Abstract
Although mesenchymal stem cells (MSCs) play pivotal supportive roles in hematopoiesis, how they interact with hematopoietic stem cells (HSCs) is not well understood. We investigated the interaction between HSCs and surrogate MSCs (C3H10T1/2 stromal cells), focusing on the molecular events induced by cell contact of these bipartite populations. C3H10T1/2 is a mesenchymal stromal cell line that can be induced to differentiate into preadipocytes (A54) and myoblasts (M1601). The stromal cell derivatives were cocultured with murine HSCs (Lineage-Sca1+), and gene expression profiles in stromal cells and HSCs were compared before and after the coculture. HSCs gave rise to cobblestone areas only on A54 cells, with ninefold more progenitors than on M1601 or undifferentiated C3H10T1/2 cells. Microarray-based screening and a quantitative reverse transcriptase directed-polymerase chain reaction showed that the levels of Notch ligands (Jagged1 and Delta-like 3) were increased in A54 cells upon interaction with HSCs. On the other hand, the expression of Notch1 and Hes1 was upregulated in the HSCs cocultured with A54 cells. A transwell assay revealed that the reciprocal upregulation was dependent on cell-to-cell contact. The result suggested that in the hematopoietic niche, HSCs help MSCs to produce Notch ligands, and in turn, MSCs help HSCs to express Notch receptor. Such a reciprocal upregulation would reinforce the downstream signaling to determine the fate of hematopoietic cell lineage. Clarification of the initiating events on cell contact should lead to the identification of specific molecular targets to facilitate HSC engraftment in transplantation therapy.
Keywords: mesenchymal stromal (stem) cell, hematopoietic stem cell, cell contact, Notch signaling
Introduction
Hematopoietic stem cells (HSCs) are capable of self-renewing and differentiating into all the blood cell lineages, and the property allows them to reconstitute adult hematopoiesis following transplantation. The growth and differentiation of HSCs is regulated by orchestrated signals from various soluble factors and the hematopoietic microenvironment, or ‘niche’. With the aid of many other cell types, osteoblasts and vascular endotherial cells maintain the balance of dormant and active HSCs in the osteoblasic and vascular niches [1-4].
The interaction between HSCs and the niche cells comprises cytokines and cell-to-cell contact. The involved cytokines include stem cell factor (SCF), stromal-derived factor 1 (SDF1), angiopoietin1 (Ang1) and osteopontin, and the functions of these factors have been studied extensively [5-9]. On the other hand, molecular events of the direct cell contact are mostly unclear. Wagner et al. investigated the behavioral and molecular changes in hematopoietic progenitors upon interaction with a stromal cell line AFT024 [10]. They found that the genes involved in the cytoskeleton reorganization, DNA stabilization and methylation were upregulated. However, molecular events in the niche cells have not vigorously explored.
Mesenchymal stem cells (MSCs) in the bone marrow play a vital role in supporting hematopoiesis, therefore they are considered as niche cells, too [11,12]. To further explore the hematopoiesis-supporting ability of MSCs, we have used a surrogate MSC line C3H10T1/2 (10T1/2) and its derivative preadipocytes (A54) and myoblasts (M1601). Among these cells, only A54 preadipocytes helped the expansion of hematopoietic progenitors with an augmented production of SCF, SDF1 and Ang1 [13,14]. In the present study, we investigated the cellular and molecular events in the interactive communication between HSCs and stromal cells using this differentiation-inducible system, particularly focusing on the changes in the stromal cells.
Materials and methods
Cells
10T1/2 cell line (from Riken Biological Resource Center, Tsukuba, Japan) was used as an inducible MSC model. A54 preadipocytes and M1601 myoblasts were established as described previously [13]. All the cell lines were cultured in Iscove's Modified Dulbecco's Medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Sigma, St. Louis, MO, USA). Bone marrow mononuclear cells were separated from C57BL/6 mouse femurs using Lympholyte-M (Cedarlane, Hornby, ON, Canada), and serially fractionated with immunomicrobeads and AutoMACS (Miltenyi Biotech, Bergisch Gradbach, Germany). Lineage-negative (Lin-) and –positive (Lin+) cells were separated with a Lineage Cell Depletion Kit (Miltenyi Biotech). Sca1-, CD45R (B220)- and CD90 (Thy1.2)-positive cells (Sca1+, B220+ and Thy1.2+ cells, respectively) were separated with appropriate microbeads (Miltenyi Biotech). Lin-Sca1+ cells were used as HSCs. Lin+B220+ cells and Lin+Thy1.2+ cells were used as B-lymphocytes and T-lymphocytes, respectively.
Coculture of HSCs and stromal cells
10T1/2, A54 and M1601 stromal cells were inoculated (5 × 103 cells/well) on a 12-well culture plate (Falcon 3043, BD, Franklin Lakes, NJ, USA). After 2 days, the near-confluent cells (ca. 2 × 104 cells/well) were gamma-irradiated (30 Gy) using a Gammacell 40 Exactor (Nordion International, Ottawa, ON, Canada) to prevent hyperproliferation. Then 1 × 104 HSCs (Lin-Sca1+ cells) were added to the irradiated stromal cells and cocultured for 5 days in α-Minimum Essential Medium (Invitrogen) supplemented with 10% FBS. A coculture was also carried out using a Cell Culture Insert (Falcon 353090, BD) on a 6-well culture plate (Falcon 3046, BD) for 5 days, with HSCs seeded in the upper wells and stromal cells in the lower wells. After coculture, cells were examined for cobblestone formation (30 or more clustered HSCs) on an inverted fluorescent microscope (Olympus IX-70, Tokyo, Japan), and the stromal cells and HSCs were separated with MACSelect Kk microbeads (Miltenyi Biotech) and AutoMACS for further analyses. The stromal cells derived from C3H mouse (H-2k) were separated as a positive fraction, and HSCs derived from C57BL/6 mouse (H-2b) were separated as a negative fraction. The separated cells were stained with a phycoerythrin-conjugated anti-mouse H-2Kk antibody and a fluorescein isothiocyanate-conjugated anti-mouse H-2Kb antibody (BD Biosciences, San Jose, CA, USA), and the purity was evaluated with BD LSR and CellQuest software (BD Immunosystems, Mountain View, CA, USA). For colony asaay after coculture, separated HSCs were cultivated (1 × 103 cells/35 mm dish) in growth factor-containing methylcellulose medium (MethoCult GF M3434, StemCell Technologies, Vancouver, BC, Canada) and colonies were counted on day 7.
Gene expression analysis
To quantify several signaling molecules, the reverse transcriptase-directed polymerase chain reaction (RT-PCR) was performed. Total RNA was extracted from the cells using an RNeasy Mini Kit (QIAGEN, Hilden, Germany), and cDNA was synthesized with a SuperScript First-Strand Synthesis Kit (Invitrogen). PCR was carried out with a QuantiTect SYBR Green PCR Kit (QIAGEN), and the incorporation of the fluorescent dye into the PCR products was monitored with ABI Prism 7700 (Applied Biosystems, Foster City, CA, USA). As internal controls, beta-actin was used for the stromal cells and glyceraldehyde-3-phosphate dehydrogenase was used for HSCs. The primers for PCR are listed in Table 1. Gene expression profiles of A54 before and after coculture with HSCs were compared using a FilgenArray Mouse 32K (Filgen, Nagoya, Japan).
Table 1.
RT-PCR primer design.
| Target | Forward primer | Reverse primer |
| Ang1 | CCAATCTAAATGGAATGTTCT | CAGAGCACCTTCAAAAGTCCA |
| beta-actin | CCATCATGAAGTGTGACGTTG | GTCCGCCTAGAAGCACTTGCG |
| BMP6 | TTCTCCCCACATCAACGACACC | AAACTCCCCACCACACAGTCC |
| Dll3 | CCAGTAGCTGCCTGAACTCC | ATTGAAGCAGGGTCCATCTG |
| GAPDH | CCTGGAGAAACCTGCCAAGTATG | AGAGTGGGAGTTGCTGTTGAAGTC |
| Hes1 | AAAGCCTATCATGGAGAAGAGGCG | GGAATGCCGGGAGCTATCTTTCTT |
| Jag1 | CCGTAATCGCATCGTACTGC | GGCCTCCACCAGCAAAGTGT |
| Notch1 | TGTTAATGAGTGCATCTCCAACCCA | CATTCGTAGCCATCAATCTTGTCC |
| SCF | ATGGACAGCCATGGCATTGC | CACCTCTTGAAATTCTCTCT |
| SDF1 | GCCCTTCAGATTGTTGCA | CGTCTGACTCACACCTCACA |
Immunostaining
A54 cells and HSCs were cocultured on Culture Slides (Falcon 354111, BD), and immunostained on day 5. The cells were fixed with 4% paraformaldehyde, and incubated with a rabbit anti-mouse Jag1 antibody (ab7771, Abcam, Cambridge, UK) and Alexa Fluor 488-conjugated goat anti-rabbit IgG (A11008, Invitrogen) for 30 minutes on ice, respectively. Slides were inspected and photographed under an IX-70 microscope.
Statistical analysis
The statistical analysis was performed using a Statcel2 (OMS Publishing, Saitama, Japan) add-on package for Microsoft Excel. Student's t-test was used for comparison between two groups, and a multiple comparison test (the Turkey-Kramer method) was used for comparison among groups of three or more. A P-value < 0.05 was considered to be significant for all analyses.
Results
Requirement of cell contact in supporting hematopoeisis
In coculure experiments with 10T1/2 or its derivatives (A54 and M1601), HSCs formed cobblestone areas only on A54 at day 5 (12.5 ± 0.6 cobblestone areas/104 cells), as we reported previously [14]. We also confirmed that the murine hematopoietic progenitors were expanded by ninefold (36.0 ± 4.0 colonies/103 cells post-coculture with A54 vs. 4.0 ± 2.0 colonies/103 cells pre-coculture, Figure 1) whereas the number of hematopoietic progenitors was not significantly increased after the coculture with parental 10T1/2 cells or M1601 myoblasts (3.3 ± 1.5 colonies/103 cells with 10T1/2, and 6.3 ± 1.5 colonies/103 cells with M1601; Figure 1). Without stromal cell support, HSCs completely lost their progenitor activity in the same medium used for coculture (‘No stroma’ in Figure 1). Indeed, only few monocyte/macrophage-like cells were found after 5-day-incubation in this population; we speculated that all the clonogenic cells fell into apoptosis because no additional cytokine was provided. As we previously reported, expression of SCF, SDF1 and Ang1 in A54 was greater than that in 10T1/2 and M1601cells [14]. Such an overexpression of SCF, SDF1 and Ang1 in A54 cells was not further augmented or cancelled by addition of interleukin-1 or coculture with HSCs (data not shown).
Figure 1.
Requirement of direct contact for preadipocytes to support hematopoiesis. 1 × 104 mouse bone marrow Lin-Sca1+ cells (HSCs) were cocultured with 2 × 104 C3H10T1/2 cells (10T1/2), 10T1/2-derived myoblasts (M1602) or preadipocytes (A54) for 5 days. After coculture, HSCs were separated with immunomagnetic beads and subjected to colony assay. HSCs were seeded on MethoCult GF M3434 (1 × 103 cells/dish) and colonies were counted on day 7. Preculture: colonies from HSCs prior to coculture. No stroma: colonies from HSCs incubated 5 days without stromal support. No stroma/Transwell: colonies from HSCs seeded on the top wells, with no stromal cells in the bottom wells. A54/Transwell: HSCs were coincubated with A54 but separated by Cell Culture Insert. *: P < 0.01 (n = 3).
Next, we addressed whether soluble factors from A54 preadipocytes were sufficient for supporting hematopoiesis. To discriminate the contribution of cell communication via direct adhesion, mouse bone marrow-derived HSCs were cultivated on a microporous membrane (1 micron Cell Culture Insert) placed in a well containing A54 cells underneath. We found that physical separation from A54 cells totally abrogated the progenitor activity of HSCs, in spite of the availability of soluble molecules from the stromal cells (Figure 1). This observation indicated that A54 cells supported hematopoiesis by cell contact in addition to secreting cytokines.
Gene expression changes in preadipocytes cocultured with HSCs
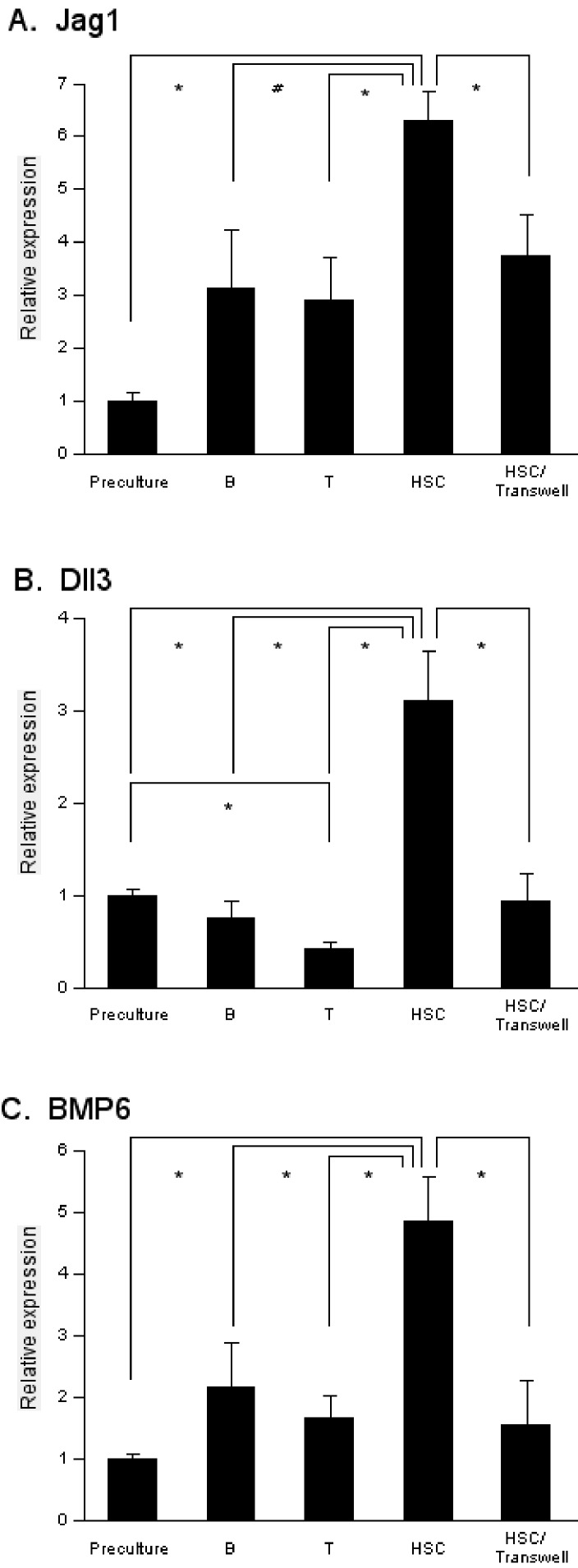
Upon learning that cell adhesion is required for A54 cells to support HSCs, we speculated that some signaling from HSCs to stromal cells might occur during coculture. This idea prompted us to search for molecules whose expression changed in A54 cells cocultured with HSCs. Total RNA was extracted from A54 cells before and after the coculture and gene expression profiles were compared by microarray. Out of 33696 genes screened, 353 genes were upregulated by twofold or more after the coculture with HSCs, and 13 genes were downregulated to the levels less than a half of starting value (data not shown). Among them, 29 genes (21 upregulated and 8 downregulated; Table 2) were implicated to be involved in hematopoiesis. Therefore we focused on these molecules for further investigation. A quantitative RTPCR analysis confirmed the upregulation of Jagged 1 (Jag1), Delta-like 3 (Dll3) and bone morphogenetic protein 6 (BMP6) in A54 cells after coculture with HSCs (Figure 2). On the other hand, upregulation of Jag1, Dll3 and BMP6 was not observed in the undifferentiated 10T1/2 cells or M1601 myoblasts after coculture with HSCs (data not shown). Expression of Jag1 was also increased in A54 cells cultured with differentiated cells such as B- and T-lymphocytes, but to a lesser extent (Figure 2A).
Table 2.
Hematopoiesis-related genes upregulated or downregulated in cocultured A54 cells.
| Upregulated or downregulated molecule | Beforea | Afterb | Ratioc |
| Upregulated | |||
| Mcl1 (Myeloid cell leukemia sequence 1) | 116 | 514 | 4.42 |
| Stom (Stomatin) | 126 | 481 | 3.83 |
| Foxk1 (Forkhead box K1) | 129 | 464 | 3.58 |
| Foxf2 (Forkhead box F2) | 133 | 455 | 3.43 |
| Bmp6 (Bone morphogenetic protein-6) | 133 | 451 | 3.38 |
| Itgbl1 (Integrin, beta-like 1) | 135 | 447 | 3.30 |
| Dll3 (Delta-like 3) | 136 | 446 | 3.29 |
| Arts1 (Adipocyte-derived leucine aminopeptidase) | 136 | 446 | 3.28 |
| Sox13 (SRY-box containing gene 13) | 136 | 444 | 3.27 |
| Cdc40 (Cell division cycle 40 homolog) | 136 | 443 | 3.26 |
| Jag1 (Jagged 1) | 137 | 443 | 3.24 |
| Shh (Sonic hedgehog) | 137 | 440 | 3.20 |
| Cdc16 (Cell division cycle 16 homolog) | 143 | 423 | 2.95 |
| Sox17 (SRY-box containing gene 17) | 146 | 416 | 2.85 |
| Cdc2l6 (Cell division cycle 2-like 6) | 144 | 408 | 2.83 |
| Col15a1 (procollagen, type XV) | 147 | 408 | 2.77 |
| Rras (Ras-related protein) | 149 | 405 | 2.72 |
| Pik3cb (phosphatidylinositol3-kinase, catalytic, b polypeptide) | 147 | 401 | 2.72 |
| Hoxa3 (Homeobox A3) | 149 | 378 | 2.54 |
| Adfp (Adipophilin, Adipose differentiation-related protein) | 150 | 361 | 2.41 |
| Cdc91l1 (cell division cycle 91-like 1) | 146 | 324 | 2.22 |
| Downregulated | |||
| Nudt2 (nucleoside diphosphate linked moiety X - type motif 2) | 3155 | 1337 | 0.42 |
| Lrp2 (LDL receptor-related protein 2) | 2210 | 963 | 0.44 |
| TPO (thrombopoietin) | 866 | 419 | 0.48 |
| Taar1 (trace amine-associated receptor 1) | 892 | 432 | 0.48 |
| Centg3 (centaurin, gamma 3) | 1932 | 942 | 0.49 |
| Ccdc66 | 777 | 382 | 0.49 |
| Olig1 (oligodendrocyte transcription factor 1) | 1293 | 639 | 0.49 |
| AA474455 | 1809 | 897 | 0.49 |
Before: Expression (net intensity) in A54 cells before coculture.
After: Expression in A54 cells after 3 days of coculture with HSCs.
Ratio = After/Before.
Figure 2.
Elevated levels of Jag1, Dll3 and BMP6 in A54 cells cocultured with HSCs. Total RNA was isolated from 2 × 104 A54 cells prior to coculture (Preculture) and after 5-day-coculture with 1 × 104 B-lymphocytes (B), 1 × 104 T-lymphocytes (T) or 1 × 104 bone marrow Lin-Sca1+ cells (HSC). Expression of Jag1 (A), Dll3 (B) and BMP6 (C) was measured by quantitative RT-PCR with the primers listed in Table 1. HSC/Transwell: A54 cells were coincubated with HSCs but separated by Cell Culture Insert. *: P < 0.01 (n = 3).
Dll3 expression in A54 was decreased after coculture with T-cells, while it was not significantly changed after coculture with B-cells (Figure 2B). BMP6 expression was mildly upregulated in A54 cells cocultured with mature lymphocytes, but the difference was not statistically significant (Figure 2C). These results suggested that the upregulation of Jag1, Dll3 and BMP6 was induced by interaction with immature hematopoietic cells. Furthermore, such elevation was cancelled out when A54 cells were separated from HSCs with a microporous membrane (Figure 2). Immunostaining with an anti-Jag1 antibody confirmed that hematopoietic progenitors stimulated Jag1 expression in the neighboring A54 preadipocytes. Jag1 signal was apparently greater in A54 cells in cobblestoned areas than those in the surrounding regions (Figure 3). Taken together, mesenchymal stromal cell-derived preadipocytes received signals from HSCs through cell contact, resulting in multiple events including upregulation of Jag1, Dll3 and BMP6.
Figure 3.
Jag1 expression in a cobblestone area. 1 × 104 murine bone marrow HSCs were incubated on 2 × 104 A54 cells for 5 days. Cells were immunostained with a rabbit anti-mouse Jag1 antibody and Alexa Fluor 488-conjugated goat anti-rabbit IgG. In the left panel (phase-contrast bright field, magnification × 400), a cobblestone area with small, rounded hematopoietic cells is indicated with a circle. In the right panel (immunofluorescent view, magnification × 400) showing the same location, several large, stretched stromal cells are positively immunostained (green), wheras stromal cells outside the cobblestone area are negative or weakly positive.
Activation of Notch signaling in HSCs
The coculture experiment showed that the expression of multiple Notch ligands (Jag1 and Dll3) was upregulated in A54 preadipocytes during their interaction with HSCs. We next addressed whether the expression of Notch receptors and downstream factors was altered in the HSCs.
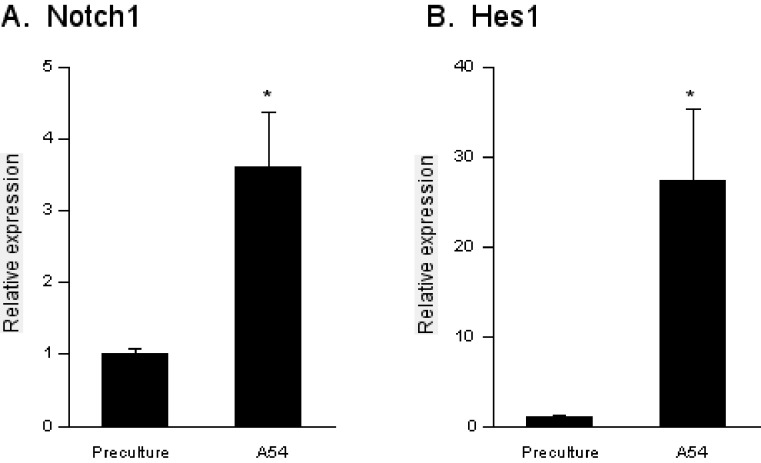
A quantitative RT-PCR analysis revealed that the message level of Notch1 was elevated in HSCs cocultured with A54 cells by 3.6-fold (Figure 4A). Simultaneously, the expression of Hairy enhancer of split-1 (Hes1), a target of Notch1, was upregulated by 27-fold, indicating that the authentic signaling pathway was actually viable (Figure 4B). The results clearly showed that Notch ligands and receptors were upregulated in the stromal cells and HSCs in a reciprocal fashion through cell contact.
Figure 4.
Expression of Notch1 and Hes1 in HSCs. Total RNA was isolated from 1 × 104 bone marrow Lin-Sca1+ cells prior to coculture and after 5-day-coculture with 2 × 104 A54 cells. Notch1 and Hes1 message levels were estimated by quantitative RT-PCR before (Preculture) and after coculture with A54 preadipocyres (A54). The primers for PCR are listed in Table 1. Both Notch1 (A) and Hes1 (B) were significantly upregulated in HSCs after coculture with A54 cells. * : P < 0.01 (n = 4).
Discussion
In the present study, we investigated the molecular events in hematopoiesis supported by mesenchymal stromal cell-derived preadipocytes. We and other investigators reported that several preadipocyte-like stromal cells were capable of supporting hematopoiesis [13–16]. While osteoblasts enhance HSC self-renewal in the hematopoietic niche [2,3], preadipocytes may have additional or different function, such as promoting the asymmetric division of HSCs and proliferation of early progenitors. Our finding that A54 preadipocytes promoted the formation of cobblestone areas and colonies supports this idea, but such function may not be completely cell-autonomous. Assuming that some instructive signals from HSCs, we investigated the gene expression profile of A54 cells in a coculture with HSCs. Among the molecules upregulated or downregulated by more than twofold in a microarray screening, an increase of Notch ligands (Jag1 and Dll3) and BMP6 was confirmed in A54 cells. This finding is intriguing, because signals from Notch and BMP receptors are integrated in osteoblasts and other cell types [17-20]. Notch is a single-pass transmembrane receptor interacting with cell-bound ligands. So far, four Notch receptors (Notch1-4) and five ligands (Jag1-2 and Dll1, 3 and 4) have been identified in mammals. As in many cell systems, Notch signaling is essential for regulating HSCs and blood cell differentiation [21,22].
Previous studies showed that Jag1 in marrow stromal cells and osteoblasts promotes HSC proliferation [3,23], Dll1 produces cobblestone areas [24]. In the present study, we demonstrated that coculturing resulted in concomitant increases in Notch ligands (Jag1 and Dll3) in A54 preadipocytes and Notch signaling in HSCs. Notably, in accordance with an elevation in levels of the Notch1 receptor, expression of the transcription factor Hes1 was also upregulated. Hes1, one of the targets of Notch signaling, regulates specific groups of genes to maintain HSCs and early progenitors [25,26]. Thus, upregulation of the Notch1-Hes1 axis in HSCs strongly suggests that this signaling pathway is actually viable with the increase in Notch ligands in neighboring preadipocytes. Since Notch activation plus cytokine receptor signaling has a combined effect on hematopoietic cells [27], production of Notch ligands and cytokines (SCF, SDF1 and Ang1) by A54 cells is likely to represent a physiological role of marrow niche cells.
Bone morphogenetic proteins (BMPs) belong to the transforming growth factor-b superfamily and have pleiotropic effects on tissue development. They have been implicated as key regulators in hematopoiesis: for example, BMP4 promotes the self-renewal of HSCs [28] and BMP6 enhances the formation of colonies [17]. Because of the redundancy of ligands/receptors and the dependence on the context and stage of differentiation of target cells, a detailed understanding of BMP signaling awaits further investigation.
The mechanisms responsible for the reciprocal upregulation of the expression of Notch ligands and receptors have yet to be clarified. We found that mature lymphocytes poorly induced Jag1, Dll3 and BMP6 expression in A54 cells, thus the stromal cells appear to receive the induction signal mainly from immature cells such as HSCs. Direct cell-to-cell contact with, or close proximity to HSCs was required for the upregulation of Jag1, Dll3 and BMP6 expression in A54 cells. Thus far several classes of molecules have been implicated to act upstream of Notch signaling. For example, the Wnt canonical pathway regulates the expression of Notch ligands in various cell types [29-31]. Sonic hedgehog (Shh) and receptors for Shh (Ptc/Smo) are expressed on primitive hematopoietic cells and marrow stromal cells and induce BMP expression [32,33]. In addition, Shh can increase levels of Notch receptors via vascular endothelial growth factor (VEGF) [34]. Of note, Wnt and Shh are lipid-modified proteins, and therefore appropriate for exerting effects on short-ranged target cells in a paracrine and/or autocrine fashion [35,36]. In the hematopoietic niche, these signaling networks must be precisely orchestrated to regulate HSC behavior, presumably through cross-talk with signals from adhesion molecules such as integrins (Figure 5). Further investigation of such communicative events will identify target molecules to improve engraftment in transplantation therapy.
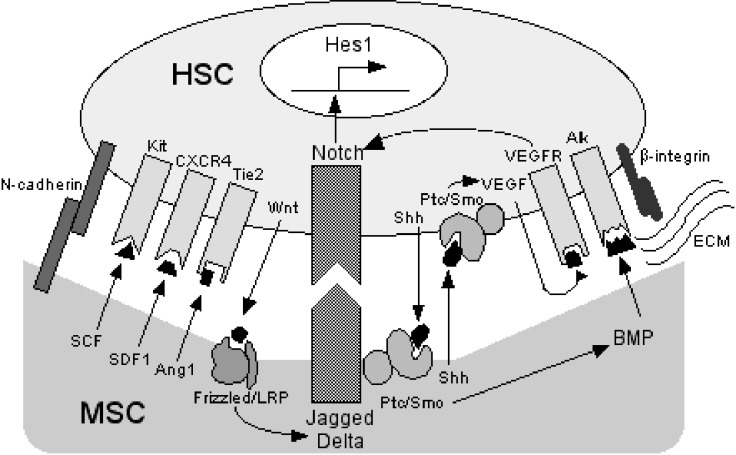
Figure 5.
Interaction of MSCs and HSCs. Various factors are involved in the cell-to-cell interaction between MSCs and HSCs with spatial and temporal complexity. HSCs attach to MSCs via adhesion molecules such as N-cadherin and b-integrins (ECM: extracellular matrix secreted from MSC-derived cells). Soluble cytokines from MSCs such as SCF, SDF1 and Ang1 support the growth and differentiation of HSCs through cognate receptors (Kit, CXCR4 and Tie2). When HSCs and MSCs are in close proximity, expression of Notch ligands (Jagged and Delta-like) is upregulated in MSCs by Wnt from HSCs, while that of Notch receptors is upregulated in HSCs by Sonic hedgehog (Shh) from MSCs and HSCs. Vascular endothelial growth factor (VEGF) can further upregulate Notch receptor expression in an autocrine or paracrine fashion. The upregulated Notch signaling pathway induces the expression of downstream targets such as Hes1. The expression of bone morphogenic protein (BMP) is upregulated in MSCs by Shh signaling. Frizzled, Ptc/Smo, VEGFR, Alk: receptors for Wnt, Shh, VEGF and BMP, respectively.
References
- 1.Schofield R. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells 1978; 4: 4-7 [PubMed] [Google Scholar]
- 2.Zhang J, Niu C, Ye L, Huang H, He X, Tong W-G, Ross J, Haug J, Johnson T, Feng JQ, Harris S, Wiedemann LM, Mishina Y, Li L. Identification of the haematopoietic stem cell niche and control of the niche size. Nature 2003; 425: 425-836 [DOI] [PubMed] [Google Scholar]
- 3.Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, Martin RP, Schipani E, Divieti P, Bringhurst FR, Milner LA, Kronenberg HM, Scadden DT. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 2003; 425: 425-841 [DOI] [PubMed] [Google Scholar]
- 4.Kiel MJ, Yilmaz ÖH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 2005; 121: 1109-1121 [DOI] [PubMed] [Google Scholar]
- 5.Heissig B, Hattori K, Dias S, Friedrich M, Ferris B, Hackett NR, Crystal RG, Besmer P, Lyden D, Moore MAS, Werb Z, Rafii S. Recruitment of stem and progenitor cells from the bone marrow niche requires MMP-9 mediated release of Kitligand. Cell 2002; 109: 109-625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ara T, Tokoyoda K, Sugiyama T, Egawa T, Kawabata K, Nagasawa T. Long-term hematopoietic stem cells require stromal cell-derived factor-1 for colonizing bone marrow during ontogeny. Immunity 2003; 19: 19-257 [DOI] [PubMed] [Google Scholar]
- 7.Hattori K, Heissig B, Tashiro K, Honjo T, Tateno M, Shieh J-H, Hackett NR, Quitoriano MS, Crystal RG, Rafii S, Moore MAS. Plasma elevation of stromal cell-derived factor-1 induces mobilization of mature and immature hematopoietic progenitor and stem cells. Blood 2001; 97: 97-3354 [DOI] [PubMed] [Google Scholar]
- 8.Arai F, Hirao A, Ohmura M, Sato H, Matsuoka S, Takubo K, Ito K, Koh GY, Suda T. Tie2/Angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell 2004; 118: 118-149 [DOI] [PubMed] [Google Scholar]
- 9.Stier S, Ko Y, Forkert R, Lutz C, Neuhaus T, Grünwald E, Chen T, Dombkowski D, Calvi LM, Rittling SR, Scadden DT. Osteopontin is a hematopoietic stem cell niche component that negatively regulates stem cell pool size. J Exp Med 2005; 201: 201-1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagner W, Saffrich R, Wirkner U, Eckstein V, Blake J, Ansorge A, Schwager C, Wein F, Milesala K, Ansorge W, Ho AD. Hematopoietic progenitor cells and cellular microenvironment: behavioral and molecular changes upon interaction. Stem Cells 2005; 37: 1180-1191 [DOI] [PubMed] [Google Scholar]
- 11.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak Multilineage potential of adult human mesenchymal stem cells. Science 1999; 284: 284-143 [DOI] [PubMed] [Google Scholar]
- 12.Wagner W, Wein F, Roderburg C, Saffrich R, Faber A, Krause U, Schubert M, Benes V, Eckstein V, Maul H, Ho AD. Adhesion of hematopoietic progenitor cells to human mesenchymal stem cells as a model for cell-cell interaction. Exp Hematol 2007; 35: 35-314 [DOI] [PubMed] [Google Scholar]
- 13.Nishikawa M, Ozawa K, Tojo A, Yoshikubo T, Okano A, Tani K, Ikebuchi k, Nakauchi H, Asano S. Changes in hematopoiesis-supporting ability of C3H10Tl/2 mouse embryo fibroblasts during differentiation. Blood 1993; 81: 1184-1192 [PubMed] [Google Scholar]
- 14.Oh I, Ozaki K, Miyazato A, Sato K, Meguro A, Muroi K, Nagai T, Mano H, Ozawa K. Screening of genes responsible for differentiation of mouse mesenchymal stromal cells by DNA micro-array analysis of C3H10T1/2 and C3H10T1/2-derived cell lines. Cytotherapy 2007; 9: 9-80 [DOI] [PubMed] [Google Scholar]
- 15.Yoshikubo T, Ozawa K, Takahashi K, Nishikawa M, Horiuchi N, Tojo A, Tani K, Kodama H, Asano S. Adhesion of NFS-60 myeloid leukemia cells to MC3T3-G2/PA6 stromal cells induces granulocyte colony-stimulating factor production. Blood 1994; 84: 84-415 [PubMed] [Google Scholar]
- 16.Nakamura M, Harigaya K, Watanabe Y. Correlation between production of colony-stimulating activity (CSA) and adipose conversion in a murine marrow-derived preadipocyte line (H-1/A). Proc Soc Exp Biol Med 1985; 179: 179-283 [DOI] [PubMed] [Google Scholar]
- 17.Detmer K, Walker AN. Bone morphogenetic proteins act synergistically with haematopoietic cytokines in the differentiation of haematopoietic progenitors. Cytokine 2002; 17: 17-36 [DOI] [PubMed] [Google Scholar]
- 18.Nobta M, Tsukazaki T, Shibata T, Xin C, Moriishi T, Sakano S, Shindo H, Yamaguchi A. Critical regulation of bone morphogenic protein-induced osteoblastic differentiation by Delta1/Jagged1-activated Notch1 signaling. J Biol Chem 2005; 280: 15842-15848 [DOI] [PubMed] [Google Scholar]
- 19.Katoh M, Katoh M. Transcriptional regulation of WNT2B based on the balance of Hedgehog, Notch, BMP and WNT signals. Int Natl J Oncol 2009; 34: 1411-1415 [PubMed] [Google Scholar]
- 20.Blank U, Karlsson S. Signaling pathways governing cell fate. Blood 2009; 111: 111-492 [DOI] [PubMed] [Google Scholar]
- 21.Chiba S. Notch signaling in stem cell systems. Stem Cells 2006; 24: 2437-2447 [DOI] [PubMed] [Google Scholar]
- 22.Weber JM, Calvi LM. Notch signaling and the bone marrow hematopoietic stem cell niche. Bone 2010; 46: 46-281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duncan AW, Rattis FM, DiMascio LN, Congdon KL, Pazianos G, Zhao C, Yoon K, Cook JM, Willert K, Gaiano N, Reya T. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nature Immunol 2005; 6: 6-314 [DOI] [PubMed] [Google Scholar]
- 24.Moore KA, Pytowski B, Witte L, Hicklin D, Lemischka I. Hematopoietic activity of a stromal cell transmembrane protein containing epidermal growth factor-like repeat motifs. Proc Natl Acad Sci USA 1997; 94: 4011-4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunisato A, Chiba S, Nakagami-Yamaguchi E, Kumano K, Saito T, Masuda S, Yamaguchi T, Osawa M, Kageyama R, Nakauchi H, Nishikawa M, Hirai H. HES-1 preserves purified hematopoietic stem cell ex vivo and accumulates side population cells in vivo. Blood 2003; 101: 1777-1783 [DOI] [PubMed] [Google Scholar]
- 26.Yu X, Alder JK, Chun JH, Friedman AD, Heimfeld S, Cheng L, Civin CI. HES1 inhibits cycling of hematopoietic progenitor cells via DNA binding. Stem Cells 2006; 24: 24-876 [DOI] [PubMed] [Google Scholar]
- 27.Varnum-Finney B, Xu L, Brashem-Stein C, Nourrigat C, Flowers D, Bakkour S, Pear WS, Bernstein ID. Pluripotent, cytokine-dependent, hematopoietic stem cells are immortalized by constitutive Notch1 signaling. Nat Med 2000; 6: 1278-1281 [DOI] [PubMed] [Google Scholar]
- 28.Bhatia M, Bonnet D, Wu D, Murdoch B, Wrana J, Gallacher L, Dick JE. Bone morphogenetic proteins regulate the developmental program of human hematopoietic stem cells. J Exp Med 1999; 189: 1139-1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murdoch B, Chadwick K, Martin M, Shojaei F, Shah KV, Gallacher L, Moon RT, Bhatia M. Wnt-5A augments repopulating capacity and primitive hematopoietic development of human blood stem cells in vivo. Proc Natl Acad Sci USA 2003; 100: 3422-3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katoh M, Katoh H. Notch ligand, JAG1, is evolutionarily conserved target of canonical WNT signaling pathway in progenitor cells. Int J Mol Med 2006; 17: 17-681 [PubMed] [Google Scholar]
- 31.Estrach S, Ambler CA, Lo Celso C, Hozumi K, Watt FM. Jagged 1 is a b-catenin target gene required for ectopic hair follicle formation in adult epidermis. Development 2006; 133: 4427-4438 [DOI] [PubMed] [Google Scholar]
- 32.Bhardwaj G, Murdoch B, Wu D, Baker DP, Williams KP, Chadwick K, Ling LE, Karanu FN, Bhatia M. Sonic hedgehog induces the proliferation of primitive hematopoietic cells via BMP regulation. Nat Immunol 2001; 2: 2-172 [DOI] [PubMed] [Google Scholar]
- 33.Katoh Y, Katoh M. Hedgehog signaling pathway and gastrointestinal stem cell signaling network (review). Int J Mol Med 2006; 18: 1019-1023 [PubMed] [Google Scholar]
- 34.Lawson ND, Vogel AM, Weinstein BM. Sonic hedgehog and vascular endothelial growth factor act upstream of the notch pathway during arterial endothelial differentiation. Dev Cell 2002; 3: 3-127 [DOI] [PubMed] [Google Scholar]
- 35.Willert K, Brown JD, Danenberg E, Duncan AW, Weissman IL, Reya T, Yates JR, III, Nusse R. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 2003; 423: 423-448 [DOI] [PubMed] [Google Scholar]
- 36.Porter JA, Young KE, Beachy PA. Cholesterol modification of Hedgehog signaling proteins in animal development. Nature 1996; 274: 274-255 [DOI] [PubMed] [Google Scholar]