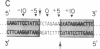Abstract
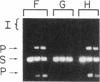
The minimal size of the recombination site required for efficient FLP recombinase-catalyzed recombination in vitro is no more than 28 base pairs, which includes parts of two 13-base-pair inverted repeats and all of an 8-base-pair spacer. The FLP recombinase cleaves the DNA at the boundaries of the spacer, becomes covalently linked to the spacer DNA via a 3' phosphate, and leaves a free 5' hydroxyl at the other end of the 8-base-pair spacer. The efficiency of recombination is reduced if the size of the spacer in a recombinant site is increased or decreased by 1 base pair, while the spacer in the second site is unaltered. Recombination between two sites with identical 1-base-pair additions or deletions in the spacer, however, is relatively unaffected. This result suggests that pairing of sequences in the spacer region is important in FLP-promoted recombination events. The sequence asymmetry utilized by the recombinase to determine the orientation of the site is located uniquely within the spacer region.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews B. J., Proteau G. A., Beatty L. G., Sadowski P. D. The FLP recombinase of the 2 micron circle DNA of yeast: interaction with its target sequences. Cell. 1985 Apr;40(4):795–803. doi: 10.1016/0092-8674(85)90339-3. [DOI] [PubMed] [Google Scholar]
- Bauer C. E., Hesse S. D., Gardner J. F., Gumport R. I. DNA interactions during bacteriophage lambda site-specific recombination. Cold Spring Harb Symp Quant Biol. 1984;49:699–705. doi: 10.1101/sqb.1984.049.01.079. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Guarascio V. R., Jayaram M. Recombination within the yeast plasmid 2mu circle is site-specific. Cell. 1982 May;29(1):227–234. doi: 10.1016/0092-8674(82)90107-6. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Hicks J. B. Replication and recombination functions associated with the yeast plasmid, 2 mu circle. Cell. 1980 Sep;21(2):501–508. doi: 10.1016/0092-8674(80)90487-0. [DOI] [PubMed] [Google Scholar]
- Cox M. M. The FLP protein of the yeast 2-microns plasmid: expression of a eukaryotic genetic recombination system in Escherichia coli. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4223–4227. doi: 10.1073/pnas.80.14.4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig N. L., Nash H. A. The mechanism of phage lambda site-specific recombination: site-specific breakage of DNA by Int topoisomerase. Cell. 1983 Dec;35(3 Pt 2):795–803. doi: 10.1016/0092-8674(83)90112-5. [DOI] [PubMed] [Google Scholar]
- Hartley J. L., Donelson J. E. Nucleotide sequence of the yeast plasmid. Nature. 1980 Aug 28;286(5776):860–865. doi: 10.1038/286860a0. [DOI] [PubMed] [Google Scholar]
- Hoess R. H., Abremski K. Mechanism of strand cleavage and exchange in the Cre-lox site-specific recombination system. J Mol Biol. 1985 Feb 5;181(3):351–362. doi: 10.1016/0022-2836(85)90224-4. [DOI] [PubMed] [Google Scholar]
- Hoess R., Abremski K., Sternberg N. The nature of the interaction of the P1 recombinase Cre with the recombining site loxP. Cold Spring Harb Symp Quant Biol. 1984;49:761–768. doi: 10.1101/sqb.1984.049.01.086. [DOI] [PubMed] [Google Scholar]
- Jayaram M. Two-micrometer circle site-specific recombination: the minimal substrate and the possible role of flanking sequences. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5875–5879. doi: 10.1073/pnas.82.17.5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce C. M., Grindley N. D. Construction of a plasmid that overproduces the large proteolytic fragment (Klenow fragment) of DNA polymerase I of Escherichia coli. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1830–1834. doi: 10.1073/pnas.80.7.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Meyer-Leon L., Senecoff J. F., Bruckner R. C., Cox M. M. Site-specific genetic recombination promoted by the FLP protein of the yeast 2-micron plasmid in vitro. Cold Spring Harb Symp Quant Biol. 1984;49:797–804. doi: 10.1101/sqb.1984.049.01.090. [DOI] [PubMed] [Google Scholar]
- Sadowski P. D., Lee D. D., Andrews B. J., Babineau D., Beatty L., Morse M. J., Proteau G., Vetter D. In vitro systems for genetic recombination of the DNAs of bacteriophage T7 and yeast 2-micron circle. Cold Spring Harb Symp Quant Biol. 1984;49:789–796. doi: 10.1101/sqb.1984.049.01.089. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Vetter D., Andrews B. J., Roberts-Beatty L., Sadowski P. D. Site-specific recombination of yeast 2-micron DNA in vitro. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7284–7288. doi: 10.1073/pnas.80.23.7284. [DOI] [PMC free article] [PubMed] [Google Scholar]