Abstract
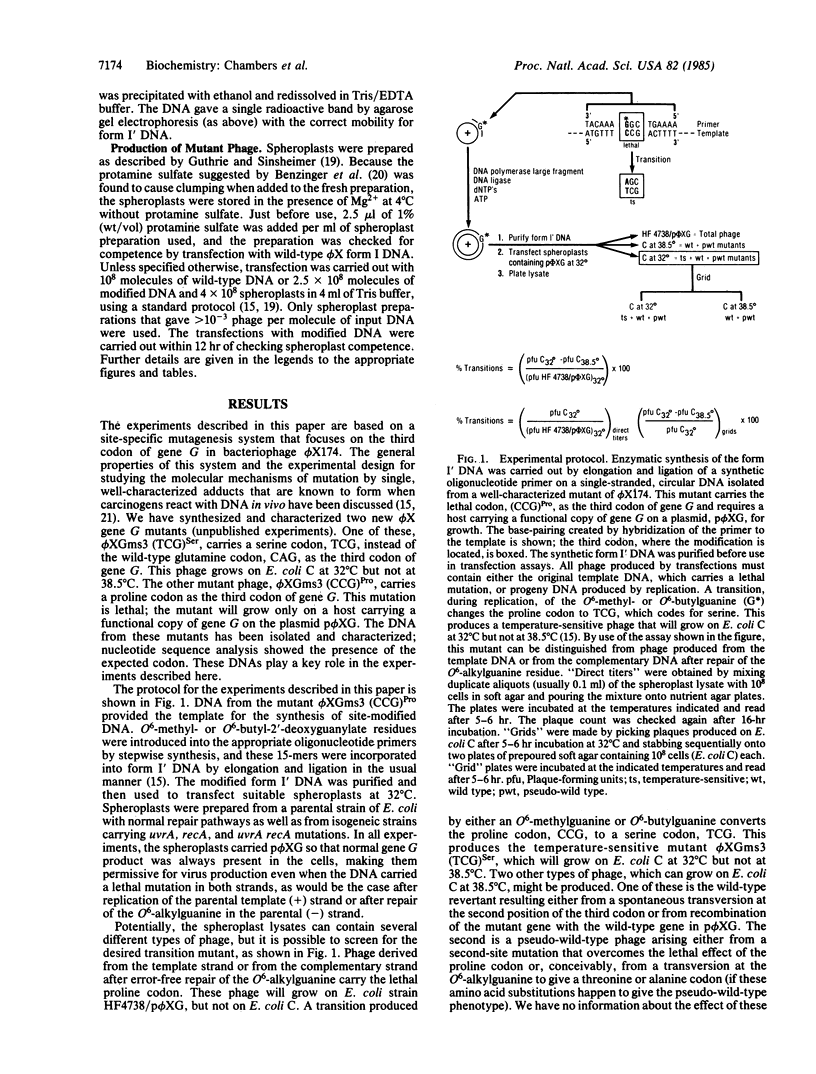
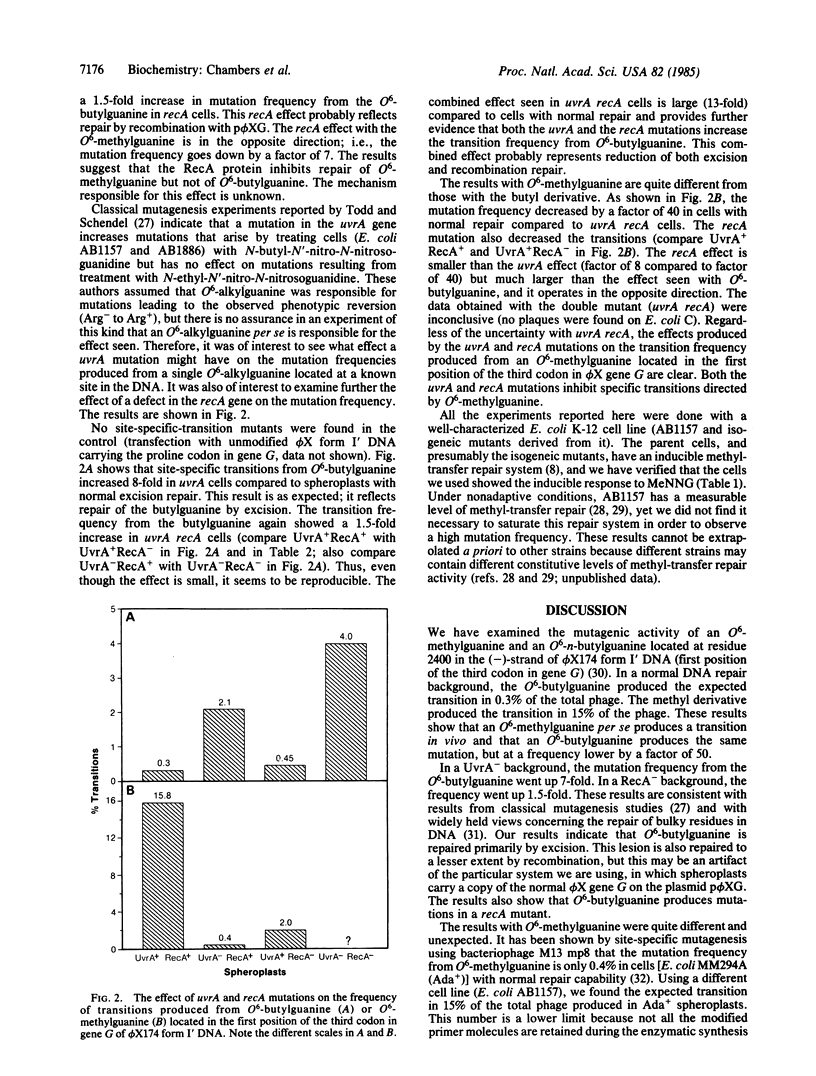
Using site-specific mutagenesis, we have examined the mutagenic activity in vivo of O6-methylguanine or O6-n-butylguanine located at a preselected site in gene G of bacteriophage phi X174. The experiments were designed so that the phage mutant produced by a targeted transition from either of these alkylated derivatives would be recognizable by a simple plaque assay. Spheroplasts derived from normal and repair-deficient cells were transfected, and the lysates were screened for mutant virus. In cells with normal repair, DNA carrying the methylguanine produced the expected transition in 15% of the total phage; DNA carrying the butylguanine produced the same mutation in 0.3% of the phage. In cells deficient in excision repair (uvrA) the transition frequency went up by a factor of 8 for O6-butylguanine and down by a factor of 40 for O6-methylguanine. In cells deficient in recombination (recA), the transition frequency increased 1.5-fold for butylguanine and decreased by a factor of 8 for methylguanine. The data show that both methyl- and butylguanine produce site-directed transitions in phi X174; the transition occurs in recA cells; the frequency of the transition is influenced by both recA and uvrA mutations; the recA and uvrA mutations alter the transition frequency for methylguanine and butylguanine in opposite directions.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benzinger R., Kleber I., Huskey R. Transfection of Escherichia coli spheroplasts. I. General facilitation of double-stranded deoxyribonucleic acid infectivity by protamine sulfate. J Virol. 1971 May;7(5):646–650. doi: 10.1128/jvi.7.5.646-650.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhanot O. S., Khan S. A., Chambers R. W. A new system for studying molecular mechanisms of mutation by carcinogens. J Biol Chem. 1979 Dec 25;254(24):12684–12693. [PubMed] [Google Scholar]
- Cairns J., Robins P., Sedgwick B., Talmud P. The inducible repair of alkylated DNA. Prog Nucleic Acid Res Mol Biol. 1981;26:237–244. doi: 10.1016/s0079-6603(08)60408-0. [DOI] [PubMed] [Google Scholar]
- Chambers R. W., Kućan I., Kućan Z. Isolation and characterization of phi X174 mutants carrying lethal missense mutations in gene G. Nucleic Acids Res. 1982 Oct 25;10(20):6465–6473. doi: 10.1093/nar/10.20.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers R. W. Site-specific mutagenesis: a new approach for studying the molecular mechanisms of mutation by carcinogens. Basic Life Sci. 1982;20:121–145. doi: 10.1007/978-1-4613-3476-7_7. [DOI] [PubMed] [Google Scholar]
- Foote R. S., Mitra S., Pal B. C. Demethylation of O6-methylguanine in a synthetic DNA polymer by an inducible activity in Escherichia coli. Biochem Biophys Res Commun. 1980 Nov 28;97(2):654–659. doi: 10.1016/0006-291x(80)90314-9. [DOI] [PubMed] [Google Scholar]
- GUTHRIE G. D., SINSHEIMER R. L. Observations on the infection of bacterial protoplasts with the deoxyribonucleic acid of bacteriophage phi X174. Biochim Biophys Acta. 1963 Jun 25;72:290–297. [PubMed] [Google Scholar]
- Gaffney B. L., Marky L. A., Jones R. A. Synthesis and characterization of a set of four dodecadeoxyribonucleoside undecaphosphates containing O6-methylguanine opposite adenine, cytosine, guanine, and thymine. Biochemistry. 1984 Nov 20;23(24):5686–5691. doi: 10.1021/bi00319a004. [DOI] [PubMed] [Google Scholar]
- Howard-Flanders P., Boyce R. P., Theriot L. Three loci in Escherichia coli K-12 that control the excision of pyrimidine dimers and certain other mutagen products from DNA. Genetics. 1966 Jun;53(6):1119–1136. doi: 10.1093/genetics/53.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humayun M. Z., Chambers R. W. Construction and characterization of an Escherichia coli plasmid bearing a functional gene G of bacteriophage phiX174. Proc Natl Acad Sci U S A. 1978 Feb;75(2):774–778. doi: 10.1073/pnas.75.2.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humayun M. Z., Chambers R. W. Construction of a site-specific, deletion-frameshift mutation in an essential gene of bacteriophage phiX174. Nature. 1979 Apr 5;278(5704):524–529. doi: 10.1038/278524a0. [DOI] [PubMed] [Google Scholar]
- Karran P., Lindahl T., Griffin B. Adaptive response to alkylating agents involves alteration in situ of O6-methylguanine residues in DNA. Nature. 1979 Jul 5;280(5717):76–77. doi: 10.1038/280076a0. [DOI] [PubMed] [Google Scholar]
- Kuzmich S., Marky L. A., Jones R. A. Specifically alkylated DNA fragments. Synthesis and physical characterization of d[CGC(O6Me)GCG] and d[CGT(O6Me)GCG]. Nucleic Acids Res. 1983 May 25;11(10):3393–3403. doi: 10.1093/nar/11.10.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loechler E. L., Green C. L., Essigmann J. M. In vivo mutagenesis by O6-methylguanine built into a unique site in a viral genome. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6271–6275. doi: 10.1073/pnas.81.20.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins P., Cairns J. Quantitation of the adaptive response to alkylating agents. Nature. 1979 Jul 5;280(5717):74–76. doi: 10.1038/280074a0. [DOI] [PubMed] [Google Scholar]
- Rupp W. D., Sancar A., Sancar G. B. Properties and regulation of the UVRABC endonuclease. Biochimie. 1982 Aug-Sep;64(8-9):595–598. doi: 10.1016/s0300-9084(82)80094-1. [DOI] [PubMed] [Google Scholar]
- Samson L., Cairns J. A new pathway for DNA repair in Escherichia coli. Nature. 1977 May 19;267(5608):281–283. doi: 10.1038/267281a0. [DOI] [PubMed] [Google Scholar]
- Sancar A., Franklin K. A., Sancar G. B. Escherichia coli DNA photolyase stimulates uvrABC excision nuclease in vitro. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7397–7401. doi: 10.1073/pnas.81.23.7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Rupp W. D. A novel repair enzyme: UVRABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell. 1983 May;33(1):249–260. doi: 10.1016/0092-8674(83)90354-9. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Friedmann T., Air G. M., Barrell B. G., Brown N. L., Fiddes J. C., Hutchison C. A., 3rd, Slocombe P. M., Smith M. The nucleotide sequence of bacteriophage phiX174. J Mol Biol. 1978 Oct 25;125(2):225–246. doi: 10.1016/0022-2836(78)90346-7. [DOI] [PubMed] [Google Scholar]
- Schendel P. F., Robins P. E. Repair of O6-methylguanine in adapted Escherichia coli. Proc Natl Acad Sci U S A. 1978 Dec;75(12):6017–6020. doi: 10.1073/pnas.75.12.6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeberg E. Multiprotein interactions in strand cleavage of DNA damaged by UV and chemicals. Prog Nucleic Acid Res Mol Biol. 1981;26:217–226. doi: 10.1016/s0079-6603(08)60406-7. [DOI] [PubMed] [Google Scholar]
- Seeberg E. Reconstitution of an Escherichia coli repair endonuclease activity from the separated uvrA+ and uvrB+/uvrC+ gene products. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2569–2573. doi: 10.1073/pnas.75.6.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha N. K., Haimes M. D. Molecular mechanisms of substitution mutagenesis. An experimental test of the Watson-Crick and topal-fresco models of base mispairings. J Biol Chem. 1981 Oct 25;256(20):10671–10683. [PubMed] [Google Scholar]
- Tanaka T., Letsinger R. L. Syringe method for stepwise chemical synthesis of oligonucleotides. Nucleic Acids Res. 1982 May 25;10(10):3249–3260. doi: 10.1093/nar/10.10.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo I., Sedgwick B., Demple B., Li B., Lindahl T. Induction of resistance to alkylating agents in E. coli: the ada+ gene product serves both as a regulatory protein and as an enzyme for repair of mutagenic damage. EMBO J. 1984 Sep;3(9):2151–2157. doi: 10.1002/j.1460-2075.1984.tb02105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd M. L., Schendel P. F. Repair and mutagenesis in Escherichia coli K-12 after exposure to various alkyl-nitrosoguanidines. J Bacteriol. 1983 Oct;156(1):6–12. doi: 10.1128/jb.156.1.6-12.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toorchen D., Topal M. D. Mechanisms of chemical mutagenesis and carcinogenesis: effects on DNA replication of methylation at the O6-guanine position of dGTP. Carcinogenesis. 1983 Dec;4(12):1591–1597. doi: 10.1093/carcin/4.12.1591. [DOI] [PubMed] [Google Scholar]
- Topal M. D., Fresco J. R. Complementary base pairing and the origin of substitution mutations. Nature. 1976 Sep 23;263(5575):285–289. doi: 10.1038/263285a0. [DOI] [PubMed] [Google Scholar]
- Van Lier J. J., Smits M. T., Buck H. M. B-Z transition in methylated DNA. A quantum-chemical study. Eur J Biochem. 1983 Apr 15;132(1):55–62. doi: 10.1111/j.1432-1033.1983.tb07324.x. [DOI] [PubMed] [Google Scholar]



