Zhu and Winans, 1999). Analysis of the protein turnover also indicated that binding of OC8HSL occurs rapidly in cells, surely during the own synthesis of TraR on polysomes (Zhu and Winans, 2001). Further crystal structures provided a mechanistic explanation for the specific interaction between TraR and OC8HSL as they revealed that the N-terminal part of TraR formed an enclosed cavity into which OC8HSL molecule could be engulfed and tightly maintained through numerous hydrophobic interactions as well as four hydrogen bounds (Vannini et al., 2002; Zhang et al., 2002b; Figure 3). To analyze the specificity of the interaction between OC8HSL and TraR, 31 analogs of OC8HSLs were tested for their abilities to activate TraR. Most of these compounds turned out to be potent antagonists of TraR under wild-type conditions of TraR expression and significant stimulators under conditions of TraR overexpression. These two features demonstrate that the specificity of the interaction between TraR and its ligand could be dependent on TraR concentration (Zhu et al., 1998). Moreover the 3-oxo function of the OC8HSL molecule seems to play important role in the interaction process as 3-oxo-C6-, 3-oxo-C7-, 3-oxo-C11-, and 3-oxo-C12-homoserine lactones (see structures in Figure 1) can also activate TraR, though with a much lower intensity than OC8HSL (Zhu et al., 1998; Luo et al., 2003b). Consistently non-conservative mutations of the threonine 129 of TraR, that was predicted to stabilize the 3-oxo group in the binding pocket, led to a strong impairment of TraR activity (Chai and Winans, 2004). In addition, alanine 49 and glutamine 58 in the N-terminal part of TraR were found to be important for the binding of the C8 acyl chain of OC8HSL since their conversion to bulkier amino acids resulted in higher affinity toward homoserine lactone derivatives with shorter acyl chain (Chai and Winans, 2004).
FIGURE 3.
Structures of the TraR–OC8HSL dimers in complex with DNA. The images were created using data from The Protein Data Bank (PDB; www.rcsb.org) (Berman et al., 2000) and the PyMOL Molecular Graphics System software. (A) PDB ID: 1H0M from Vannini et al. (2002). (B) PDB ID: 1L3L from Zhang et al. (2002b).
INTERACTION BETWEEN OC8HSL AND TraR FACILITATES FORMATION OF ACTIVE HOMODIMERS
The observation that C-terminal deletion mutants of TraR exerted strong dominant negativity over their wild-type counterparts led to the hypothesis that TraR–OC8HSL complexes had to multimerize to be active (Luo and Farrand, 1999). Thereafter, size exclusion chromatography techniques revealed that purified active OC8HSL–TraR complexes formed homodimers, and hybrid expression reporter systems demonstrated that OC8HSL was required for this dimerization to take place (Qin et al., 2000). The existence of active OC8HSL–TraR homodimers was further supported by analysis of crystal structures which also suggested that these dimers were significantly asymmetric (Vannini et al., 2002; Zhang et al., 2002b). Two dimerization domains were identified in TraR sequence, one in the N-terminal part of the protein, partially overlapping with the OC8HSL-binding domain and another, less extensive, in the C-terminal part (Luo et al., 2003a). Several findings illustrated the role of OC8HSL binding in the maturation and dimerization process of TraR. In absence of OC8HSL, TraR proteins were intrinsically unstructured, insoluble in cells and rapidly degraded by proteases. On the opposite, presence of OC8HSL directed the release of active TraR into cytosol and enhanced the resistance of the protein against proteolysis (Qin et al., 2000; Zhu and Winans, 2001; Pinto and Winans, 2009). Additionally the proper folding of TraR and acquisition of mature ternary structure following the interaction with OC8HSL was shown to be mediated by the chaperone GroESL (Chai and Winans, 2009).
TraR–OC8HSL HOMODIMERS SPECIFICALLY RECOGNIZES tra BOXES
As mentioned above, tra boxes are 18 bp-inverted repeat operator sequences with a pronounced dyad symmetry, found in the two classes of TraR-regulated promoters (Fuqua and Winans, 1996a). The crystallization of TraR–OC8HSL complexes in presence of the tra box I sequence strongly suggested that each subunit of TraR–OC8HSL dimer binds to half of the tra box via C-terminal helix-turn-helix DNA binding motifs, thereby leading to an extensive DNA–protein interaction (Vannini et al., 2002; Zhang et al., 2002b; Figure 3). However, it was later demonstrated that six nucleotides at the center of the tra boxes did not interact with TraR and that yet these nucleotides contributed to proper activation of transcription, presumably by creating a flexible DNA bend (White and Winans, 2007). In parallel different screenings of TraR mutants resulted in the identification of three regions located in the N- and C-terminal part of the protein, which are critical for transactivation function but not for accumulation or DNA binding ability (Qin et al., 2004a, 2009; White and Winans, 2005). This finding suggested that these regions could cooperatively modulate the recruitment of the RNA polymerase and thereby differently control the expressions of TraR-regulated genes. Consistently some TraR mutants defective in transactivation of the traI promoter could still activate the traM promoter (Costa et al., 2009).
TraM-MEDIATED INACTIVATION OF TraR IS DUE TO OLIGOMERIC ASSOCIATION
In an effort to better understand how TraM could deactivate TraR, two crystal structures of TraM were obtained. They showed that the TraM protein can form homodimers with one unit linked to the other by an extensive hydrophobic interface (Chen et al., 2004; Vannini et al., 2004). The importance of this interface and the dimerization properties of TraM were also assessed using deletion mutants (Qin et al., 2004b). In addition, purifications of inactive TraR/TraM complexes carried out by different groups and with different biochemical techniques led to the conclusion that the inactive complexes were composed of two TraR–OC8HSL dimers and two TraM dimers both in vitro and in vivo (Chen et al., 2004; Vannini et al., 2004; Qin et al., 2007). Several domains important for this oligomerization and the resulting inhibitory effect were identified both in TraR and TraM sequences (Luo et al., 2000; Swiderska et al., 2001; Qin et al., 2007). Moreover, to explain the way TraM could inactivate DNA-bound TraR–OC8HSL dimers, a study convincingly proposed a stepwise mechanism according to which the apparition of inactive TraR–OC8HSL/TraM complexes was preceded by a nucleoprotein intermediate comprising one dimer of each protein in association with DNA (Qin et al., 2007). Interestingly the biochemical and structural properties of the TraR/TraM complexes were also investigated in the Rhizobium sp. strain NGR234 and led to similar conclusions regarding the mechanisms by which TraM can negatively impact TraR functions (Chen et al., 2007).
PLANT FACTORS ASSOCIATED TO A. tumefaciens QS
ROLE OF THE OPINES: MASTER CONTROL AND FINE-TUNING OF QS REGULATION
Opines are the small organic compounds which are produced during development of crown gall disease in transformed plant cells through the action of synthesis genes present on the T-DNA. All A. tumefaciens Ti plasmids harbor operons specialized in the uptake and assimilation of the opines they contribute to produce (Dessaux et al., 1992, 1998; Platt et al., 2012b). The two types most investigated in laboratories are the octopine- and the nopaline-type. Moreover, specific opines, called conjugal opines, are strictly required to enable conjugation of the A. tumefaciens Ti plasmid (Kerr et al., 1977; Petit et al., 1978). Therefore the finding, at the beginning of the 1990s, that this phenomenon was also dependent on the TraR/TraI QS system (Zhang and Kerr, 1991), sparked off significant interest and a number of studies aimed at understanding how these regulatory steps could be related. Successive genetic analysis, sequence determination and promoter dissections ultimately allowed the complete elucidation of the signaling pathway, clearly establishing the prominent role played by the conjugal opines for traR expression and QS initiation.
In the case of nopaline-type Ti plasmids, agrocinopines A and B which are a mixture of two non-nitrogenous phosphodiesters of sugars serve as conjugal opines (Ellis et al., 1982). These molecules can provoke, presumably by direct inhibitory interaction, the release of the transcriptional repression exerted by AccR, a member of the FucR family of transcriptional regulator (Beck von Bodman et al., 1992). In turn this derepression causes the expression of two divergently oriented operons: the acc (agrocinopines catabolism) and arc (agrocinopine regulation of the conjugation) operons of the Ti plasmid. The acc operon encodes seven proteins involved in internalization and degradation of agrocinopines plus the repressor AccR (Kim and Farrand, 1997) while the arc operon encodes five proteins, the fourth being TraR (Piper et al., 1999). In contrast, in octopine-type Ti plasmids, traR is the last of 14 genes of the occ operon which codes for functions associated with octopine assimilation (Fuqua and Winans, 1996b). Octopine molecules are formed in transformed plant cells from arginine and pyruvate. Octopine is a conjugal opine as it binds to OccR, a transcriptional activator of the LysR family, thereby eliciting transcription of the occ operon including traR (Habeeb et al., 1991; Cho and Winans, 1993). Remarkably, the absence of the conjugal opines totally prevents QS-mediated conjugation of both nopaline- and octopine-type Ti plasmids. Moreover, despite the differences in traR location, the structures of the TraR-regulated operons are well conserved between the nopaline- and octopine-type Ti plasmids (Cho and Winans, 2007). This feature actually supports the view that traR and TraR-regulated genes constitute a functional unit, subjected to multiple and fortuitous recombination events in the course of A. tumefaciens evolution, and whose integration under the strict control of an opine regulon may have resulted in an important selective advantage for the bacteria (Piper et al., 1999; Oger and Farrand, 2001). In this sense the fact that such different molecules as agrocinopines and octopine can regulate traR expression in different Ti plasmids is remarkable.
Apart from the master control depicted above, opines are also involved in at least two other fine-tuning QS regulatory mechanisms. The first one was described in the A. tumefaciens strain R10 that harbors an octopine-type Ti plasmid. In this strain, the existence of a TraR antiactivator encoded by the Ti plasmid and different from TraM, named TrlR, was evidenced. Interestingly, TrlR expression was inducible by the opine mannopine (Oger et al., 1998). TrlR strongly resembles TraR but lacks its DNA-binding domain (Zhu and Winans, 1998). Experimental data provided evidence that TrlR could block TraR activity by forming inactive TrlR:TraR dimers (Chai et al., 2001). However, the impact of TrlR on QS implementation, especially in vivo, remains poorly understood. A second example of QS fine-tuning by opines is documented. In the nopaline-type A. tumefaciens C58 strain, expression of the Ti plasmid gene aiiB was shown to be induced by the agrocinopines, the same opines which are required for QS initiation (Haudecoeur et al., 2009b). Curiously aiiB codes for the AiiB lactonase that is highly similar to the AiiA lactonase from Bacillus sp. These proteins belong to a large family of Zn-hydrolases that encompasses lactonases of Arthrobacter, Bacillus, Klebsiella, Mesorhizobium, Photorhabdus, and Rhizobium. Biochemical and structural properties of AiiB were investigated. The AiiB protein is able to cleave the lactone rings of a large range of homoserine lactone derivatives, with a general preference for non-3-oxo-substituted molecules and substrates with an acyl chain longer than four carbons (Liu et al., 2007). Further conjugation experiments demonstrated the capacity of this lactonase to modulate A. tumefaciens QS responses both in vitro and in planta (Haudecoeur et al., 2009b). Globally the characteristics of trlR and aiiB (specific to octopine- and nopaline-type, respectively, and close homologs to traR and aiiA, respectively) suggest that these two genes could have arisen from gene duplication (for trlR) and horizontal gene transfer (for aiiB). On the other hand the conservation of an opine dependent regulation of their expression implies that there would be – somehow paradoxically – an advantage for A. tumefaciens cells to dampen QS communication at moments when opines, including conjugal opines, accumulate in tumors.
THE EXPRESSION OF THE OC8HSL-DEGRADING BlcC (FORMERLY AttM) LACTONASE IS INDUCED BY PLANT METABOLITES
As AiiB, the BlcC protein is a member of the AiiA lactonase family. Different studies have shown that BlcC degrades various homoserine lactone derivatives, including gamma-butyrolactone (GBL, see structure in Figure 1) and OC8HSLs. The blcC gene is part of the three-gene blcABC operon which codes for the catabolic pathway converting GBL to succinate, through gamma-hydroxybutyrate (GHB) and succinic semialdehyde (SSA) intermediates (Chai et al., 2007). Remarkably BlcC confers to Agrobacterium the ability to grow with GBL as sole source of carbon, but it does not with OC8HSLs (Carlier et al., 2004). The expression of the blcABC operon is tightly controlled by the transcriptional repressor BlcR. Carbon and nitrogen starvation, GBL, GHB, and SSA can all release the repression exerted by BlcR, hence allowing the expression of the blcABC genes (Zhang et al., 2002a; Carlier et al., 2004). The plant metabolite gamma-amino butyric acid (GABA), through conversion to SSA (Chevrot et al., 2006; Wang et al., 2006b), and the plant defense signaling hormone salicylic acid, through an unknown mechanism (Yuan et al., 2008), can also induce blcC expression. Based on the observations that GABA induces the expression of the blcABC operon and that GABA accumulates in tumors, it was proposed that the BlcC activity could coincide with QS communication during interactions between A. tumefaciens and plant hosts. However, in tomato tumors, the effect of BlcC on QS-dependent Ti plasmid conjugation was weak and transient (Khan and Farrand, 2009), suggesting that plant tumor tissues could exert a negative control on the expression of the BlcC expression.
The capacity of A. tumefaciens to take up GABA was extensively investigated in the last years. Studies revealed the involvement of two distinct transport systems. The gene atu2422, located on the circular chromosome is widely conserved within the Agrobacterium genus and codes for a periplasmic GABA-binding protein that controls GABA import through the bra ABC transporter (Planamente et al., 2010). Interestingly the GABA import by atu2422 is strongly antagonized by proline, alanine, and valine, suggesting that these compounds which accumulate in tumors could also indirectly modulate the overall BlcC lactonase activity in the bacterial cells (Haudecoeur et al., 2009a). In comparison, the periplasmic binding protein encoded by the linear chromosome gene atu4243 appears highly specific for GABA (Planamente et al., 2012). Strikingly, the expression of atu4243 is totally repressed by atu4232-encoded protein and mechanisms of derepression are so far unknown (Planamente et al., 2012). Collectively these data illustrate the complexity of factors coming into play when searching to determine the impact of BlcC on A. tumefaciens QS. Of special interest would be the critical examination of plant metabolism to evaluate how the GABA, GBL, GHB, and SSA produced in the tumors may activate BlcC in colonizing A. tumefaciens cells. Such studies might reveal that the role of BlcC varies according to the metabolic status of the plant hosts.
INTERACTIONS BETWEEN THE Ti AND AT PLASMIDS IN THE PLANT TUMOR
Another interesting feature of the blcC gene lies in its location on the companion At plasmid. This makes it the only component involved in A. tumefaciens QS that is not present on the Ti plasmid. Ecologically this characteristic raises interesting questions and notably that to know whether the dissociation of the At and Ti plasmids could result in a QS deregulation. To date very little is known about the maintenance of the At plasmid in A. tumefaciens populations. If no gene essential for the survival of A. tumefaciens C58 is present on the At plasmid (Goodner et al., 2001; Wood et al., 2001), the carriage of this At plasmid imposes in vitro high fitness costs to A. tumefaciens host cells (Morton et al., 2013). On the other hand, the At plasmid encodes several functions which confer or may confer a fitness advantage to agrobacteria in plant tumors (Haudecoeur et al., 2009b). Besides the degradation of butyrolactones and their derivatives mentioned above, the At plasmid is involved in the assimilation of some opines of Amadori compounds (Vaudequin-Dransart et al., 1998; Baek et al., 2005). The At plasmid also seems to have a positive impact on the virulence capacity of A. tumefaciens (Matthysse et al., 2008), although this point is debatable as it was recently shown that a large deletion in the At plasmid resulted in increase of the bacterial virulence (Morton et al., 2013). In conclusion, one can reasonably assume that, as for Ti plasmids, the tumor compartment is an appropriate environment for the dissemination of the At plasmid. Remarkably it was recently demonstrated that in A. tumefaciens C58, the conjugations of At and Ti plasmids are related events controlled by the agrocinopines-responsive regulator AccR and it was suggested that this mechanism of co-regulation could be instrumental in the conservation of the reciprocally beneficial functions carried by the two replicons (Lang et al., 2013).
OC8HSL-ASSOCIATED PLANT RESPONSES
The interactions between A. tumefaciens and plant hosts are mediated by several factors, from the phenolic compounds accumulated at wound sites that induce the expression of the Ti plasmid vir genes, to the opines produced in the tumor niche that control horizontal transfer of bacterial plasmids. It is therefore tempting to speculate on a possible implication of QS signal molecules in this generic trans-kingdom association, especially as several lines of evidence showed that N-acyl-homoserine lactone molecules could induce specific responses in eukaryote cells (Williams, 2007). For instance, in axenic plant systems, exogenous supply of different homoserine lactone derivatives was found to modulate plant immunity and development although the outcomes drastically differed according to the nature of the tested QS molecules (Klein et al., 2009; Hartmann and Schikora, 2012).
To our knowledge only three studies investigated the impact of OC8HSL on plants. In the first one, authors devised an inducible gene expression system based on TraR-OC8HSL activity which they introduced in Arabidopsis thaliana plants (You et al., 2006). To verify that induction with OC8HSL of the transferred gene did not affect the transcriptome of the transformed plants, the authors extracted RNA from 12-day-old seedlings treated or not by foliar application with 1 mM of OC8HSL for 24 h and carried out microarray experiments using Agilent technology. Processing of the data prompted them to conclude that no gene was differentially expressed by presence of the QS signal. In a second paper, a proteome analysis of Arabidopsis thaliana roots grown for 24 h in a hydroponic system in the presence or not of 10 μM of OC8HSL revealed that the levels of 53 proteins involved in the metabolism of carbohydrate and energy, protein biosynthesis, defense responses, and cytoskeleton remodeling, were significantly affected by the QS signal (Miao et al., 2012). The modest number of proteins differentially affected in this study suggests that plants sense A. tumefaciens QS signals only in a very restricted way. Noteworthy, in the two above-mentioned experiments, the used concentrations of homoserine lactone derivatives were in the micromolar and millimolar range while the concentrations at which QS molecules are active in A. tumefaciens are usually rather in the nanomolar range. Finally Arabidopsis thaliana defense responses upon exposure to OC8HSL-producing Rhizobium etli were recently analyzed. The results established that this condition had no impact on the plant defense (Zarkani et al., 2013), thereby strengthening the notion that plants are immune to OC8HSLs.
IMPLICATIONS AND SELECTIVE ADVANTAGES OF THE TIGHTLY REGULATED QS SYSTEM IN A. tumefaciens
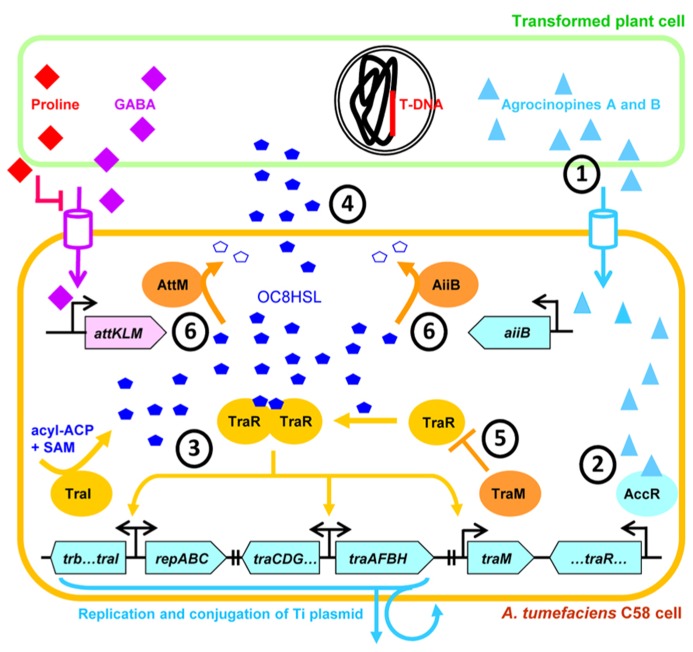
Taken together the findings presented above described a very sophisticated system in which A. tumefaciens QS action is not only placed under the strict control of the conjugal opine regulon but is also modulated by various adjacent components like antiactivator or lactonases (Figure 4). Now we will discuss the implications of such hierarchical regulatory cascades and speculate about the selective advantages they may confer to A. tumefaciens.
FIGURE 4.
Representation of the sophisticated hierarchical QS regulation in A. tumefaciens strain C58. QS-dependent conjugation and copy-number amplification of the Ti plasmid is initiated when OC8HSL and TraR reach appropriate concentration and form transcriptional active dimers. QS-signaling is positively regulated by the conjugal opines agrocinopines which are produced by the transformed plant cells (1) and induce production of TraR by releasing AccR repressive action (2). Then, active TraR-OC8HSL dimers activate the production of TraI, thereby triggering a positive feedback in the synthesis of OC8HSLs (3) which are diffusible molecules (4). The QS activation is delayed by the TraR-antagonist TraM (5), as well as OC8HSL-cleaving lactonases AiiB and AttM (6) whose expressions are controlled by agrocinopines and GABA, respectively. SAM, S-adenosylmethionine; ACP, acyl carrier protein. The Ti plasmid genes and the At plasmid genes are in blue and pink boxes, respectively.
CONJUGATION OF Ti PLASMID IN OPINE-PRODUCING TUMORS
As mentioned previously, the expression of traR gene requires the presence of conjugal opines. Therefore the QS system of A. tumefaciens functions only in host plants and only after transformed tissues have accumulated sufficient amount of conjugal opines. This restriction suggests that mature tumors are the most conducive environments for Ti plasmid dissemination and that, in these plant tumors, the selective advantages conferred to A. tumefaciens by a functional Ti plasmid would overcome the associated costs of maintenance. Supporting these notions, it has been demonstrated that Ti plasmid imposed a high fitness cost under conditions reminiscent of tumorigenesis but not anymore when opines were fully supplied (Platt et al., 2012a). It has also been observed that large proportion of A. tumefaciens cells present in mature tumors were devoid of Ti plasmids or harbored a mutated Ti plasmid (Fortin et al., 1993; Belanger et al., 1995). Thus the master control by conjugal opines could allow a large dissemination of functional Ti plasmids in an A. tumefaciens population characterized by a high proportion of potential recipient cells. The resulting selective advantages would be manifold. By amplifying the number of genes involved in opine assimilation, this mechanism could increase the colonizing fitness of the A. tumefaciens population, especially in older tumors where nutritive resources are scarcer. Multiplication of vir genes may also enhance aggressiveness of the bacteria. In relation, several reports already correlated an impairment of A. tumefaciens QS communication with a diminution of the crown gall symptoms (Haudecoeur et al., 2009b; Planamente et al., 2010, 2012). At last dissemination of Ti plasmids would increase the potential of migratory agrobacterial cells to initiate new infections. Interestingly Ti plasmid transfers to other bacterial species present in plant tumors may also occur, a feature that would favor genetic biodiversity. In this regard it is unfortunate that, even if the plant tumors are generally considered as privileged entry points for other bacteria, no information on plant tumor microbiomes are available at the moment.
DOES A. tumefaciens QS REALLY MEASURE A QUORUM OF DONOR CELLS?
Since the finding that A. tumefaciens QS controlled Ti plasmid conjugation, a “nagging” question remained to understand the relevance of a system in which donor cells could only monitor the density of other donors that already harbor a Ti-plasmid. Indeed as conjugation cannot happen in a cell already containing a resident Ti plasmid (Cho et al., 2009), the risk of uselessly activating, at the quorum concentration, the horizontal transfer machinery in the absence of sufficiently numerous recipient cells seems elevated. Nonetheless, as evoked previously, the master control of QS by conjugal opines might provide a way to circumvent this difficulty by allowing the conjugation of Ti plasmid only in mature tumors, i.e., in environments where the proportion of recipient cells would have extended. In such a context, the adjustment of the activation of the tra regulon according to a quorum of donor cells should maximize the efficiency of Ti plasmid dissemination and would be fully sensible. Under laboratory conditions, all the collected data firmly sustain the notion that A. tumefaciens QS functions as a cell density-dependent process. However, these conditions, using most of the time cell cultures and constant concentration of conjugal opines to initiate QS, may not reflect natural conditions. In V. fischeri the quorum nature of the system is defined by a production of LuxR at relatively high basal level and by a concentration of OC6HSL which increases as a function of cell density until reaching the threshold of LuxR activation (Miller and Bassler, 2001). In contrast, in A. tumefaciens, production of an active TraR regulator is subordinated to the presence of conjugal opines and to that of the antiactivator TraM. Taking full consideration of this characteristic implies that QS can be partly dissociated from solely functioning as a measure of population density. Another element of complexity may be brought by the non-linear accumulation of OC8HSL in tumors. Indeed plant tumors are not homogenous structures; they emerged from wound sites and underwent neoplastic expansion (Aloni et al., 1995; Veselov et al., 2003). In these complex environments colonizing A. tumefaciens shall form different clusters of cells more or less isolated one from the other and located in surface or intercellular spaces where diffusion rates are different as well as temporally changing. It therefore appears unlikely that the OC8HSL concentration which can be measured in a tumor or a part of the tumor does strictly mirror the cell density of the pathogen in this environment. Interestingly when they simulated the QS-induced transition in liquid cell cultures or biofilm, Goryachev et al. (2005) noticed that the first condition required a much higher threshold density than the second. They consequently came to the conclusion that A. tumefaciens QS served as a detector of biofilm formation rather than a sensor of cell concentration. If a growing attention has been given in the last years to mechanisms of biofilm formation in A. tumefaciens (Tomlinson et al., 2010; Hibbing and Fuqua, 2012), no data so far have related them to QS and very little is known about the formation of biofilms in the context of the agrobacterial interactions with plant host. However, it would definitely be relevant for the bacteria to place the coordination of Ti plasmid conjugation upon biofilm perception since the cell aggregates would constitute a very appropriate context for activation of the horizontal transfer machinery, either by minimizing the distances between donor and recipient cells or by acting as a shield against all kinds of physical or biological perturbations.
RELATIONSHIP BETWEEN QS REGULATION, Ti PLASMID CONJUGATION, AND A. tumefaciens HOST CELL
In the above discussion, the question of the QS-dependent dissemination of Ti plasmids was addressed only according to the selective advantages this dissemination may confer to agrobacterial cells. However, another perspective would be to consider Ti plasmids as selfish elements which somehow hijack A. tumefaciens cells in order to disseminate their genetic backgrounds. In this framework Ti plasmids would take advantage of the opine and QS regulations to optimize the efficiency of their conjugations. It is furthermore important to note that the tumor conditions where the selective advantage conferred to A. tumefaciens cells by the Ti plasmids is the strongest coincide with the conditions where the dissemination of these Ti plasmids is the most important. The recent discovery in A. tumefaciens C58 that the conjugations of both Ti and At plasmids are exacerbated by conjugal opines (Lang et al., 2013) further supports the notion that Ti and At plasmids may collaborate to transform avirulent A. tumefaciens cells into virulent in order to perpetuate and disseminate their genetic traits.
CONCLUSION
In this review, we described the A. tumefaciens TraI/TraR QS system and showed how it exquisitely regulated the dissemination of Ti plasmids.
The QS systems of LuxI/LuxR type are generally thought to have originated early in evolution of Gram-negative Proteobacteria, with functional pairs of autoinducer synthases and receptors coevolving as regulatory cassettes, although in many cases these cassettes could also be inherited horizontally (Gray and Garey, 2001). In A. tumefaciens, the TraI/TraR system and the related QS-regulated genes are well conserved in all nopaline- and octopine-type strains studied to date, suggesting that this regulatory mechanism has been anciently selected. The target genes of A. tumefaciens QS are involved in the dissemination of Ti plasmids, both by replication and conjugation, but also in positive and negative feedback controls with the OC8HSL-synthesis TraI enzyme and the TraM antiactivator. Different studies demonstrated that this last protein plays a critical role in the implementation of the QS, even if it is not clear yet whether TraM is more relevant in delaying QS activation or in stabilizing and limiting QS activity.
At the molecular level, the A. tumefaciens QS communication has been largely deciphered. Two crystal structures have notably been obtained for TraR, in association with OC8HSL and DNA, providing a first class access to the interaction specificities of the system. Thorough biochemical investigations of active and inactive complexes also allowed to better understand multimerization processes of the QS components.
Consistent with the particular phytopathogenic lifestyle of the bacteria, A. tumefaciens QS system displays an original scheme including several differently acquired regulatory elements. The most important of these elements, common to all A. tumefaciens strains, are the conjugal opines which accumulate in tumors as a consequence of plant transformation and are strictly required for traR expression and hence for QS initiation. In parallel, only specific to some A. tumefaciens strains, lactonases such as AiiB and BlcC or supplementary anti-activator like TrlR can also modulate QS responses. This complex network of horizontal and lateral regulation suggests that there would be an advantage for A. tumefaciens to restrain as much as possible the window of QS activation.
Assessing reasons why a biological system has been selected is always challenging because this selection hinges on a trade-off between advantages and drawbacks which cannot be fully appreciated under laboratory conditions. By perusing different possibilities, we nonetheless hypothesized that the tight regulation of A. tumefaciens QS surely allowed the bacteria to disseminate the Ti plasmid in an environment where carrying the replicon would be clearly advantageous and at a moment when the energetic and physical factors would be ideal.
For the future, some important questions still remain to be answered to complete our understanding of A. tumefaciens QS functioning during the interactions with the host plant. For instance how do conjugal opines and TraM cooperate to produce active TraR-OC8HSL dimers? Precise dosage of conjugal opines in the course of tumor development as well as advances in knowledge of traM regulation might help solve this question. It would also be very interesting to better determine how the BlcC lactonase interferes with OC8HSL levels in tumors induced on different plants hosts and what are the ecological implications regarding horizontal transfers of both At and Ti plasmids. At last, analysis of bacterial populations found in natural tumors could deliver exciting results regarding abundance of potential Ti plasmid recipient cells. This kind of data might also unveil the extent of competition between the phytopathogen and other bacterial species present in plant tumors, hence leading to a novel appreciation of A. tumefaciens QS activity.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Julien Lang was supported by a CNRS PhD-grant, Denis Faure by CNRS and the two ANR-Blanc SENSOR (ANR-12-BSV8-0003-01) and ECORUM (ANR-11-SVSE7). We are grateful to Dr. Yves Dessaux (ISV/CNRS, Gif-sur-Yvette, France) for the critical reading of the manuscript and to Armelle Vigouroux (LEBS/CNRS, Gif-sur-Yvette, France) for the processing of the structure data.
REFERENCES
- Aloni R., Pradel K., Ullrich C. (1995). The three-dimensional structure of vascular tissues in Agrobacterium tumefaciens-induced crown galls and in the host stems of Ricinus communis L. Planta 196 597–605 10.1007/BF00203661 [DOI] [Google Scholar]
- Baek C. H., Farrand S. K., Park D. K., Lee K. E., Hwang W., Kim K. S. (2005). Genes for utilization of deoxyfructosyl glutamine (DFG), an amadori compound, are widely dispersed in the family Rhizobiaceae. FEMS Microbiol. Ecol. 53 221–233 10.1016/j.femsec.2004.12.008 [DOI] [PubMed] [Google Scholar]
- Beck von Bodman S., Hayman G. T., Farrand S. K. (1992). Opine catabolism and conjugal transfer of the nopaline Ti plasmid pTiC58 are coordinately regulated by a single repressor. Proc. Natl. Acad. Sci. U.S.A. 89 643–647 10.1073/pnas.89.2.643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger C., Canfield M. L., Moore L. W., Dion P. (1995). Genetic analysis of nonpathogenic Agrobacterium tumefaciens mutants arising in crown gall tumors. J. Bacteriol. 177 3752–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., et al. (2000). The Protein Data Bank. Nucleic Acids Res. 28 235–242 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier A., Chevrot R., Dessaux Y., Faure D. (2004). The assimilation of gamma-butyrolactone in Agrobacterium tumefaciens C58 interferes with the accumulation of the N-acyl-homoserine lactone signal. Mol. Plant Microbe Interact. 17 951–957 10.1094/MPMI.2004.17.9.951 [DOI] [PubMed] [Google Scholar]
- Chai Y., Tsai C. S., Cho H., Winans S. C. (2007). Reconstitution of the biochemical activities of the AttJ repressor and the AttK, AttL, and AttM catabolic enzymes of Agrobacterium tumefaciens. J. Bacteriol. 189 3674–3679 10.1128/JB.01274-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y., Winans S. C. (2004). Site-directed mutagenesis of a LuxR-type quorum-sensing transcription factor: alteration of autoinducer specificity. Mol. Microbiol. 51 765–776 10.1046/j.1365-2958.2003.03857.x [DOI] [PubMed] [Google Scholar]
- Chai Y., Winans S. C. (2009). The chaperone GroESL enhances the accumulation of soluble, active TraR protein, a quorum-sensing transcription factor from Agrobacterium tumefaciens. J. Bacteriol. 191 3706–3711 10.1128/JB.01434-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y., Zhu J., Winans S. C. (2001). TrlR, a defective TraR-like protein of Agrobacterium tumefaciens, blocks TraR function in vitro by forming inactive TrlR:TraR dimers. Mol. Microbiol. 40 414–421 10.1046/j.1365-2958.2001.02385.x [DOI] [PubMed] [Google Scholar]
- Chen G., Jeffrey P. D., Fuqua C., Shi Y., Chen L. (2007). Structural basis for antiactivation in bacterial quorum sensing. Proc. Natl. Acad. Sci. U.S.A. 104 16474–16479 10.1073/pnas.0704843104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Malenkos J. W., Cha M. R., Fuqua C., Chen L. (2004). Quorum-sensing antiactivator TraM forms a dimer that dissociates to inhibit TraR. Mol. Microbiol. 52 1641–1651 10.1111/j.1365-2958.2004.04110.x [DOI] [PubMed] [Google Scholar]
- Chevrot R., Rosen R., Haudecoeur E., Cirou A., Shelp B. J., Ron E., et al. (2006). GABA controls the level of quorum-sensing signal in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. U.S.A. 103 7460–7464 10.1073/pnas.0600313103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H., Pinto U. M., Winans S. C. (2009). Transsexuality in the rhizosphere: quorum sensing reversibly converts Agrobacterium tumefaciens from phenotypically female to male. J. Bacteriol. 191 3375–3383 10.1128/JB.01608-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H., Winans S. C. (2005). VirA and VirG activate the Ti plasmid repABC operon, elevating plasmid copy number in response to wound-released chemical signals. Proc. Natl. Acad. Sci. U.S.A. 102 14843–14848 10.1073/pnas.0503458102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H., Winans S. C. (2007). TraA, TraC and TraD autorepress two divergent quorum-regulated promoters near the transfer origin of the Ti plasmid of Agrobacterium tumefaciens. Mol. Microbiol. 63 1769–1782 10.1111/j.1365-2958.2007.05624.x [DOI] [PubMed] [Google Scholar]
- Cho K., Winans S. C. (1993). Altered-function mutations in the Agrobacterium tumefaciens OccR protein and in an OccR-regulated promoter. J. Bacteriol. 175 7715–7719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D. M., Li P. L., Ruchaud F., Padden S., Farrand S. K. (1997). Ti plasmid conjugation is independent of vir: reconstitution of the tra functions from pTiC58 as a binary system. J. Bacteriol. 179 1291–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa E. D., Chai Y., Winans S. C. (2012). The quorum-sensing protein TraR of Agrobacterium tumefaciens is susceptible to intrinsic and TraM-mediated proteolytic instability. Mol. Microbiol. 84 807–815 10.1111/j.1365-2958.2012.08037.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa E. D., Cho H., Winans S. C. (2009). Identification of amino acid residues of the pheromone-binding domain of the transcription factor TraR that are required for positive control. Mol. Microbiol. 73 341–351 10.1111/j.1365-2958.2009.06755.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessaux Y., Petit A., Farrand S. K., Murphy P. J. (1998). “Opines and opine-like molecules involved in plant–Rhizobiaceae interactions,” in The Rhizobiaceae, Molecular Biology of Model Plant-associated Bacteria eds Spaink H. P., Kondorosi A., Hooykaas P. J. J. (Dordrecht: Kluwer Academic Publisher) 173–197 [Google Scholar]
- Dessaux Y., Petit A., Tempe J. (1992). “Opines in Agrobacterium biology,” in Molecular Signals in Plant-microbe Communications ed. Vermaed D. P. S. (Boca Raton, FL: CRC Press) 109–136 [Google Scholar]
- Eberhard A. (1972). Inhibition and activation of bacterial luciferase synthesis. J. Bacteriol. 109 1101–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J. G., Kerr A., Petit A., Tempe J. (1982). Conjugal transfer of nopaline and agropine Ti-plasmids: the role of agrocinopines. Mol. Gen. Genet. 186 269–274 10.1007/BF00331861 [DOI] [Google Scholar]
- Farrand S. K., Hwang I., Cook D. M. (1996). The tra region of the nopaline-type Ti plasmid is a chimera with elements related to the transfer systems of RSF1010, RP4, and F. J. Bacteriol. 178 4233–4247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin C., Marquis C., Nester E. W., Dion P. (1993). Dynamic structure of Agrobacterium tumefaciens Ti plasmids. J. Bacteriol. 175 4790–4799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederix M., Downie A. J. (2011). Quorum sensing: regulating the regulators. Adv. Microb. Physiol. 58 23–80 10.1016/B978-0-12-381043-4.00002-7 [DOI] [PubMed] [Google Scholar]
- Fuqua C., Burbea M., Winans S. C. (1995). Activity of the Agrobacterium Ti plasmid conjugal transfer regulator TraR is inhibited by the product of the TraM gene. J. Bacteriol. 177 1367–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua C., Winans S. C. (1996a). Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J. Bacteriol. 178 435–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua C., Winans S. C. (1996b). Localization of OccR-activated and TraR-activated promoters that express two ABC-type permeases and the traR gene of Ti plasmid pTiR10. Mol. Microbiol. 20 1199–1210 10.1111/j.1365-2958.1996.tb02640.x [DOI] [PubMed] [Google Scholar]
- Fuqua C., Winans S. C., Greenberg E. P. (1996). Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50 727–751 10.1146/annurev.micro.50.1.727 [DOI] [PubMed] [Google Scholar]
- Fuqua W. C., Winans S. C. (1994). A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J. Bacteriol. 176 2796–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua W. C., Winans S. C., Greenberg E. P. (1994). Quorum sensing in bacteria: the LuxR–LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176 269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelencser Z., Choudhary K. S., Coutinho B. G., Hudaiberdiev S., Galbats B., Venturi V., et al. (2012). Classifying the topology of AHL-driven quorum sensing circuits in proteobacterial genomes. Sensors 12 5432–5444 10.3390/s120505432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodner B., Hinkle G., Gattung S., Miller N., Blanchard M., Qurollo B., et al. (2001). Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294 2323–2328 10.1126/science.1066803 [DOI] [PubMed] [Google Scholar]
- Goryachev A. B., Toh D. J., Wee K. B., Lee T., Zhang H. B., Zhang L. H. (2005). Transition to quorum sensing in an Agrobacterium population: a stochastic model. PLoS Comput. Biol. 1:e37 10.1371/journal.pcbi.0010037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould T. A., Schweizer H. P., Churchill M. E. (2004). Structure of the Pseudomonas aeruginosa acyl-homoserinelactone synthase LasI. Mol. Microbiol. 53 1135–1146 10.1111/j.1365-2958.2004.04211.x [DOI] [PubMed] [Google Scholar]
- Gray K. M., Garey J. R. (2001). The evolution of bacterial LuxI and LuxR quorum sensing regulators. Microbiology 147 2379–2387 [DOI] [PubMed] [Google Scholar]
- Habeeb L. F., Wang L., Winans S. C. (1991). Transcription of the octopine catabolism operon of the Agrobacterium tumor-inducing plasmid pTiA6 is activated by a LysR-type regulatory protein. Mol. Plant Microbe Interact. 4 379–385 10.1094/MPMI-4-379 [DOI] [PubMed] [Google Scholar]
- Hartmann A., Schikora A. (2012). Quorum sensing of bacteria and trans-kingdom interactions of N-acyl homoserine lactones with eukaryotes. J. Chem. Ecol. 38 704–713 10.1007/s10886-012-0141-7 [DOI] [PubMed] [Google Scholar]
- Haudecoeur E., Planamente S., Cirou A., Tannieres M., Shelp B. J., Morera S., et al. (2009a). Proline antagonizes GABA-induced quenching of quorum-sensing in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. U.S.A. 106 14587–14592 10.1073/pnas.0808005106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haudecoeur E., Tannieres M., Cirou A., Raffoux A., Dessaux Y., Faure D. (2009b). Different regulation and roles of lactonases AiiB and AttM in Agrobacterium tumefaciens C58. Mol. Plant Microbe Interact. 22 529–537 10.1094/MPMI-22-5-0529 [DOI] [PubMed] [Google Scholar]
- Hibbing M. E., Fuqua C. (2012). Inhibition and dispersal of Agrobacterium tumefaciens biofilms by a small diffusible Pseudomonas aeruginosa exoproduct(s). Arch. Microbiol. 194 391–403 10.1007/s00203-011-0767-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I., Cook D. M., Farrand S. K. (1995). A new regulatory element modulates homoserine lactone-mediated autoinduction of Ti plasmid conjugal transfer. J. Bacteriol. 177 449–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I., Li P. L., Zhang L., Piper K. R., Cook D. M., Tate M. E., et al. (1994). TraI, a LuxI homologue, is responsible for production of conjugation factor, the Ti plasmid N-acylhomoserine lactone autoinducer. Proc. Natl. Acad. Sci. U.S.A. 91 4639–4643 10.1073/pnas.91.11.4639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I., Smyth A. J., Luo Z. Q., Farrand S. K. (1999). Modulating quorum sensing by antiactivation: TraM interacts with TraR to inhibit activation of Ti plasmid conjugal transfer genes. Mol. Microbiol. 34 282–294 10.1046/j.1365-2958.1999.01595.x [DOI] [PubMed] [Google Scholar]
- Kerr A., Manigault P., Tempe J. (1977). Transfer of virulence in vivo and in vitro in Agrobacterium. Nature 265 560–561 10.1038/265560a0 [DOI] [PubMed] [Google Scholar]
- Khan S. R., Farrand S. K. (2009). The BlcC (AttM) lactonase of Agrobacterium tumefaciens does not quench the quorum-sensing system that regulates Ti plasmid conjugative transfer. J. Bacteriol. 191 1320–1329 10.1128/JB.01304-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Farrand S. K. (1997). Characterization of the acc operon from the nopaline-type Ti plasmid pTiC58, which encodes utilization of agrocinopines A and B and susceptibility to agrocin 84. J. Bacteriol. 179 7559–7572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein I., Von Rad U., Durner J. (2009). Homoserine lactones: do plants really listen to bacterial talk? Plant Signal. Behav. 4 50–51 10.4161/psb.4.1.7300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang J., Planamente S., Mondy S., Dessaux Y., Morera S., Faure D. (2013). Concerted transfer of the virulence Ti plasmid and companion At plasmid in the Agrobacterium tumefaciens-induced plant tumor. Mol. Microbiol. 90 1178–1189 10.1111/mmi.12423 [DOI] [PubMed] [Google Scholar]
- Li P. L., Everhart D. M., Farrand S. K. (1998). Genetic and sequence analysis of the pTiC58 trb locus, encoding a mating-pair formation system related to members of the type IV secretion family. J. Bacteriol. 180 6164–6172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P. L., Farrand S. K. (2000). The replicator of the nopaline-type Ti plasmid pTiC58 is a member of the repABC family and is influenced by the TraR-dependent quorum-sensing regulatory system. J. Bacteriol. 182 179–188 10.1128/JB.182.1.179-188.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Thomas P. W., Momb J., Hoang Q. Q., Petsko G. A., Ringe D., et al. (2007). Structure and specificity of a quorum-quenching lactonase (AiiB) from Agrobacterium tumefaciens. Biochemistry 46 11789–11799 10.1021/bi7012849 [DOI] [PubMed] [Google Scholar]
- Luo Z. Q., Farrand S. K. (1999). Signal-dependent DNA binding and functional domains of the quorum-sensing activator TraR as identified by repressor activity. Proc. Natl. Acad. Sci. U.S.A. 96 9009–9014 10.1073/pnas.96.16.9009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z. Q., Qin Y., Farrand S. K. (2000). The antiactivator TraM interferes with the autoinducer-dependent binding of TraR to DNA by interacting with the C-terminal region of the quorum-sensing activator. J. Biol. Chem. 275 7713–7722 10.1074/jbc.275.11.7713 [DOI] [PubMed] [Google Scholar]
- Luo Z. Q., Smyth A. J., Gao P., Qin Y., Farrand S. K. (2003a). Mutational analysis of TraR. Correlating function with molecular structure of a quorum-sensing transcriptional activator. J. Biol. Chem. 278 13173–13182 10.1074/jbc.M210035200 [DOI] [PubMed] [Google Scholar]
- Luo Z. Q., Su S., Farrand S. K. (2003b). In situ activation of the quorum-sensing transcription factor TraR by cognate and noncognate acyl-homoserine lactone ligands: kinetics and consequences. J. Bacteriol. 185 5665–5672 10.1128/JB.185.19.5665-5672.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse A. G., Jaeckel P., Jeter C. (2008). attG and attC mutations of Agrobacterium tumefaciens are dominant negative mutations that block attachment and virulence. Can. J. Microbiol. 54 241–247 10.1139/W08-005 [DOI] [PubMed] [Google Scholar]
- Miao C., Liu F., Zhao Q., Jia Z., Song S. (2012). A proteomic analysis of Arabidopsis thaliana seedling responses to 3-oxo-octanoyl-homoserine lactone, a bacterial quorum-sensing signal. Biochem. Biophys. Res. Commun. 427 293–298 10.1016/j.bbrc.2012.09.044 [DOI] [PubMed] [Google Scholar]
- Miller M. B., Bassler B. L. (2001). Quorum sensing in bacteria. Annu. Rev. Microbiol. 55 165–199 10.1146/annurev.micro.55.1.165 [DOI] [PubMed] [Google Scholar]
- More M. I., Finger L. D., Stryker J. L., Fuqua C., Eberhard A., Winans S. C. (1996). Enzymatic synthesis of a quorum-sensing autoinducer through use of defined substrates. Science 272 1655–1658 10.1126/science.272.5268.1655 [DOI] [PubMed] [Google Scholar]
- Morton E. R., Merritt P. M., Bever J. D., Fuqua C. (2013). Large deletions in the pAtC58 megaplasmid of Agrobacterium tumefaciens can confer reduced carriage cost and increased expression of virulence genes. Genome Biol. Evol. 5 1353–1364 10.1093/gbe/evt095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nealson K. H., Platt T., Hastings J. W. (1970). Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 104 313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oger P., Farrand S. K. (2001). Co-evolution of the agrocinopine opines and the agrocinopine-mediated control of TraR, the quorum-sensing activator of the Ti plasmid conjugation system. Mol. Microbiol. 41 1173–1185 10.1046/j.1365-2958.2001.02584.x [DOI] [PubMed] [Google Scholar]
- Oger P., Kim K. S., Sackett R. L., Piper K. R., Farrand S. K. (1998). Octopine-type Ti plasmids code for a mannopine-inducible dominant-negative allele of traR, the quorum-sensing activator that regulates Ti plasmid conjugal transfer. Mol. Microbiol. 27 277–288 10.1046/j.1365-2958.1998.00671.x [DOI] [PubMed] [Google Scholar]
- Pappas K. M. (2008). Cell-cell signaling and the Agrobacterium tumefaciens Ti plasmid copy number fluctuations. Plasmid 60 89–107 10.1016/j.plasmid.2008.05.003 [DOI] [PubMed] [Google Scholar]
- Pappas K. M., Winans S. C. (2003a). A LuxR-type regulator from Agrobacterium tumefaciens elevates Ti plasmid copy number by activating transcription of plasmid replication genes. Mol. Microbiol. 48 1059–1073 10.1046/j.1365-2958.2003.03488.x [DOI] [PubMed] [Google Scholar]
- Pappas K. M., Winans S. C. (2003b). The RepA and RepB autorepressors and TraR play opposing roles in the regulation of a Ti plasmid repABC operon. Mol. Microbiol. 49 441–455 10.1046/j.1365-2958.2003.03560.x [DOI] [PubMed] [Google Scholar]
- Parsek M. R., Val D. L., Hanzelka B. L., Cronan J. E., Jr., Greenberg E. P. (1999). Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. U.S.A. 96 4360–4365 10.1073/pnas.96.8.4360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira C. S., Thompson J. A., Xavier K. B. (2013). AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 37 156–181 10.1111/j.1574-6976.2012.00345.x [DOI] [PubMed] [Google Scholar]
- Petit A., Tempe J., Kerr A., Holsters M., Van Montagu M., Schell J. (1978). Substrate induction of conjugative activity of Agrobacterium tumefaciens Ti plasmids. Nature 271 570–572 10.1038/271570a0 [DOI] [Google Scholar]
- Pinto U. M., Pappas K. M., Winans S. C. (2012). The ABCs of plasmid replication and segregation. Nat. Rev. Microbiol. 10 755–765 10.1038/nrmicro2882 [DOI] [PubMed] [Google Scholar]
- Pinto U. M., Winans S. C. (2009). Dimerization of the quorum-sensing transcription factor TraR enhances resistance to cytoplasmic proteolysis. Mol. Microbiol. 73 32–42 10.1111/j.1365-2958.2009.06730.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper K. R., Beck Von Bodman S., Farrand S. K. (1993). Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature 362 448–450 10.1038/362448a0 [DOI] [PubMed] [Google Scholar]
- Piper K. R., Beck Von Bodman S., Hwang I., Farrand S. K. (1999). Hierarchical gene regulatory systems arising from fortuitous gene associations: controlling quorum sensing by the opine regulon in Agrobacterium. Mol. Microbiol. 32 1077–1089 10.1046/j.1365-2958.1999.01422.x [DOI] [PubMed] [Google Scholar]
- Piper K. R., Farrand S. K. (2000). Quorum sensing but not autoinduction of Ti plasmid conjugal transfer requires control by the opine regulon and the antiactivator TraM. J. Bacteriol. 182 1080–1088 10.1128/JB.182.4.1080-1088.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planamente S., Mondy S., Hommais F., Vigouroux A., Morera S., Faure D. (2012). Structural basis for selective GABA binding in bacterial pathogens. Mol. Microbiol. 86 1085–1099 10.1111/mmi.12043 [DOI] [PubMed] [Google Scholar]
- Planamente S., Vigouroux A., Mondy S., Nicaise M., Faure D., Morera S. (2010). A conserved mechanism of GABA binding and antagonism is revealed by structure–function analysis of the periplasmic binding protein Atu2422 in Agrobacterium tumefaciens. J. Biol. Chem. 285 30294–30303 10.1074/jbc.M110.140715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T. G., Bever J. D., Fuqua C. (2012a). A cooperative virulence plasmid imposes a high fitness cost under conditions that induce pathogenesis. Proc. Biol. Sci. 279 1691–1699 10.1098/rspb.2011.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T. G., Fuqua C., Bever J. D. (2012b). Resource and competitive dynamics shape the benefits of public goods cooperation in a plant pathogen. Evolution 66 1953–1965 10.1111/j.1558-5646.2011.01571.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y., Keenan C., Farrand S. K. (2009). N- and C-terminal regions of the quorum-sensing activator TraR cooperate in interactions with the alpha and sigma-70 components of RNA polymerase. Mol. Microbiol. 74 330–346 10.1111/j.1365-2958.2009.06865.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y., Luo Z. Q., Farrand S. K. (2004a). Domains formed within the N-terminal region of the quorum-sensing activator TraR are required for transcriptional activation and direct interaction with RpoA from Agrobacterium. J. Biol. Chem. 279 40844–40851 10.1074/jbc.M405299200 [DOI] [PubMed] [Google Scholar]
- Qin Y., Smyth A. J., Su S., Farrand S. K. (2004b). Dimerization properties of TraM, the antiactivator that modulates TraR-mediated quorum-dependent expression of the Ti plasmid tra genes. Mol. Microbiol. 53 1471–1485 10.1111/j.1365-2958.2004.04216.x [DOI] [PubMed] [Google Scholar]
- Qin Y., Luo Z. Q., Smyth A. J., Gao P., Beck Von Bodman S., Farrand S. K. (2000). Quorum-sensing signal binding results in dimerization of TraR and its release from membranes into the cytoplasm. EMBO J. 19 5212–5221 10.1093/emboj/19.19.5212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y., Su S., Farrand S. K. (2007). Molecular basis of transcriptional antiactivation. TraM disrupts the TraR–DNA complex through stepwise interactions. J. Biol. Chem. 282 19979–19991 10.1074/jbc.M703332200 [DOI] [PubMed] [Google Scholar]
- Seet Q., Zhang L. H. (2011). Antiactivator QslA defines the quorum sensing threshold and response in Pseudomonas aeruginosa. Mol. Microbiol. 80 951–965 10.1111/j.1365-2958.2011.07622.x [DOI] [PubMed] [Google Scholar]
- Stevens A. M., Schuster M., Rumbaugh K. P. (2012). Working together for the common good: cell–cell communication in bacteria. J. Bacteriol. 194 2131–2141 10.1128/JB.00143-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Khan S. R., Farrand S. K. (2008). Induction and loss of Ti plasmid conjugative competence in response to the acyl-homoserine lactone quorum-sensing signal. J. Bacteriol. 190 4398–4407 10.1128/JB.01684-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiderska A., Berndtson A. K., Cha M. R., Li L., Beaudoin G. M., III, Zhu J., et al. (2001). Inhibition of the Agrobacterium tumefaciens TraR quorum-sensing regulator. Interactions with the TraM anti-activator. J. Biol. Chem. 276 49449–49458 10.1074/jbc.M107881200 [DOI] [PubMed] [Google Scholar]
- Tomlinson A. D., Ramey-Hartung B., Day T. W., Merritt P. M., Fuqua C. (2010). Agrobacterium tumefaciens ExoR represses succinoglycan biosynthesis and is required for biofilm formation and motility. Microbiology 156 2670–2681 10.1099/mic.0.039032-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini A., Volpari C, Di Marco S. (2004). Crystal structure of the quorum-sensing protein TraM and its interaction with the transcriptional regulator TraR. J. Biol. Chem. 279 24291–24296 10.1074/jbc.M401855200 [DOI] [PubMed] [Google Scholar]
- Vannini A., Volpari C., Gargioli C., Muraglia E., Cortese R., De Francesco R., et al. (2002). The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 21 4393–4401 10.1093/emboj/cdf459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaudequin-Dransart V. R., Petit A., Chilton W. S., Dessaux Y. (1998). The cryptic plasmid of Agrobacterium tumefaciens cointegrates with the Ti plasmid and cooperates for opine degradation. Mol. Plant Microbe Interact. 11 583–591 10.1094/MPMI.1998.11.7.583 [DOI] [Google Scholar]
- Veselov D., Langhans M., Hartung W., Aloni R., Feussner I., Gotz C., et al. (2003). Development of Agrobacterium tumefaciens C58-induced plant tumors and impact on host shoots are controlled by a cascade of jasmonic acid, auxin, cytokinin, ethylene and abscisic acid. Planta 216 512–522 [DOI] [PubMed] [Google Scholar]
- Wang C., Zhang H. B., Chen G., Chen L., Zhang L. H. (2006a). Dual control of quorum sensing by two TraM-type antiactivators in Agrobacterium tumefaciens octopine strain A6. J. Bacteriol. 188 2435–2445 10.1128/JB.188.7.2435-2445.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Zhang H. B., Wang L. H., Zhang L. H. (2006b). Succinic semialdehyde couples stress response to quorum-sensing signal decay in Agrobacterium tumefaciens. Mol. Microbiol. 62 45–56 10.1111/j.1365-2958.2006.05351.x [DOI] [PubMed] [Google Scholar]
- Watson W. T., Murphy F. V. T., Gould T. A., Jambeck P., Val D. L., Cronan J. E., et al. (2001). Crystallization and rhenium MAD phasing of the acyl-homoserinelactone synthase EsaI. Acta Crystallogr. D. Biol. Crystallogr. 57 1945–1949 10.1107/S0907444901014512 [DOI] [PubMed] [Google Scholar]
- White C. E., Winans S. C. (2005). Identification of amino acid residues of the Agrobacterium tumefaciens quorum-sensing regulator TraR that are critical for positive control of transcription. Mol. Microbiol. 55 1473–1486 10.1111/j.1365-2958.2004.04482.x [DOI] [PubMed] [Google Scholar]
- White C. E., Winans S. C. (2007). The quorum-sensing transcription factor TraR decodes its DNA binding site by direct contacts with DNA bases and by detection of DNA flexibility. Mol. Microbiol. 64 245–256 10.1111/j.1365-2958.2007.05647.x [DOI] [PubMed] [Google Scholar]
- Williams P. (2007). Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology 153(Pt 12) 3923–3938 10.1099/mic.0.2007/012856-0 [DOI] [PubMed] [Google Scholar]
- Wood D. W., Setubal J. C., Kaul R., Monks D. E., Kitajima J. P., Okura V. K., et al. (2001). The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294 2317–2323 10.1126/science.1066804 [DOI] [PubMed] [Google Scholar]
- You Y. S., Marella H., Zentella R., Zhou Y., Ulmasov T., Ho T. H., et al. (2006). Use of bacterial quorum-sensing components to regulate gene expression in plants. Plant Physiol. 140 1205–1212 10.1104/pp.105.074666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z. C., Haudecoeur E., Faure D., Kerr K. F., Nester E. W. (2008). Comparative transcriptome analysis of Agrobacterium tumefaciens in response to plant signal salicylic acid, indole-3-acetic acid and gamma-amino butyric acid reveals signalling cross-talk and Agrobacterium – plant co-evolution. Cell. Microbiol. 10 2339–2354 10.1111/j.1462-5822.2008.01215.x [DOI] [PubMed] [Google Scholar]
- Zarkani A. A., Stein E., Rohrich C. R., Schikora M., Evguenieva-Hackenberg E., Degenkolb T., et al. (2013). Homoserine lactones influence the reaction of plants to rhizobia. Int. J. Mol. Sci. 14 17122–17146 10.3390/ijms140817122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. B., Wang L. H., Zhang L. H. (2002a). Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. U.S.A. 99 4638–4643 10.1073/pnas.022056699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R. G., Pappas K. M., Brace J. L., Miller P. C., Oulmassov T., Molyneaux J. M., et al. (2002b). Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature 417 971–974 10.1038/nature00833 [DOI] [PubMed] [Google Scholar]
- Zhang L., Kerr A. (1991). A diffusible compound can enhance conjugal transfer of the Ti plasmid in Agrobacterium tumefaciens. J. Bacteriol. 173 1867–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Murphy P. J., Kerr A., Tate M. E. (1993). Agrobacterium conjugation and gene regulation by N-acyl-L-homoserine lactones. Nature 362 446–448 10.1038/362446a0 [DOI] [PubMed] [Google Scholar]
- Zhu J., Beaber J. W., More M. I., Fuqua C., Eberhard A., Winans S. C. (1998). Analogs of the autoinducer 3-oxooctanoyl-homoserine lactone strongly inhibit activity of the TraR protein of Agrobacterium tumefaciens. J. Bacteriol. 180 5398–5405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Winans S. C. (1998). Activity of the quorum-sensing regulator TraR of Agrobacterium tumefaciens is inhibited by a truncated, dominant defective TraR-like protein. Mol. Microbiol. 27 289–297 10.1046/j.1365-2958.1998.00672.x [DOI] [PubMed] [Google Scholar]
- Zhu J., Winans S. C. (1999). Autoinducer binding by the quorum-sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proc. Natl. Acad. Sci. U.S.A. 96 4832–4837 10.1073/pnas.96.9.4832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Winans S. C. (2001). The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proc. Natl. Acad. Sci. U.S.A. 98 1507–1512 10.1073/pnas.98.4.1507 [DOI] [PMC free article] [PubMed] [Google Scholar]