Abstract
This study aimed to identify new peptide antigens from Chlamydia (C.) trachomatis in a proof of concept approach which could be used to develop an epitope-based serological diagnostic for C. trachomatis related infertility in women. A bioinformatics analysis was conducted examining several immunodominant proteins from C. trachomatis to identify predicted immunoglobulin epitopes unique to C. trachomatis. A peptide array of these epitopes was screened against participant sera. The participants (all female) were categorized into the following cohorts based on their infection and gynecological history; acute (single treated infection with C. trachomatis), multiple (more than one C. trachomatis infection, all treated), sequelae (PID or tubal infertility with a history of C. trachomatis infection), and infertile (no history of C. trachomatis infection and no detected tubal damage). The bioinformatics strategy identified several promising epitopes. Participants who reacted positively in the peptide 11 ELISA were found to have an increased likelihood of being in the sequelae cohort compared to the infertile cohort with an odds ratio of 16.3 (95% c.i. 1.65–160), with 95% specificity and 46% sensitivity (0.19–0.74). The peptide 11 ELISA has the potential to be further developed as a screening tool for use during the early IVF work up and provides proof of concept that there may be further peptide antigens which could be identified using bioinformatics and screening approaches.
Keywords: Chlamydia, Infertility, ELISA, HtrA, Peptide
Highlights
-
•
A bioinformatics analysis of known C. trachomatis immunodominant proteins successfully predicted unique immunoglobulin peptide epitopes.
-
•
Serological screening of these peptides demonstrated identification of antigens specific to women with C. trachomatis disease sequelae.
-
•
Further development of one peptide as an ELISA demonstrated high specificity to diagnose women with C. trachomatis infertility and sequelae.
1. Introduction
Chlamydia (C.) trachomatis is the most prevalent sexually transmitted bacterial infection worldwide, with 1,307,893 infections reported in the USA in 2010 [1]. The infection is frequently asymptomatic and thus actual case numbers are higher than those notified [2,3]. The major burden of Chlamydia infections relates to the serious sequelae, such as pelvic inflammatory disease (PID), infertility, and ectopic pregnancy in women. The nature of the infection and the detection of the sequelae much later in life mean that it is difficult to predict the number of untreated infections which progress to serious sequelae. A retrospective study of 43,715 women in Sweden over a 10–15 year time frame, using health data repositories, indicated that for the women who tested positive to C. trachomatis infection; 5.6% had PID, 2.7% had ectopic pregnancy, and 6.7% had infertility [4]. A review of published studies by Land and co-workers found that diagnosed lower genital tract infection can progress to PID in 0.43–31% of cases, to tubal infertility in 0.02–4.6% of cases, and PID progresses to tubal infertility in 11.4–20% of cases [5].
The pathogenic mechanism, host genetic factors, and other epidemiological factors which lead to serious sequelae in a proportion of infected women are not well understood, although it is clear that infertility in women is associated with tubal damage resulting in fallopian tube occlusion. Additionally, minor tubal adhesions or damage to the cilia lining the fallopian tubes are also considered to be C. trachomatis infection related sequelae which impact on fertility. This has been further supported by a recent finding by Coppus and co-workers that participants attending a fertility clinic with high C. trachomatis serum antibody titers by MIF or ELISA (>1:32 or 1.1 respectively) with patent tubes by hysterosalpingography (HSG) or laparoscopy, had a 33% lower ongoing pregnancy rate [6].
C. trachomatis antibody titers (CAT) are currently the gold standard for diagnosis of Chlamydia related infertility in women. There are two main types of tests which most commonly used; immunofluorescence tests (MIF, IF, or WIF) or ELISA. There are several different commercially available ELISA tests, and they are typically based on major outer membrane protein (MOMP) peptides or LPS. The sensitivity and specificity of these tests varies widely between studies for the diagnosis of tubal infertility. A recent meta-analysis by Broeze and colleagues confirmed this considerable variation even when the same commercial tests were used between different studies. The ELISA (mainly Medac, Biomerieux, Labsystems) sensitivity varied from 12% to 69%, and specificity varied from 62% to 100% [7]. The immunofluorescence methods were more sensitive (50–91%) but had lower specificity (35–85%) [7].
There are also differences in clinical practice regarding the use of CAT as a routine part of infertility work up. Many clinics prefer to use HSG or laparoscopy to diagnose tubal infertility in women without CAT. The guidelines regarding use of CAT in the preliminary infertility work up also vary. The Dutch Society for Obstetrics and Gynecology recommends CAT during initial fertility work up and suggest a fixed cut off of IgG MIF 1:32, or ELISA >1.1, as positive cut off for CAT [8], whereas CAT is not mentioned in Australian diagnosis recommendations. More accurate CAT implemented during infertility workup has the potential to reduce time to successful pregnancy and patient risk by indicating definitive treatment regimes with the goal of removing the need for invasive diagnosis by HSG or laparoscopy.
Despite these differences in clinical acceptance, it remains possible that new assays targeting different chlamydial antigens could be used to develop more accurate (sensitive and specific) CATs to predict female infertility due to C. trachomatis past infection which could be implemented with greater clinical efficacy. This application of a new CAT assay has led to proteomic investigations within the field to identify chlamydial antigens for the development of a more sensitive and specific CAT to predict Chlamydia-related infertility in women. A genome wide study using an expression array identified that a combination of CT443 (OmcB) and CT381 (ArtJ) had a 67.5% sensitivity and 100% specificity [9]. A proteomic approach searching for B and T cell antigens recognized by patients with a high MIF titer frequently identified MOMP, HtrA, OmcB, TART, GroEL, LCR-E, and CT662 [10]. In another study using a western blot strategy on 2-D gels of the Chlamydia proteome MOMP, PmpD, and OmcB were found to be the most specific to infection positive participants (not solely infertility) [11]. HtrA as a full length protein was found to be immunoreactive from C. trachomatis infected participants [12].
In this current study we used a bioinformatics approach to predict single antibody epitopes with unique sequence specificity to C. trachomatis from a selected subset of the most frequently identified antigens from these previously published proteomic investigations. A library of peptides based on these epitopes was used in ELISAs with patient sera to characterize performance (sensitivity and odds ratio) with 95% specificity for Chlamydia related sequelae as a proof of concept for identifying accurate epitopes for Chlamydia related sequelae. Using this approach it is possible that additional antigens be identified that will advance the use of CAT for diagnosis of Chlamydia related infertility in women.
2. Materials and methods
2.1. Bioinformatics analysis to predict peptides with C. trachomatis specificity and putative B cell antigenicity
C. trachomatis proteins were selected for this study which had previously been identified in multiple studies as highly immunoreactive, when screened against serological positive females with C. trachomatis infection or C. trachomatis-related tubal factor infertility in more than one previous investigations. These proteins were: HtrA, cHSP60, Ct443, and Ct381 [9–12]. A protein unique to Chlamydia pneumoniae (Cpn0236) was also included in the study as a negative control [13].
The protein sequences were analysed using a series of in silico bioinformatic analyses to identify potential linear B cell epitopes, and the specificity of these epitopes to C. trachomatis. The sequences of the proteins of interest were screened for predicted linear B cell epitopes using BepiPred algorithm software [14], confirmed with antigenicity prediction software (Kolaskar & Tongaonkar Antigenicity) [15], and hydrophilic domains identified to avoid transmembrane domains (Parker Hydrophilicity Prediction) [16]. Candidate C. trachomatis HtrA, cHSP60, CT443, and CT381 linear B cell epitope peptides were screened for sequence specificity against the NR database where BLAST E values were used as criteria; (1) E values <0.004 for C. trachomatis specificity, and (2) E values >0.1 for C. pneumoniae to remove nonspecific epitopes. Regions that did not meet criteria were excluded from further study. The predicted epitopes that met criteria were then used to design a series of 12-mer peptides (Table 1) which include partial or complete sequences of these epitopes. Predicted peptide epitopes larger than 12 amino acids were split into overlapping epitopes. These included epitopes for HtrA (peptides 7–9; Table 1) and HSP60 (peptides 15–18, 23–28, 32–34, and 37–43; Table 1), two of the most highly reported immunogenic antigens for Chlamydia.n= 11)
Table 1.
Results of screening of peptide array against different participant cohorts. For each generated peptide sequence, the predicted epitope is depicted in bold. All peptides have been screened bioinformatically for homology with other bacteria.
| # | Peptide sequence | Mean absorbance at 450 nm (standard deviation) by cohort |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| BLAST |
Sequelae | Infertility controls |
Multiple (n= 6) | Acute (n= 6) | |||||
| C. trach | C. pn | All (n= 16) | C. pn+ (n= 7) | C. pn– (n= 9) | |||||
| HtrA generated peptides (1–14) | |||||||||
| 1 | SPMLGYSASKKD | 0.003 | 2.4 | 0.172 (0.059) | 0.163 (0.074) | 0.145 (0.079) | 0.176 (0.071) | 0.163 (0.040) | 0.169 (0.08) |
| 2 | ADICLAVSSGDQ | 0.005 | 1.2 | 0.157 (0.057) | 0.139 (0.052) | 0.123 (0.049) | 0.151 (0.052) | 0.152 (0.039) | 0.169 (0.085) |
| 3 | AVSSGDQEVSQE | 0.01 | 0.87 | 0.171 (0.065) | 0.154 (0.062) | 0.139 (0.069) | 0.165 (0.058) | 0.175 (0.0369) | 0.183 (0.074) |
| 4 | EQQRPQQRDAVR | 0.0002 | 1.7 | 0.169 (0.061) | 0.155 (0.064) | 0.140 (0.071) | 0.167 (0.059) | 0.173 (0.047) | 0.164 (0.067) |
| 5 | QKYTAKIVGLDP | 0.002 | 0.006 | 0.163 (0.060) | 0.150 (0.063) | 0.126 (0.063) | 0.168 (0.060) | 0.168 (0.039) | 0.176 (0.087) |
| 6 | AISLMMPGTRVI | 0.0006 | 0.12 | 0.155 (0.058) | 0.146 (0.064) | 0.126 (0.067) | 0.160 (0.062) | 0.164 (0.032) | 0.163 (0.079) |
| 7 | VTVTQIPTEDGV | 0.003 | 0.036 | 0.169 (0.073) | 0.160 (0.073) | 0.134 (0.069) | 0.179 (0.072) | 0.171 (0.033) | 0.165 (0.065) |
| 8 | EDGVSALQKMGV | 0.004 | 0.01 | 0.153 (0.054) | 0.157 (0.058) | 0.139 (0.050) | 0.171 (0.061) | 0.181 (0.050) | 0.189 (0.088) |
| 9 | VSALQKMGVRVQ | 0.003 | 0.006 | 0.176 (0.060) | 0.163 (0.069) | 0.15 (0.063) | 0.173 (0.075) | 0.176 (0.060) | 0.164 (0.066) |
| 10 | MGVRVQNITGLA | 0.36 | 0.23 | 0.149 (0.045) | 0.141 (0.080) | 0.118 (0.053) | 0.157 (0.095) | 0.143 (0.034) | 0.148 (0.069) |
| 11 | ADTRGILVVAVE | 0.001 | 0.11 | 0.174 (0.062) | 0.158 (0.046) | 0.16 (0.044) | 0.157 (0.049) | 0.166 (0.037) | 0.199 (0.097) |
| 12 | EAGSPAASAGVA | 0.024 | 0.015 | 0.156 (0.094) | 0.226 (0.053) | 0.236 (0.064) | 0.216 (0.041) | 0.126 (0.046) | 0.131 (0.028) |
| 13 | AVNRGRVASVEE | 0.49 | 0.12 | 0.159 (0.055) | 0.160 (0.075) | 0.128 (0.069) | 0.184 (0.074) | 0.172 (0.044) | 0.171 (0.074) |
| 14 | VNRGRVASVEEL | 0.003 | 0.26 | 0.165 (0.062) | 0.152 (0.072) | 0.132 (0.070) | 0.166 (0.072) | 0.166 (0.036) | 0.170 (0.076) |
| HSP60 generated peptides (15–48) | |||||||||
| 15 | YHILSRIELSDP | 0.0006 | 0.87 | 0.156 (0.058) | 0.155 (0.077) | 0.126 (0.072) | 0.175 (0.076) | 0.167 (0.049) | 0.172 (0.079) |
| 16 | IELSDPFERIGV | 0.001 | 3.5 | 0.156 (0.051) | 0.149 (0.073) | 0.127 (0.078) | 0.165 (0.068) | 0.172 (0.046) | 0.164 (0.067) |
| 17 | FERIGVYFARSL | 0.0008 | 7.1 | 0.171 (0.055) | 0.162 (0.045) | 0.153 (0.035) | 0.169 (0.052) | 0.194 (0.050) | 0.194 (0.11) |
| 18 | RSLAKRIHKRHA | 0.002 | 0.32 | 0.283 (0.014) | 0.289 (0.17) | 0.243 (0.15) | 0.322 (0.18) | 0.229 (0.095) | 0.244 (0.13) |
| 19 | ADGVISSVILLR | 0.01 | 0.065 | 0.170 (0.062) | 0.144 (0.064) | 0.129 (0.069) | 0.155 (0.062) | 0.146 (0.036) | 0.170 (0.073) |
| 20 | LLRAFLKASIPF | 0.003 | 0.44 | 0.180 (0.060) | 0.150 (0.072) | 0.133 (0.079) | 0.162 (0.068) | 0.164 (0.035) | 0.171 (0.073) |
| 21 | LKASIPFIDQGL | 0.003 | 0.86 | 0.161 (0.061) | 0.143 (0.079) | 0.123 (0.074) | 0.158 (0.082) | 0.168 (0.033) | 0.162 (0.077) |
| 22 | ASIPFIDQGLSP | 0.003 | 0.86 | 0.155 (0.055) | 0.161 (0.089) | 0.150 (0.099) | 0.170 (0.085) | 0.165 (0.031) | 0.174 (0.084) |
| 23 | ASALASQKEAVC | 0.014 | 3.5 | 0.164 (0.052) | 0.154 (0.069) | 0.137 (0.059) | 0.167 (0.075) | 0.175 (0.052) | 0.173 (0.074) |
| 24 | AYLHSHSFLLKD | 0.001 | 2.4 | 0.159 (0.051) | 0.149 (0.063) | 0.130 (0.065) | 0.162 (0.061) | 0.176 (0.036) | 0.170 (0.081) |
| 25 | KDASKVLGLIRS | 0.014 | 2.4 | 0.164 (0.058) | 0.160 (0.070) | 0.139 (0.076) | 0.176 (0.064) | 0.185 (0.045) | 0.176 (0.069) |
| 26 | LIRSHLPDPLIG | 0.001 | 1.7 | 0.164 (0.057) | 0.150 (0.082) | 0.137 (0.092) | 0.160 (0.076) | 0.147 (0.034) | 0.161 (0.078) |
| 27 | GEAFAEAVAYTG | 0.008 | 0.17 | 0.176 (0.063) | 0.159 (0.082) | 0.134 (0.077) | 0.177 (0.084) | 0.167 (0.038) | 0.184 (0.091) |
| 28 | VAYTGHEGAVAL | 0.008 | 5 | 0.172 (0.075) | 0.150 (0.079) | 0.123 (0.072) | 0.170 (0.081) | 0.155 (0.038) | 0.181 (0.099) |
| 29 | SQRSGSTLHLFC | 0.93 | 0.62 | 0.173 (0.059) | 0.155 (0.073) | 0.132 (0.075) | 0.173 (0.070) | 0.159 (0.031) | 0.180 (0.092) |
| 30 | TLHLKGIQTQKG | 0.01 | 1.2 | 0.168 (0.063) | 0.149 (0.078) | 0.128 (0.078) | 0.165 (0.079) | 0.180 (0.041) | 0.171 (0.080) |
| 31 | TQKGYRVPSFFP | 0.0006 | 2.4 | 0.166 (0.055) | 0.158 (0.069) | 0.142 (0.043) | 0.170 (0.083) | 0.176 (0.036) | 0.183 (0.082) |
| 32 | FPHDSFHENPIV | 0.0006 | 0.44 | 0.168 (0.058) | 0.152 (0.076) | 0.129 (0.065) | 0.169 (0.082) | 0.180 (0.064) | 0.138 (0.079) |
| 33 | NPIVAPKIFVTD | 0.0001 | 1.7 | 0.157 (0.056) | 0.155 (0.069) | 0.133 (0.069) | 0.173 (0.067) | 0.146 (0.076) | 0.197 (0.086) |
| 34 | FVTDQKIHCLFP | 0.001 | 1.2 | 0.16 (0.057) | 0.144 (0.064) | 0.131 (0.059) | 0.154 (0.069) | 0.146 (0.038) | 0.166 (0.094) |
| 35 | DHAIHNAEDETS | 0.001 | 2.4 | 0.169 (0.060) | 0.156 (0.071) | 0.138 (0.076) | 0.170 (0.069) | 0.178 (0.045) | 0.179 (0.072) |
| 36 | ETSRKLLKKRKH | 0.003 | 1.7 | 0.265 (0.108) | 0.252 (0.123) | 0.222 (0.083) | 0.274 (0.15) | 0.195 (0.069) | 0.224 (0.105) |
| 37 | RKHRLENSIAII | 0.001 | 0.32 | 0.155 (0.049) | 0.138 (0.055) | 0.126 (0.045) | 0.146 (0.060) | 0.169 (0.047) | 0.173 (0.080) |
| 38 | SIAIIPVKQDTA | 0.004 | 1.2 | 0.152 (0.055) | 0.141 (0.053) | 0.13 (0.05) | 0.149 (0.052) | 0.168 (0.040) | 0.19 (0.092) |
| 33 | QDTAPLHELALK | 0.003 | 0.87 | 0.163 (0.051) | 0.153 (0.079) | 0.133 (0.076) | 0.168 (0.081) | 0.174 (0.050) | 0.191 (0.086) |
| 40 | ALKTLNSTQESG | 0.014 | 0.091 | 0.170 (0.045) | 0.157 (0.075) | 0.142 (0.069) | 0.167 (0.080) | 0.179 (0.055) | 0.187 (0.081) |
| 41 | ESGFVLGGGAAL | 0.038 | 0.54 | 0.178 (0.064) | 0.165 (0.069) | 0.149 (0.074) | 0.176 (0.067) | 0.186 (0.03) | 0.197 (0.081) |
| 42 | AALLYATQSLSS | 0.02 | 1.7 | 0.163 (0.062) | 0.128 (0.059) | 0.110 (0.063) | 0.141 (0.055) | 0.146 (0.036) | 0.179 (0.055) |
| 43 | LSSSPEHSQEEQ | 0.003 | 0.62 | 0.180 (0.064) | 0.160 (0.084) | 0.140 (0.079) | 0.174 (0.089) | 0.165 (0.045) | 0.190 (0.097) |
| 44 | EEQAAVQILQTA | 0.004 | 0.048 | 0.173 (0.062) | 0.163 (0.071) | 0.143 (0.063) | 0.177 (0.076) | 0.163 (0.041) | 0.224 (0.087) |
| 45 | DKLCSLGTPSLG | 0.008 | 0.026 | 0.167 (0.058) | 0.149 (0.080) | 0.121 (0.065) | 0.170 (0.087) | 0.155 (0.043) | 0.189 (0.084) |
| 46 | YGPAYSSSSKDF | 0.0005 | 2.3 | 0.199 (0.072) | 0.237 (0.077) | 0.245 (0.032) | 0.231 (0.099) | 0.202 (0.087) | 0.209 (0.052) |
| 47 | VFSSPPFSNKPP | 0.0003 | 0.86 | 0.208 (0.14) | 0.277 (0.097) | 0.300 (0.042) | 0.250 (0.14) | 0.157 (0.043) | 0.339 (0.039) |
| 48 | SLSSSPEHSQEE | 0.001 | 0.23 | 0.116 (0.030) | 0.098 (0.022) | 0.105 (0.019) | 0.093 (0.024) | 0.111 (0.038) | 0.097 (0.014) |
| Ct443 generated peptides (49–51) | |||||||||
| 49 | VDRKEVAPVHES | 0.0005 | 12 | 0.107 (0.047) | 0.111 (0.045) | 0.119 (0.04) | 0.106 (0.048) | 0.190 (0.24) | 0.069 (0.006) |
| 50 | PVSFSGPTKGTIT | 0.0005 | 12 | 0.193 (0.109) | 0.267 (0.13) | 0.253 (0.070) | 0.276 (0.17) | 0.186 (0.093) | 0.222 (0.068) |
| 51 | LTVPVSDTENTH | 0.0005 | 0.033 | 0.140 (0.042) | 0.110 (0.036) | 0.110 (0.031) | 0.109 (0.041) | 0.163 (0.079) | 0.185 (0.033) |
| Ct381 generated peptide (52) | |||||||||
| 52 | VGIGVASDRPAL | 0.001 | 0.11 | 0.136 (0.034) | 0.108 (0.036) | 0.114 (0.029) | 0.104 (0.040) | 0.164 (0.075) | 0.153 (0.057) |
| OmpB generated peptide (53) | |||||||||
| 53 | AVVSSGSDNELA | 0.0005 | 2 | 0.127 (0.039) | 0.130 (0.046) | 0.137 (0.026) | 0.125 (0.057) | 0.178 (0.089) | 0.170 (0.063) |
| C. pneumoniaeCp0236 generated peptides (54–55) | |||||||||
| 54 | EHFSPEPPNEPL | 0.23 | 0.00005 | 0.060 (0.0073) | 0.063 (0.072) | 0.063 (0.012) | 0.063 (0.008) | 0.068 (0.011) | 0.061 (0.006) |
| 55 | GSSLRTKEGNTI | 2.2 | 0.002 | 0.057 (0.0052) | 0.0099 (0.022) | 0.060 (0.006) | 0.062 (0.011) | 0.059 (0.0076) | 0.056 (0.005) |
2.2. Analysis of epitope array to identify epitopes and development of ELISA
The designed peptide epitopes were commercially synthesized using solid phase synthesis onto a Biotin-SGSG motif (Mimotopes, Melbourne, Australia). All peptides were solubilized in 50% isopropanol overnight on a gentle rocker at room temperature. The initial screening ELISAs were conducted using the complete peptide epitope array against 39 participants that sampled all cohorts. The ELISAs were conducted using streptavidin coated plates (Reacti-Bind Streptavidin High Binding Capacity Coated 96-well Plates, ThermoScientific, Australia) where peptides were coated at approximately 0.15 μg/well for 1 h at room temperature in PBS/0.1% Tween 20. The plates were then washed four times in PBS 0.1% Tween 20, and blocked overnight in SuperBlock buffer (Pierce, Australia) with 0.1% Tween 20 at 4 °C. The participant sera was added to the wells at dilutions of 1/200 and 1/1000 (in SuperBlock buffer in PBS with 0.1% Tween 20) and incubated for 1 h. The plates were washed five times in 2× PBS with 0.1% Tween 20. The secondary antibody used was goat anti-human IgG-HRP (Invitrogen, Australia) at a dilution of 1/15,000 in SuperBlock buffer PBS with 0.1% Tween 20. The plates were developed by the addition of 100 μg/ml 3,3,5,5,-Tetramethyl-benzadine (TMB) (Sigma-Aldrich, Australia) in DMSO dissolved in phosphate citrate buffer with sodium perborate (Sigma-Aldrich, Australia), incubated for 10 min at room temperature and stopped by the addition of 1.0 M H2SO4. ELISA plates were read on a Bio-Rad xMark Microplate Spectrophotometer at 450 nm. These ELISAs were conducted by screening the whole array against each participant sera with primary and secondary antibody only controls on each plate. The peptides that fit criteria for further development in the initial analysis were then subjected to further analysis. Optimized assays were attempted, which included further serological dilution series, and a variety of alteration in washing conditions. The optimized peptide 11 ELISA was conducted as per the above protocol except that 0.2 μg of peptide was coated to each well.
2.3. Participant/cohort collection and definitions
Participants (female) were recruited as a part of the Queensland Chlamydia Research Network which includes all Queensland Health Sexual Health Clinics, QUT Health Services Medical Centre, and The Wesley Reproductive Medicine and Gynecological surgery unit. The participants were fully informed and consented prior to participation in the study. Ethical approval for the study was obtained via the following Human Research Ethics Committees: QUT Human Research Ethics Committee approval number 080000268, Prince Charles Hospital Human Research Ethics Committee approval number EC2809, Ipswich and West Moreton Health Services District Human Research Ethics committee approval number (10-09), Gold Coast Hospital District Human Research Ethics Committee approval number (200893), Cairns Sexual Health Clinic (HREC/09/QCH/4-554), and The Wesley Hospital Human Research Ethics Committee (2008/02). Clinical data was collected from all participants including: age, number of sexual partners, fertility status (if known), self reported and patient chart reported history of sexually transmitted infections, current and previous C. trachomatis infections, contraceptive use and type. Participants sourced from the Wesley Reproductive Medicine and Gynecological surgery unit were also asked to allow their gynecologist to provide additional data relating to their infertility status and infertility cause. Participants from this clinic were all tested for C. trachomatis infection history using both the MEDAC and Bioclone C. trachomatis ELISAs (sourced from Bioclone, Sydney, Australia and MEDAC sourced from Biocene, Sydney, Australia). Potentially confounding participants were excluded where they were found to be both seropositive to C. trachomatis and had a diagnosed cause of infertility that was not tubal factor. These participants were excluded due to the potential for their infertility to still relate to C. trachomatis immunopathology.
Participants (female) recruited from the Queensland Chlamydia Research Network were divided into four C. trachomatis cohorts: sequelae, acute, multiple and infertility controls (subdivided by C. pneumoniae serological status). The sequelae cohort included women who had a history of PID, tubal ectopic pregnancy, or tubal factor infertility (laparoscopy diagnosis) and also a history of C. trachomatis infection (either by recorded participant medical chart recorded PCR positive previous infection, or C. trachomatis positive serology by MEDAC and Bioclone assays). Acute participants were those who had a history of single treated C. trachomatis infection (PCR diagnosis) with no reported sequelae, and the multiple infections cohort included participants who had a history of more than one treated C. trachomatis infection (PCR diagnosis) with no reported sequelae. These two cohorts were generally recruited at the Sexual Health Clinics. The infertility control cohort included women attending the IVF clinic who were infertile but did not have tubal factor infertility; these participants were all negative for C. trachomatis serology by the MEDAC and Bioclone tests.
Participants (female) were also recruited from patients attending the Gynecology outpatient department at the Sarfdarjung Hospital, New Delhi, India. Participants were informed and consented to participate in the study. The study was approved by the Sarfdarjung Hospital Human Research Ethics Committee. Clinical data collected included: age, regularity of menstrual cycle, years of infertility (if infertile), clinical details such as cervicitis, bacterial vaginosis, Mycoplasma sp. culture, Ureaplasma sp. by culture, bacterial vaginosis, and C. trachomatis infection status by direct fluorescence assay and PCR from swabs. Participants from this study were divided into cohorts based on infertility (primary or secondary; any cause), cervicitis and current C. trachomatis genital infection status (assayed by PCR and DFA).
3. Results
3.1. Screening new peptide antigens using ELISA confirms bioinformatic predictions and identifies potential new antigens for Chlamydia antibody testing
The peptide array (55 unique peptides) was developed using a series of bioinformatic analyses of the most commonly reported proteins to have a serological response in C. trachomatis participants. Proteins previously frequently identified during screening or proteomic studies when screening with participant sera from C. trachomatis positive participants were selected (HtrA, Hsp60, CT443, and CT381), along with a C. pneumoniae specific protein as a negative control (CP0236). MOMP was not included for this study given the existing peptide ELISAs based on this protein and the serovar specific sequence variability in this protein. These proteins were then searched bioinformatically for B cell epitopes, as outlined in Section 2. Predicted epitopes were then searched by BLAST to reduce the pool of potential epitopes to only those which had high sequence specificity to C. trachomatis. An initial screening assay was conducted to identify peptides which may be useful detectors of the sequelae disease cohort. The peptide array was screened in duplicate against 39 participant sera belonging to the four cohorts: sequelae, acute, multiple, and infertility controls (subdivided by C. pneumoniae serological status). The raw values for each epitope from each participant in each cohort were then analyzed (Table 1). The data was analyzed by ANOVA, comparing the data for each cohort to either sequelae or infertility control groups. Only peptide 47 showed a significant difference (P< 0.05, sequelae c.f. acute) when the results were tested for statistical significance between the cohorts during this initial screening ELISA. Specific criteria for sequelae specificity was then applied to select peptide epitopes for further analysis.
In order to identify which peptides have the potential for further assay optimization for specific detection of the sequelae cohort a set of criteria were used to select the peptides to develop further. The three criteria were: (1) achieve a higher average absorbance in the sequelae cohort compared to the combined infertility controls (a difference off of >0.015 was chosen), (2) to avoid false positive detection due to prior C. pneumoniae infection (<0.015 difference between the C. pneumoniae subdivided infertility controls), and (3) distinguish sequelae from acute participants (>0.015 different in absorbance between the two cohorts) (Table 1). These three criteria identified the following four peptides (protein source of the peptide is indicated): 11 (HtrA: MGVRVQNITGLA), 48 (HSP60: SLSSSPEHSQEE), 51 (Ct443: LTVPVSDTENTH), and 52 (Ct381: VGIGVASDRPAL).
3.2. Peptide 11 ELISA is an effective serological diagnostic for chlamydial tubal factor infertility
In order to optimize the serological assay, performance of the different assay conditions trialed with the different sera dilutions for each of the four peptides was assessed by area under the curve analysis. Specificity was controlled at ≥95% to establish absorbance (450 nm) thresholds reflecting a diagnostic use to detect those women who have C. trachomatis sequelae from C. trachomatis infection or negative cohorts. Peptide 11 showed the most rigorous performance and hence only the data from this peptide is shown here. Fig. 1 shows the receiver operator characteristic curve for the sera at a 1/200 dilution comparing participants from sequelae and infertility control cohorts. The tested dilution series did not significantly change the accuracy of the test as there was negligible difference in area under the curve. The selection of 1/200 dilution was one of practicability, as the absorbance threshold was to be 0.184 at a 1/800 dilution and 0.296 at a 1/200 dilution which achieves a better signal to noise threshold for assay development.
Fig. 1.
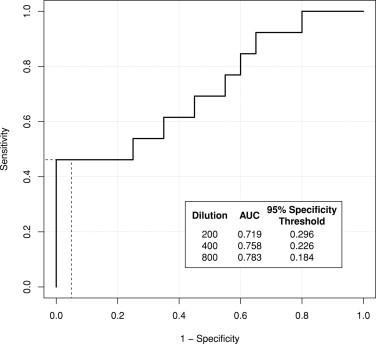
Receiver operator characteristic curve for the sera dilution of 1/200 comparing participants from sequelae and infertility control cohorts. In the box (bottom right) area under the curve (AUC) for 1/200, 1/400 and 1/800 sera dilutions are shown with calculated absorbance (450 nm) thresholds for a 95% specificity cut off. Dashed line indicates the 95% specificity (x-axis) cut off which corresponds to 46% sensitivity (y-axis).
The 1/200 dilution peptide 11 assay with a 0.296 absorbance threshold was found to be able to statistically discriminate participants in the sequelae cohort from infertility controls and from acute cohorts (Table 2). The comparison of sequelae to infertility controls yielded an odds ratio of 16.3 with a 95% confidence interval 1.63–160 (P = 0.005). The bioinformatic prediction that the test will not be influenced by C. pneumoniae serological status was also supported by its performance against the infertilty controls, which includes 7 C. pneumoniae seropositive participants. The test was also able to produce a positive odds ratio (5.14; 95% confidence interval 1.12–23.5) for sequelae compared to the acute infection cohort, as per the third design criteria. The optimised peptide 11 assay was not able to statistically discriminate sequelae from the multiple infection cohort at the 0.296 absorbance threshold (P = 0.091), but did yield a positive odds ratio (4; 95% confidence interval 0.763–21.0) for the comparison. These results are supportive of the potential utility of such a test during the initial infertility investigation where women with sequelae are clearly distinguished from women with a previous acute infection or women who are negative.
Table 2.
Performance of peptide 11 ELISA.
| Comparison | 450 nm Absorbance threshold for comparison | Specificity (95% confidence interval) | Sensitivity (95% confidence interval) | Odds ratio (95% confidence interval) | P value (chi-squared) for difference in observed proportions |
|---|---|---|---|---|---|
| Sequelae vs. infertily controls | 0.296 | 0.95 (0.75–1.00) | 0.46 (0.19–0.74) | 16.3 (1.65–160) | 0.005 |
| Multiple vs. infertiliy controls | 0.296 | 0.95 (0.75–1.00) | 0.176 (0.038–0.434) | 4.07 (0.38–43.4) | 0.217 |
| Acute vs. infertilty controls | 0.296 | 0.95 (0.75–99.9) | 0.143 (0.040–0.327) | 3.17 (0.33–30.7) | 0.299 |
| Sequelae vs. multiple | 0.296 | 0.82 (0.57–0.96) | 0.46 (0.19–0.75) | 4 (0.763–21.0) | 0.091 |
| Sequelae vs. acute | 0.296 | 0.86 (0.67–0.96) | 0.46 (0.19–0.75) | 5.14 (1.12–23.5) | 0.027 |
| Multiple vs. acute | 0.296 | 0.86 (0.67–0.96) | 0.18 (0.04–0.43) | 1.29 (0.25–5.6) | 0.760 |
| Sequelae vs. acute | 0.408 | 0.96 (0.81–1.00) | 0.23 (0.05–0.53) | 8.10 (0.75–87.2) | 0.050 |
| Multiple vs. acute | 0.408 | 0.96 (0.82–1.00) | 0.06 (0–0.29) | 1.68 (0.110–24.6) | 0.715 |
In order to further analyse if it might be possible to develop a test to distinguish between women potentially at risk of developing sequelae (such as women with multiple exposures) from women who have had an infection with no evidence of sequelae a different analysis was conducted using a different absorbance cut off. The specificity for detecting multiple infections and sequelae from acute could be further improved by increasing the absorbance threshold to 0.408, but resulted in a decrease of statistical significance from the 0.296 absorbance threshold for the sequelae vs. acute comparison (P(Abs = 0.296) = 0.012 to P(Abs = 0.408) = 0.050; Table 2). Whilst these analyses were not statistically significant it indicates that this an approach were the assay may have utility at the GP clinic for women who wish to determine their future infertility risk could be developed from this peptide–epitope strategy.
3.3. Testing the peptide 11 assay as a serological diagnostic using cohort of women attending a gynecology clinic in India
The peptide 11 ELISA was then tested against participants recruited by the Sarfdarjung Hospital Gynaecology Clinic, South Delhi. The peptide 11 ELISA was tested on 129 participant's sera attending the clinic using the optimized 1/200 dilution of sera with the designed 95% specificity absorbance threshold of 0.296. Nine women tested positive to the peptide 11 ELISA (Table 3) which included 7 women with cervicitis and primary infertility and 2 women with primary infertility and current C. trachomatis infection. These proportions were significantly different to those who tested negative (P = 0.039). The women who tested positive in the peptide 11 ELISA compared to those who tested negative were analyzed for any potential confounders which may have influenced the result. There was no statistical significance in age of women who tested positive and those who tested negative (P = 0.3458, 28.43 average years for peptide 11 positive c.f. 30.16 average years for peptide 11 negative), or in the patient reported years of infertility (P = 0.680, 6.47 compared to 7.11 years for peptide 11 positive group). Furthermore, there were no significant differences in those women who were positive in the peptide 11 ELISA and whether or not they had current genital infections (Mycoplasma, or Ureaplasma). In comparison, the MEDAC MOMP ELISA detected 5 women (absorbance above 1.1) including; 2 women with C. trachomatis and cervicitis, 2 women with primary infertility, and 1 primary infertility with C. trachomatis infection, who was also detected by the peptide 11 assay. These proportions were not significantly different from those who were negative to the MEDAC MOMP ELISA (P = 0.529).
Table 3.
Comparison of women attending the Sarfdarjung Hospital Gyneacology clinic who were positive in either the peptide 11 or MEDAC MOMP assays by infertility, cervicitis and current C. trachomatis infection status.
| Cervicitis | Infertility | C. trach PCR/DFA statusa | Total number of participants | Peptide 11 positive | MEDAC MOMP positive |
|---|---|---|---|---|---|
| + | − | + | 3 | 0 | 2 |
| − | − | − | 15 | 0 | 0 |
| − | Primary | − | 25 | 0 | 2 |
| + | Primary | − | 31 | 7 | 0 |
| − | Primary | + | 35 | 2 | 1 |
| − | Secondary | − | 11 | 0 | 0 |
| − | Secondary | + | 9 | 0 | 0 |
C. trachomatis infection status was assessed by positive reactions in both PCR and DFA. Fisher's exact test for count data shows a significant difference in observed cohort proportions between peptide 11 positive and negative (P = 0.039; negative = total – positive). No significant difference in observed cohort proportions was demonstrated between MEDAC MOMP positive and negative women (Fisher's exact test for count data; P = 0.529).
All of the women who were peptide 11 positive were women with primary infertility (Table 3). Those women who were positive in the peptide 11 ELISA had an odds ratio of 8.8 (95% confidence interval 0.5–156.29) of having primary infertility and not cervicitis or secondary infertility (irrespective of C. trachomatis status), but the comparison was not statistically significant (P = 0.075). A positive MEDAC MOMP ELISA result had a lower odds ratio of 1.82 (95% confidence interval 0.29–11.28) for the same comparison and was also not statistically significant (P = 0.240). Raw absorbance values for the peptide 11 ELISA did not correlate with MEDAC MOMP ELISA (R2 = 0.001, P = 0.677) supporting the independence of these tests and differing rates of detection. Whilst a large number of the women recruited at this clinic were positive for a current C. trachomatis infection (47 positive by both urine DFA and PCR), 2 of the 9 women who tested positive using the peptide 11 test also had a current infection further supporting that the design of the test does not bias towards detection women with acute infections in the context of other pathology. Sensitivity and specificity of peptide 11 or MEDAC performance relative to the gold standard of laparoscopy or HSG was not possible on these patients.
4. Discussion
This paper reports for the first time that a peptide epitope from C. trachomatis HtrA has the potential to be further developed into a specific diagnostic to detect serious sequelae from this infection. Women who reacted positively in the peptide 11 ELISA developed during this study had a 16.3 odds ratio of having C. trachomatis sequelae (tubal infertility, ectopic pregnancy, or PID) (specificity 95% and sensitivity 46%). The test also showed the potential to distinguish these women with sequelae from women with a history of C. trachomatis single treated infection (P = 0.027, Table 2) or multiple infections (P = 0.091, Table 2), although the cohort numbers were not large enough in this study to provide statistical validity. This is an important finding as very few studies reporting new antigens for diagnosis of tubal factor infertility report the performance of the test against infection cohorts (such as acute or multiple infections). Certainly cohort definitions in studies such as these are difficult, as patients may have had previous untreated infections. However, given that some of the acute and multiple infection participants included in this study have been sourced at sexual health clinics it is likely that this cohort includes some participants who may have more infections than diagnosed and yet the assay was still specific to the sequelae cohort (Table 2). The poor sensitivity observed in this study may be improved by future optimization of peptide concentrations or multiple peptide combination assay formats. Whilst the sensitivity and technical parameters (absorbance, signal to noise ratio) of the assay are clearly not adequate for implementation as a diagnostic in it's current format, these results support that using bioinformatics can help to eliminate issues with specificity (i.e. cross reactivity due to C. pneumonia sero-positive status), which has been a perceived barrier to implementation of CAT. This study has been limited to proteins previously identified as highly immunoreactive for Chlamydia thus, it provides proof of concept that this strategy, along with further assay development (such as assessing assay performance against standard serological tests and improved technical parameters), could be successfully implemented to identify new specific antigens for CAT diagnosis of Chlamydia tubal factor infertility. The potential application of this assay during the initial infertility investigation is clearly the major focus, hence the need for high specificity so that few false positives are diagnosed even though with the current low sensitivity some true positives will not be diagnosed. However, the analysis using a different absorbance cut off to distinguish between the acute and multiple and acute and sequelae cohorts also supports that with further technical development an assay of this format may also have utility at the GP clinic where women may be seeking to examine their fertility risks. Therefore, the overall strategy of using a combination of previous proteomic data on immunogenic antigens and bioinformatic tools to select epitopes for peptide ELISA data was supported by the findings in this study.
The peptide 11 ELISA in the current format was tested in another setting and it was found that 9 out of 129 women attending a gynecology clinic in India would be predicted to have tubal infertility using this assay. Interestingly, all of these women had primary infertility and none of the women with secondary infertility were positive by this assay (no significant age difference). The results of the peptide 11 ELISA and MEDAC MOMP assays were different, however, whilst MEDAC MOMP titre above 1.1 is recommended in the Dutch fertility guidelines as a diagnostic for C. trachomatis sequelae this assay is actually marketed by the manufacturer as test for infection history. Hence, the different performance of these two assays is not necessarily unexpected. There were 47 women with a current C. trachomatis infection of the 129 women recruited for this study (36.4%). This prevalence is slightly higher than a previously reported prevalence of C. trachomatis positive women at another clinic in Delhi, however there is potential for a recruitment bias in our study as the clinicians were aware that this is a Chlamydia research study (24–30%) (PCR diagnosis) [17]. C. trachomatis infection prevalence in populations in India (generally sexual health clinic, gynecology clinic or sex worker screening studies) varies from 7% to 30% [18,19]. It is more difficult to estimate the likely percent of infertility that relates to Chlamydia-mediated tubal factor infertility. However, we can be guided by two different prospective studies where women attending IVF clinics were recruited, and the percent of participants with tubal infertility and a positive antibody testing for a history of Chlamydia infection calculated to be the prevalence of Chlamydia related tubal infertility. In a study based in Denmark this prospective screening approach identified that that 6.9% of infertile women attending the IVF clinic was due to C. trachomatis tubal infertility [20]. For a similar study conducted in China 7.6% of women attending the IVF clinic had C. trachomatis tubal infertility [21]. The participants attending the clinic in India for this current investigation do not have access to laparoscopy or HSG technologies, meaning it is not possible to validate the results of the peptide 11 assay in this second cohort. However, 7.0% of the participants attending the clinic, all with primary infertility, were positive by this assay.
Overall, this study has provided proof of concept that new peptide based ELISAs which are highly specific for detection of women with C. trachomatis sequelae such as tubal factor infertility can be developed. The assay is under further development to improve the sensitivity by including multiple markers within the assay which has shown increased sensitivity [9]. Increased sensitivity may make this assay a useful diagnostic for initial fertility work up, and these potential improvements may also be useful for implementation as a screening tool in epidemiological studies, or within general practice as an early warning system for women to plan their families.
Acknowledgements
The authors wish to thank all of the clinical staff, and participants who made this study possible. In particular the authors wish to acknowledge the help of Dr. Joseph Debattista for organizing clinic participation in the study. The authors would also like to particularly thank Dr. Janet Allan for assistance with clinic involvement in the study. The authors acknowledge technical input from Sarina Gloeckl, Helen Iconomou, and Alcides Garcia Orjuela. The technical assistance and hospitality of staff and team members within the Mittal Lab is gratefully acknowledged. This project has been funded by a support grants from The Wesley Research Institute, NHMRC Project Grant (553020), NHMRC Peter Doherty Fellowship (awarded to WHuston) and the Queensland Smart State NIRAP scheme.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.CDC. www.cdc.gov/std/chlamydia/stdfact-chlamydia.htm; 2011.
- 2.Biro FM, Reising SF, Doughman JA, Kollar LM, Rosenthal SL. A comparison of diagnostic methods in adolescent girls with and without symptoms of Chlamydia urogenital infection. Pediatrics. 1994;93:476–480. [PubMed] [Google Scholar]
- 3.Hu D, Hook EW, Goldie SJ. The impact of natural history parameters on the cost-effectiveness of Chlamydia trachomatis screening strategies. Sexually Transmitted Diseases. 2006;33:428–436. doi: 10.1097/01.olq.0000200577.46261.b0. [DOI] [PubMed] [Google Scholar]
- 4.Low N, Egger M, Sterne JA, Harbord RM, Ibrahim F, Lindblom B. Incidence of severe reproductive tract complications associated with diagnosed genital chlamydial infection: the Uppsala Women's Cohort Study. Sexually Transmitted Infections. 2006;82:212–218. doi: 10.1136/sti.2005.017186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Land JA, Van Bergen JE, Morre SA, Postma MJ. Epidemiology of Chlamydia trachomatis infection in women and the cost-effectiveness of screening. Human Reproduction Update. 2010;16:189–204. doi: 10.1093/humupd/dmp035. [DOI] [PubMed] [Google Scholar]
- 6.Coppus SF, Land JA, Opmeer BC, Steures P, Eijkemans MJ, Hompes PG. Chlamydia trachomatis IgG seropositivity is associated with lower natural conception rates in ovulatory subfertile women without visible tubal pathology. Human Reproduction. 2011;26:3061–3067. doi: 10.1093/humrep/der307. [DOI] [PubMed] [Google Scholar]
- 7.Broeze KA, Opmeer BC, Coppus SF, Van Geloven N, Alves MF, Anestad G. Chlamydia antibody testing and diagnosing tubal pathology in subfertile women: an individual patient data meta-analysis. Human Reproduction Update. 2011;17:301–310. doi: 10.1093/humupd/dmq060. [DOI] [PubMed] [Google Scholar]
- 8.Swart P, Mol BW, van der Even F, van Beurden M, Redekop WK, Bossuyt PM. The accuracy of hysterosalpingography in the diagnosis of tubal pathology: a meta-analysis. Fertility and Sterility. 1995;64:486–491. doi: 10.1016/s0015-0282(16)57781-4. [DOI] [PubMed] [Google Scholar]
- 9.Rodgers AK, Budrys NM, Gong S, Wang J, Holden A, Schenken RS. Genome-wide identification of Chlamydia trachomatis antigens associated with tubal factor infertility. Fertility and Sterility. 2011;96:715–721. doi: 10.1016/j.fertnstert.2011.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finco O, Frigimelica E, Buricchi F, Petracca R, Galli G, Faenzi E. Approach to discover T- and B-cell antigens of intracellular pathogens applied to the design of Chlamydia trachomatis vaccines. Proceedings of the National Academy of Sciences. 2011;108:9969–9974. doi: 10.1073/pnas.1101756108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchez-Campillo M, Bini L, Comanducci M, Raggiaschi R, Marzocchi B, Pallini V. Identification of immunoreactive proteins of Chlamydia trachomatis by Western blot analysis of a two-dimensional electrophoresis map with patient sera. Electrophoresis. 1999;20:2269–2279. doi: 10.1002/(SICI)1522-2683(19990801)20:11<2269::AID-ELPS2269>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 12.Huston WM, Armitage CW, Lawrence A, Gloeckl S, Bell SJ, Debattista J. HtrA, RseP, and Tsp proteins do not elicit a pathology-related serum IgG response during sexually transmitted infection with Chlamydia trachomatis. Journal of Reproductive Immunology. 2010;85:168–171. doi: 10.1016/j.jri.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 13.Wolf K, Plano GV, Fields KA. A protein secreted by the respiratory pathogen Chlamydia pneumoniae impairs IL-17 signaling via interaction with human Act1. Cellular Microbiology. 2009;11:769–779. doi: 10.1111/j.1462-5822.2009.01290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larsen JE, Lund O. Improved method for predicting linear B-cell epitopes. Immunome Research. 2006;2:2. doi: 10.1186/1745-7580-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolaskar AS, Tongaonkar PC. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Letters. 1990;276:172–174. doi: 10.1016/0014-5793(90)80535-q. [DOI] [PubMed] [Google Scholar]
- 16.Parker JM, Guo D, Hodges RS. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry. 1986;25:5425–5432. doi: 10.1021/bi00367a013. [DOI] [PubMed] [Google Scholar]
- 17.Patel AL, Sachdev D, Nagpal P, Chaudhry U, Sonkar SC, Mendiratta SL. Prevalence of Chlamydia infection among women visiting a gynaecology outpatient department: evaluation of an in-house PCR assay for detection of Chlamydia trachomatis. Annals of Clinical Microbiology and Antimicrobials. 2010;9:24. doi: 10.1186/1476-0711-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dwibedi B, Pramanik JM, Sahu P, Kar SK, Moharana T. Prevalence of Chlamydia infection in females attending an Obstetrics and Gynaecology out patient department in Orissa. Indian Journal of Dermatology, Venereology and Leprology. 2009;75:614. doi: 10.4103/0378-6323.57730. [DOI] [PubMed] [Google Scholar]
- 19.Choudhry S, Ramachandran VG, Das S, Bhattacharya SN, Mogha NS. Pattern of sexually transmitted infections and performance of syndromic management against etiological diagnosis in patients attending the sexually transmitted infection clinic of a tertiary care hospital. Indian Journal of Sexually Transmitted Diseases and AIDS. 2010;31:104–108. doi: 10.4103/2589-0557.74998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Svenstrup HF, Fedder J, Kristoffersen SE, Trolle B, Birkelund S, Christiansen G. Mycoplasma genitalium, Chlamydia trachomatis, and tubal factor infertility—a prospective study. Fertility and Sterility. 2008;90:513–520. doi: 10.1016/j.fertnstert.2006.12.056. [DOI] [PubMed] [Google Scholar]
- 21.Mei B, Luo Q, Du K, Huo Z, Wang F, Yu P. Association of MICA gene polymorphisms with Chlamydia trachomatis infection and related tubal pathology in infertile women. Human Reproduction. 2009;24:3090–3095. doi: 10.1093/humrep/dep339. [DOI] [PubMed] [Google Scholar]


