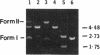
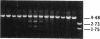
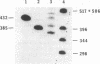
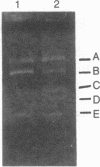
Abstract
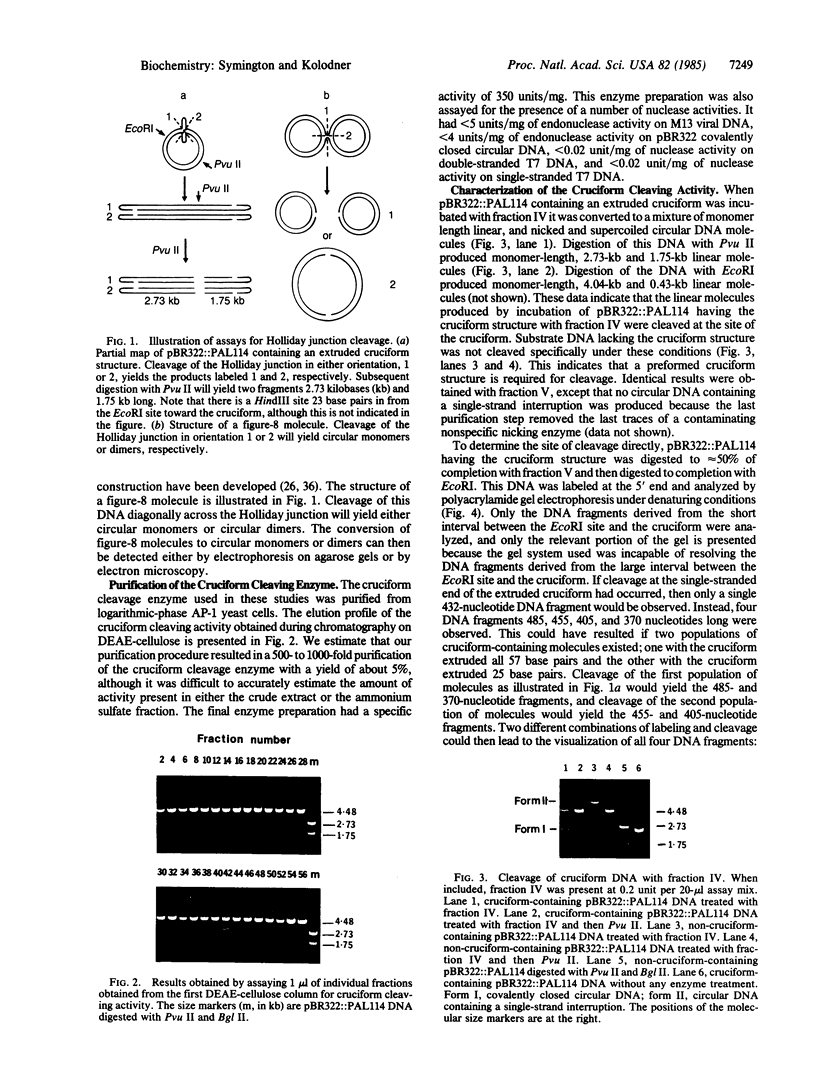
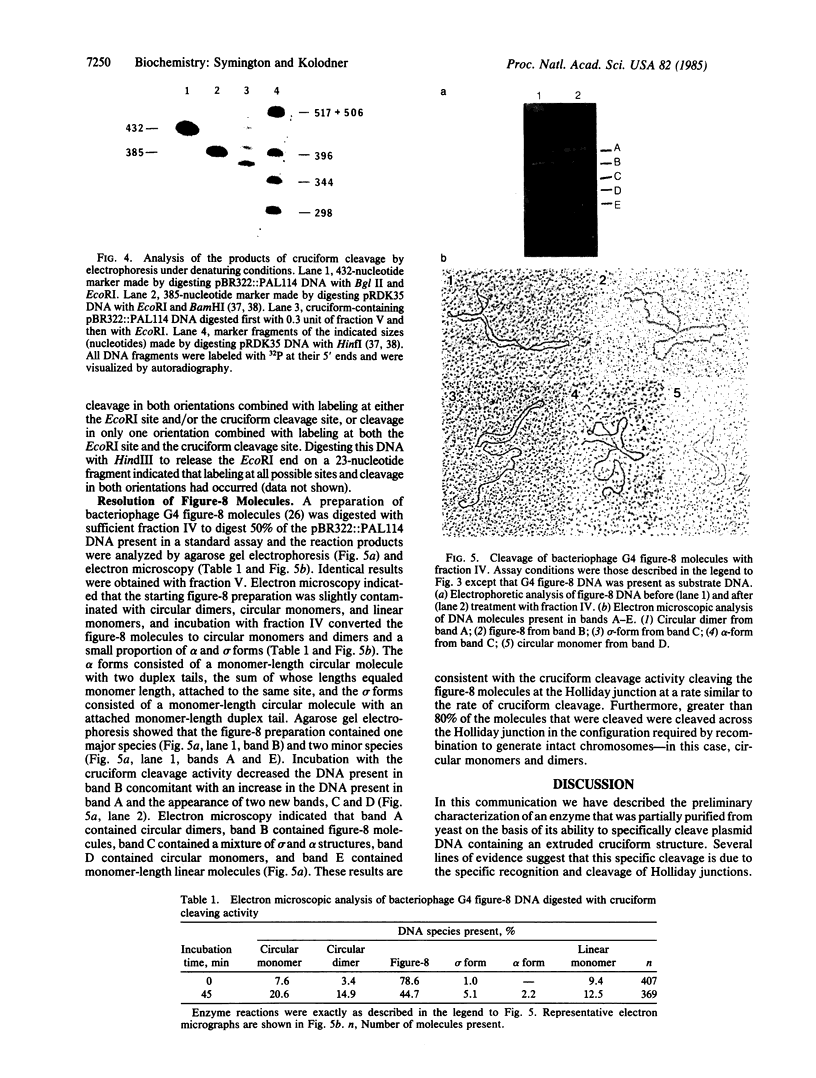
An enzyme from Saccharomyces cerevisiae that cleaves Holliday junctions was partially purified approximately 500- to 1000-fold by DEAE-cellulose chromatography, gel filtration on Sephacryl S300, and chromatography on single-stranded DNA-cellulose. The partially purified enzyme did not have any detectable nuclease activity when tested with single-stranded or double-stranded bacteriophage T7 substrate DNA and did not have detectable endonuclease activity when tested with bacteriophage M13 viral DNA or plasmid pBR322 covalently closed circular DNA. Analysis of the products of the cruciform cleavage reaction by electrophoresis on polyacrylamide gels under denaturing conditions revealed that the cruciform structure was cleaved at either of two sites present in the stem of the cruciform and was not cleaved at the end of the stem. The cruciform cleavage enzyme was able to cleave the Holliday junction present in bacteriophage G4 figure-8 molecules. Eighty percent of these Holliday junctions were cleaved in the proper orientation to generate intact chromosomes during genetic recombination.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bell L. R., Byers B. Homologous association of chromosomal DNA during yeast meiosis. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):829–840. doi: 10.1101/sqb.1983.047.01.095. [DOI] [PubMed] [Google Scholar]
- Bell L., Byers B. Occurrence of crossed strand-exchange forms in yeast DNA during meiosis. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3445–3449. doi: 10.1073/pnas.76.7.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benbow R. M., Zuccarelli A. J., Sinsheimer R. L. Recombinant DNA molecules of bacteriophage phi chi174. Proc Natl Acad Sci U S A. 1975 Jan;72(1):235–239. doi: 10.1073/pnas.72.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B., Goetsch L. Reversible pachytene arrest of Saccharomyces cerevisiae at elevated temperature. Mol Gen Genet. 1982;187(1):47–53. doi: 10.1007/BF00384382. [DOI] [PubMed] [Google Scholar]
- Center M. S., Richardson C. C. An endonuclease induced after infection of Escherichia coli with bacteriophage T7. I. Purification and properties of the enzyme. J Biol Chem. 1970 Dec 10;245(23):6285–6291. [PubMed] [Google Scholar]
- Cunningham R. P., Wu A. M., Shibata T., DasGupta C., Radding C. M. Homologous pairing and topological linkage of DNA molecules by combined action of E. coli RecA protein and topoisomerase I. Cell. 1981 Apr;24(1):213–223. doi: 10.1016/0092-8674(81)90517-1. [DOI] [PubMed] [Google Scholar]
- Doherty M. J., Morrison P. T., Kolodner R. Genetic recombination of bacterial plasmid DNA. Physical and genetic analysis of the products of plasmid recombination in Escherichia coli. J Mol Biol. 1983 Jul 5;167(3):539–560. doi: 10.1016/s0022-2836(83)80097-7. [DOI] [PubMed] [Google Scholar]
- Doniger J., Warner R. C., Tessma I. Role of circular dimer DNA in the primary recombination mechanism of bacteriophage S13. Nat New Biol. 1973 Mar 7;242(114):9–12. doi: 10.1038/newbio242009a0. [DOI] [PubMed] [Google Scholar]
- Gellert M., O'Dea M. H., Mizuuchi K. Slow cruciform transitions in palindromic DNA. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5545–5549. doi: 10.1073/pnas.80.18.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T. S., Wang J. C. Thermodynamic properties of superhelical DNAs. Biochemistry. 1975 Feb 11;14(3):527–535. doi: 10.1021/bi00674a011. [DOI] [PubMed] [Google Scholar]
- Hsu P. L., Landy A. Resolution of synthetic att-site Holliday structures by the integrase protein of bacteriophage lambda. Nature. 1984 Oct 25;311(5988):721–726. doi: 10.1038/311721a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James A. A., Morrison P. T., Kolodner R. Genetic recombination of bacterial plasmid DNA. Analysis of the effect of recombination-deficient mutations on plasmid recombination. J Mol Biol. 1982 Sep 25;160(3):411–430. doi: 10.1016/0022-2836(82)90305-9. [DOI] [PubMed] [Google Scholar]
- Kemper B., Brown D. T. Function of gene 49 of bacteriophage T4. II. Analysis of intracellular development and the structure of very fast-sedimenting DNA. J Virol. 1976 Jun;18(3):1000–1015. doi: 10.1128/jvi.18.3.1000-1015.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper B., Janz E. Function of gene 49 of bacteriophage T4. I. Isolation and biochemical characterization of very fast-sedimenting DNA. J Virol. 1976 Jun;18(3):992–999. doi: 10.1128/jvi.18.3.992-999.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiec E. B., Kroeger P. E., Brougham M. J., Holloman W. K. Topological linkage of circular DNA molecules promoted by Ustilago rec 1 protein and topoisomerase. Cell. 1983 Oct;34(3):919–929. doi: 10.1016/0092-8674(83)90549-4. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lee D., Sadowski P. D. Genetic recombination of bacteriophage T7 in vivo studied by use of a simple physical assay. J Virol. 1981 Dec;40(3):839–847. doi: 10.1128/jvi.40.3.839-847.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Meselson M. S., Radding C. M. A general model for genetic recombination. Proc Natl Acad Sci U S A. 1975 Jan;72(1):358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K., Kemper B., Hays J., Weisberg R. A. T4 endonuclease VII cleaves holliday structures. Cell. 1982 Jun;29(2):357–365. doi: 10.1016/0092-8674(82)90152-0. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Panet A., van de Sande J. H., Loewen P. C., Khorana H. G., Raae A. J., Lillehaug J. R., Kleppe K. Physical characterization and simultaneous purification of bacteriophage T4 induced polynucleotide kinase, polynucleotide ligase, and deoxyribonucleic acid polymerase. Biochemistry. 1973 Dec 4;12(25):5045–5050. doi: 10.1021/bi00749a003. [DOI] [PubMed] [Google Scholar]
- Potter H., Dressler D. On the mechanism of genetic recombination: electron microscopic observation of recombination intermediates. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3000–3004. doi: 10.1073/pnas.73.9.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C. C. The 5'-terminal nucleotides of T7 bacteriophage deoxyribonucleic acid. J Mol Biol. 1966 Jan;15(1):49–61. doi: 10.1016/s0022-2836(66)80208-5. [DOI] [PubMed] [Google Scholar]
- Sigal N., Alberts B. Genetic recombination: the nature of a crossed strand-exchange between two homologous DNA molecules. J Mol Biol. 1972 Nov 28;71(3):789–793. doi: 10.1016/s0022-2836(72)80039-1. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Symington L. S., Fogarty L. M., Kolodner R. Genetic recombination of homologous plasmids catalyzed by cell-free extracts of Saccharomyces cerevisiae. Cell. 1983 Dec;35(3 Pt 2):805–813. doi: 10.1016/0092-8674(83)90113-7. [DOI] [PubMed] [Google Scholar]
- Symington L. S., Morrison P. T., Kolodner R. Genetic recombination catalyzed by cell-free extracts of Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 1984;49:805–814. doi: 10.1101/sqb.1984.049.01.091. [DOI] [PubMed] [Google Scholar]
- Symington L. S., Morrison P., Kolodner R. Plasmid recombination intermediates generated in a Saccharomyces cerevisiae cell-free recombination system. Mol Cell Biol. 1985 Sep;5(9):2361–2368. doi: 10.1128/mcb.5.9.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Thompson B. J., Camien M. N., Warner R. C. Kinetics of branch migration in double-stranded DNA. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2299–2303. doi: 10.1073/pnas.73.7.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B. J., Escarmis C., Parker B., Slater W. C., Doniger J., Tessman I., Warner R. C. Figure-8 configuration of dimers of S13 and phiX174 replicative form DNA. J Mol Biol. 1975 Feb 5;91(4):409–419. doi: 10.1016/0022-2836(75)90269-7. [DOI] [PubMed] [Google Scholar]
- Thuring R. W., Sanders J. P., Borst P. A freeze-squeeze method for recovering long DNA from agarose gels. Anal Biochem. 1975 May 26;66(1):213–220. doi: 10.1016/0003-2697(75)90739-3. [DOI] [PubMed] [Google Scholar]
- Tsujimoto Y., Ogawa H. Intermediates in genetic recombination of bacteriophage T7 DNA. J Mol Biol. 1977 Jan 25;109(3):423–426. doi: 10.1016/s0022-2836(77)80021-1. [DOI] [PubMed] [Google Scholar]
- Valenzuela M. S., Inman R. B. Visualization of a novel junction in bacteriophage lambda DNA. Proc Natl Acad Sci U S A. 1975 Aug;72(8):3024–3028. doi: 10.1073/pnas.72.8.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G. J., Green R. L. Comparison of physical and genetic properties of palindromic DNA sequences. J Bacteriol. 1985 Mar;161(3):1103–1111. doi: 10.1128/jb.161.3.1103-1111.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss B., Jacquemin-Sablon A., Live T. R., Fareed G. C., Richardson C. C. Enzymatic breakage and joining of deoxyribonucleic acid. VI. Further purification and properties of polynucleotide ligase from Escherichia coli infected with bacteriophage T4. J Biol Chem. 1968 Sep 10;243(17):4543–4555. [PubMed] [Google Scholar]
- de Massy B., Studier F. W., Dorgai L., Appelbaum E., Weisberg R. A. Enzymes and sites of genetic recombination: studies with gene-3 endonuclease of phage T7 and with site-affinity mutants of phage lambda. Cold Spring Harb Symp Quant Biol. 1984;49:715–726. doi: 10.1101/sqb.1984.049.01.081. [DOI] [PubMed] [Google Scholar]