Abstract
The ATP-binding cassette transporter Rv1747 is required for the growth of Mycobacterium tuberculosis in mice and in macrophages. Its structure suggests it is an exporter. Rv1747 forms a two-gene operon with pknF coding for the serine/threonine protein kinase PknF, which positively modulates the function of the transporter. We show that deletion of Rv1747 or pknF results in a number of transcriptional changes which could be complemented by the wild type allele, most significantly up-regulation of the iniBAC genes. This operon is inducible by isoniazid and ethambutol and by a broad range of inhibitors of cell wall biosynthesis and is required for efflux pump functioning. However, neither the Rv1747 or pknF mutant showed increased susceptibility to a range of drugs and cell wall stress reagents including isoniazid and ethambutol, cell wall structure and cell division appear normal by electron microscopy, and no differences in lipoarabinomannan were found. Transcription from the pknF promoter was not induced by a range of stress reagents. We conclude that the loss of Rv1747 affects cell wall biosynthesis leading to the production of intermediates that cause induction of iniBAC transcription and implicates it in exporting a component of the cell wall, which is necessary for virulence.
Keywords: mycobacteria, virulence, serine/threonine protein kinase, transcriptomics, DNA microarray, isoniazid
Introduction
The increase in multidrug and extensively drug-resistant Mycobacterium tuberculosis strains has made the search for new TB drugs ever more important. Deciphering the function of important M. tuberculosis proteins is a key strategy to identify potential new drug targets. Rv1747 is an ATP-binding cassette (ABC) transporter that is required for the growth of M. tuberculosis in macrophages, dendritic cells and mice (Curry et al., 2005). A total of 37 ABC transporters have been identified in M. tuberculosis; 16 have been categorised as importers and 21 as exporters (Braibant et al., 2000).
ABC transporters are integral membrane proteins comprising two transmembrane domains and two cytoplasmic nucleotide-binding domains; they bind and hydrolyse ATP providing energy for uptake or export of substrates across cell membranes. Functions include the uptake of nutrients into the cells and the export of virulence factors and toxins (Holland et al., 2003). Bacterial ABC importers are typically formed from four polypeptide chains that are often separately encoded (Saurin et al., 1999) and require an external binding protein, which functions to deliver the substrate to the transporter (Dawson et al., 2007). In contrast, bacterial exporters are produced as one polypeptide where a single gene usually encodes both the transmembrane domain and a nucleotide-binding domain (Saurin et al., 1999). The presence of fused nucleotide-binding and transmembrane domains is a strong indicator of an ABC exporter (Dawson et al., 2007; Davidson et al., 2008). Based on its amino acid sequence, Rv1747 belongs to the G subfamily of ABC transporters; this family consists of half-transporters, which oligomerise to form the functional transporter. Furthermore, this protein encodes both the transmembrane and nucleotide-binding domains in one polypeptide and is thus probably an exporter. Rv1747 forms a two-gene operon with its upstream adjacent gene, the serine–threonine protein kinase (STPK) pknF (Spivey et al., 2011).
Interestingly, Rv1747 also contains two forkhead-associated (FHA) domains; these are modular phosphopeptide recognition motifs, and their presence is taken to indicate that the protein is likely to interact with a phosphorylated protein partner (Durocher & Jackson, 2002; Spivey et al., 2011). Rv1747 exhibits ATPase activity and is a substrate for PknF in vitro; furthermore, both FHA domains of Rv1747 are required for this interaction in a yeast two-hybrid assay (Molle et al., 2004; Curry et al., 2005). More recently, we have identified specific threonine residues on Rv1747 that are phosphorylated, at least in vitro, by PknF and which have in vivo modulatory effects on the function of the Rv1747 ABC transporter (Spivey et al., 2011).
Bacterial ABC exporters transport many different substances including cell surface components such as lipopolysaccharides, lipids, proteins involved in pathogenesis, peptides, drugs and siderophores (Dassa, 2003). In M. tuberculosis, one ABC transporter, DrrABC and an RND family transmembrane protein, MmpL7, are required for the translocation to the cell surface of phthiocerol dimycocerosates (PDIMs), complex lipids required for virulence (Cox et al., 1999; Camacho et al., 2001); interestingly, MmpL7 is a potential substrate of another STPK, PknD (Pérez et al., 2006). Rv1747 could export any one of these molecules, which would make the function of the transporter important for growth in vivo. Rv1747 falls into a subclass of M. tuberculosis ABC transporters, which have an unknown function (Braibant et al., 2000). Similarity was found to the White protein from Drosophila melanogaster, a permease necessary for the transport of pigment precursors responsible for eye colour, and to NodI from Rhizobium strains, a protein implicated in the nodulation process by export of a polysaccharide (Braibant et al., 2000).
We have investigated the phenotypic consequences of the loss of the Rv1747 and PknF proteins in deletion mutants. Significantly, using transcriptional profiling, we demonstrate that the expression of the iniBAC operon is up-regulated in both mutants.
Materials and methods
Bacterial strains and growth conditions
Mycobacterium tuberculosis H37Rv and Escherichia coli K-12 strains are described in Table 1. Growth conditions have been described previously (Spivey et al., 2011).
Table 1.
Bacterial strains and plasmids
| Strains or plasmids | Genotype or description | Source or reference |
|---|---|---|
| E. coli strains | ||
| E. coli TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG; used for general cloning | Invitrogen |
| M. tb. strains | ||
| H37Rv | M. tuberculosis WT strain | Oatway & Steenken (1936) |
| ΔpknF | H37Rv with deletion of pknF constructed by homologous recombination with targeting construct pRW51 | Spivey et al. (2011) |
| pknF complement | ΔpknF containing complementing plasmid pRW95 | Spivey et al. (2011) |
| ΔRv1747 | H37Rv with deletion of Rv1747 constructed by homologous recombination with targeting construct pRW69 | Curry et al. (2005) |
| Rv1747 complement | ΔRv1747 containing complementing plasmid pRW76 | Curry et al. (2005) |
| M. tuberculosis shuttle plasmids | ||
| p2Nil | Suicide gene delivery vector, oriE, KanR | Hinds et al. (1999) |
| pKP186 | Integrase negative derivative of the integrating vector pMV306, KanR | Papavinasasundaram et al. (2001) |
| pBS-Int | Suicide vector containing integrase, AmpR | Springer et al. (2001) |
| pEJ414 | pMV306 derivative containing a promoterless E. coli lacZ reporter gene, KanR | Papavinasasundaram et al. (2001) |
| pRW69 | p2Nil containing a 2-kb region of H37Rv DNA flanking each side of the Rv1747 gene, HygR | Curry et al. (2005) |
| pRW76 | Rv1747 complementing plasmid. pKP186 derivative containing 609 bp Rv1745c,pknF and Rv1747, KanR HygR | Curry et al. (2005) |
| pRW51 | p2Nil containing a 1.5-kb region of H37Rv DNA flanking each side of the pknF gene | Spivey et al. (2011) |
| pRW95 | pknF complementing plasmid. pKP186 derivative containing 609 bp Rv1745c,pknF and 20 bp of Rv1747, KanR | Spivey et al. (2011) |
| pVS_01 | pEJ414 containing pknF promoter region, KanR | This work |
Generation of the pknF and Rv1747 deletion and complementing strains
The construction of the pknF (Rv1746) null strain (ΔpknF) was described previously (Spivey et al., 2011); this has an in-frame unmarked deletion to avoid downstream polar effects on the Rv1747 gene. For complementation of the pknF deletion, the genes pknF, 609 bp of Rv1745c and 20 bp of Rv1747 were amplified by PCR (Spivey et al., 2011) and the product cloned into the vector pKP186 (Rickman et al., 2005), a pMV306 (Kong & Kunimoto, 1995) derivative lacking the integrase gene and electroporated into the ΔpknF mutant along with the mycobacterial suicide vector, pPS-Int containing the integrase gene (Springer et al., 2001; Curry et al., 2005). Construction of the hygromycin-marked Rv1747 deletion mutant was described previously (Curry et al., 2005). For complementation of the Rv1747 deletion, the genes Rv1747,Rv1746 (pknF), and 609 bp of Rv1745c were amplified by PCR (Curry et al., 2005), cloned into the vector pKP186 (Rickman et al., 2005) and transformed into the ΔRv1747 mutant.
cDNA labelling and microarray analysis
RNA isolation from M. tuberculosis liquid cultures was described previously (Spivey et al., 2011). Whole genome DNA microarrays of M. tuberculosis (version 2) were provided by the BμG@S group (St. George's, University of London). cDNA labelling and RNA–DNA microarray hybridisations were described previously (Rickman et al., 2005). Microarray slides were scanned as previously (Hunt et al., 2008), grids were fitted using bluefuse software and analysed using genespring, version 10 (Agilent). Three biological replicates were performed for each condition, carried out in duplicate for dye swaps. The genes described only include those whose differential gene regulation was restored to wild type (WT) in complemented mutants. The array design is available in BμG@Sbase (Accession No. A-BUGS-23; http://bugs.sgul.ac.uk/A-BUGS-23) and ArrayExpress (Accession No. A-BUGS-23). Fully annotated microarray data have been deposited in BμG@Sbase (accession number E-BUGS-149; http://bugs.sgul.ac.uk/E-BUGS-149) and ArrayExpress (accession number E-BUGS-149).
Quantitative real-time PCR (qRT-PCR)
cDNA was generated from 1 μg RNA using the Quantitect reverse transcription kit (Qiagen). Primers (Table 2) were designed using primer express 3.0 (Applied Biosystems). Real-time quantitative PCR analysis on this cDNA was performed using the ABI Prism 7500 using the Fast SYBR green master mix (Applied Biosystems). Data were normalised to sigA expression.
Table 2.
Primers used for qRT-PCR
| Primer name | Sequence (5′-3′) |
|---|---|
| pknF F | CACGAACGTCGGCTGTTG |
| pknF R | GACGATCAGGTGAATCAGGATTG |
| Rv1747 F | TACGGTCGACCTGATCAAATTG |
| Rv1747 R | GCGCTGGCGGTGTGA |
| iniA F | TCATCGCAGTCTCATCACTGTTG |
| iniA R | TTGGACTCTTCGTTGAGCTCTTT |
| iniB F | TTATCGATTACATCCTGAGCCTGTT |
| iniB R | CGGAGCGGCAACGAA |
| iniC F | ACTCCGAATGCTAAGCCTTTTG |
| iniC R | CAGCGACGCGATTTCGT |
| ethA F | GCAAGCCCATCCTCGAGTAC |
| ethA R | CGGATATGCCTGTCGATTCC |
| pknD F | CAACGGACAGTTCTTTGTCGAA |
| pknD R | TGTTTCAATAGGGCGCGTAAA |
| sigA F | TCGGTTCGCGCCTACCT |
| sigA R | GGCTAGCTCGACCTCTTCCT |
Other methods are described in Supporting information, Appendix S1.
Results
Transcriptional microarray analysis of the ΔpknF and ΔRv1747 mutants
Transcriptional microarray analysis was performed to compare gene expression in WT H37Rv vs. ΔRv1747, WT vs. ΔpknF, WT vs. the Rv1747 complemented mutant and WT vs. the pknF-complemented mutant. The top 10 most highly regulated genes whose differential expression was restored by complementation are shown in Table 3. As expected, expression of Rv1747 was 29-fold down-regulated in the mutant (compared with WT). In the Rv1747 complemented strain, expression of Rv1747 was 1.3-fold up-regulated compared with WT, confirming restoration of gene expression.
Table 3.
Microarray data for the 10 Mycobacterium tuberculosis genes most highly up- and down-regulated upon Rv1747 deletion
| Rv number | Gene name | Gene product | Fold regulated | P-value |
|---|---|---|---|---|
| Rv0005 | gyrB | DNA gyrase subunit B | 2.0 up | 0.001 |
| Rv0046c | ino1 | Myo-inositol-1-phosphate synthase Ino1 | 2.0 up | 3.00E−04 |
| Rv0047c | Rv0047c | Conserved hypothetical protein | 2.2 up | 4.10E−05 |
| Rv0341 | iniB | Isoniazid-inducible gene protein IniB | 3.8 up | 2.40E−04 |
| Rv0342 | iniA | Isoniazid-inducible gene protein IniA | 3.2 up | 8.10E−05 |
| Rv0822c | Rv0822c | Conserved hypothetical protein | 2.0 up | 4.00E−05 |
| Rv1040c | PE8 | PE family protein | 2.1 down | 0.013 |
| Rv1380 | pyrB | Probable aspartate carbamoyltransferase PyrB | 2.2 down | 0.001 |
| Rv1382 | Rv1382 | Probable export or membrane protein | 2.2 down | 6.20E−04 |
| Rv1747 | Rv1747 | Probable ABC transporter | 29.2 down | 8.90E−06 |
| Rv1999c | Rv1999c | Probable conserved integral membrane protein | 2.2 down | 0.004 |
| Rv2007c | fdxA | Probable ferredoxin FdxA | 2.1 up | 0.005 |
| Rv2265 | Rv2265 | Possible conserved integral membrane protein | 2.1 down | 0.005 |
| Rv2396 | PE_PGRS41 | PE-PGRS family protein | 2.0 up | 7.00E−04 |
| Rv2415c | Rv2415c | Conserved hypothetical protein | 2.3 down | 7.90E−04 |
| Rv2528c | mrr | Probable restriction system protein Mrr | 2.1 down | 1.90E−04 |
| Rv2577 | Rv2577 | Conserved hypothetical protein | 2.1 down | 0.015 |
| Rv2814c | Rv2814c | Probable transposase | 2.2 down | 0.011 |
| Rv3140 | fadE23 | Probable acyl-CoA dehydrogenase FadE23 | 2.1 up | 0.001 |
| Rv3854c | ethA | Monooxygenase EthA | 2.0 up | 0.003 |
The gene most up-regulated (3.8-fold) in the ΔRv1747 strain was the isoniazid-inducible gene iniB, whilst the second most up-regulated was iniA (3.2-fold). iniB forms an operon with iniA and iniC, which was up-regulated 1.6-fold. Other genes in the top 10 list of up-regulated genes included the probable acyl-CoA dehydrogenase fadE23 (2.1-fold) and a probable ferredoxin, fdxA (2.1-fold). Genes with a 2.0-fold up-regulation in the mutant were gyrB (DNA gyrase subunit B), PE_PGRS41 (Rv2396; a PE_PGRS family protein), ethA (whose gene product activates the prodrug ethionamide), ino1 [involved in the phosphatidyl-myo-inositol (PI) biosynthetic pathway] and two conserved hypothetical proteins, Rv0047c and Rv0822c. Of the STPKs, pknF (1.9-fold) and pknD (1.8-fold) were up-regulated. pknF expression was 2.0-fold up-regulated in the Rv1747 complement strain, probably because the complementing plasmid contains an intact copy of pknF.
Comparison of the WT and ΔpknF strains yielded only 12 genes differentially regulated at least twofold, all down-regulated in the mutant. This number increased to 72 with a 1.5-fold cut-off, of which only 12 were up-regulated. The microarray data for the 10 most up- and down-regulated genes in the pknF deletion strain are presented (Table 4). The pknF gene itself did not appear in the microarray results list because it did not pass the filtering stages within the analysis.
Table 4.
Microarray data for the 10 Mycobacterium tuberculosis genes most highly up- and down-regulated upon pknF deletion
| Rv number | Gene name | Gene product | Fold regulated | P-value |
|---|---|---|---|---|
| Rv0175 | Rv0175 | Probable conserved Mce-associated membrane protein | 1.5 up | 0.002 |
| Rv0341 | iniB | Isoniazid-inducible gene protein IniB | 1.8 up | 0.001 |
| Rv0796 | Rv0796 | Putative transposase for insertion sequence element IS6110 | 2.0 down | 0.035 |
| Rv1370c | Rv1370c | Putative transposase for insertion sequence element IS6110 | 2.0 down | 0.042 |
| Rv1371 | Rv1371 | Probable conserved membrane protein | 2.4 down | 0.042 |
| Rv1372 | Rv1372 | Conserved hypothetical protein | 2.2 down | 0.037 |
| Rv1738 | Rv1738 | Conserved hypothetical protein | 1.6 up | 0.005 |
| Rv2007c | fdxA | Probable ferredoxin fdxA | 1.8 up | 0.023 |
| Rv2106 | Rv2106 | Probable transposase | 2.0 down | 0.029 |
| Rv2167c | Rv2167c | Probable transposase | 2.0 down | 0.015 |
| Rv2480c | Rv2480c | Possible transposase for insertion sequence element IS6110 | 2.4 down | 0.003 |
| Rv2515c | Rv2515c | Conserved hypothetical protein | 2.0 down | 0.034 |
| Rv2815c | Rv2815c | Probable transposase | 2.0 down | 0.044 |
| Rv3640c | Rv3640c | Probable transposase | 2.0 down | 0.036 |
| Rv3727 | Rv3727 | Possible oxidoreductase | 1.5 up | 0.008 |
| Rv3728 | Rv3728 | Probable conserved two domain membrane protein | 1.5 up | 0.006 |
| Rv3842c | glpQ1 | Probable glycerophosphoryl diester phosphodiesterase GlpQ1 | 1.8 up | 0.006 |
| Rv3850 | Rv3850 | Conserved hypothetical protein | 1.7 up | 0.002 |
| Rv3854c | ethA | Monooxygenase EthA | 1.7 up | 0.006 |
| Rv3864 | espE | Esx-1 secretion-associated protein EspE | 1.6 up | 0.003 |
Interestingly, the gene most up-regulated in the ΔpknF strain was also iniB (1.8-fold). iniA was also up-regulated 1.5-fold in the pknF deletion strain. Other genes present in the ΔpknF list that were also up-regulated in the Rv1747 null strain were fdxA (1.8-fold) and ethA (1.7-fold). Other genes up-regulated in the ΔpknF mutant were glpQ1, a probable glycerophosphoryl diester phosphodiesterase (1.8-fold), the conserved hypothetical proteins Rv3850 (1.7-fold) and Rv1738 (espE), an Esx1 secretion-associated protein (1.6-fold), Rv3727 (probably involved in cellular metabolism), Rv3728 (probably involved in an efflux system) and Rv0175 (a probable conserved Mce-associated membrane protein; all 1.5-fold). The top 10 list of genes down-regulated in the ΔpknF strain comprised seven possible transposases; Rv2480c (2.4-fold), and Rv1380,Rv2106,Rv3640c,Rc2815c,Rv2167c,Rv0796 (all 2.0-fold). The remaining three down-regulated genes were Rv1371 (2.4-fold; conserved membrane protein) and Rv1372 (2.2-fold) and Rv2515c (2.0-fold), both annotated as conserved hypothetical proteins.
Confirmation of the microarray results for selected genes was obtained using qRT-PCR. The results (Fig. 1) confirmed that expression of Rv1747 in the deletion mutant was virtually undetectable and transcription was restored to WT levels in the complementing strain. Expression of pknF did not increase in the ΔRv1747 mutant, but did increase in the complementing strain, as mentioned above. Unlike the microarray data, the level of pknF transcript did not increase in the ΔRv1747 strain when determined by qRT-PCR. The transcriptional profiles of iniA,iniB,iniC,ethA and pknD all follow the same expression pattern as shown by the microarray results: transcript levels were increased in the Rv1747 mutant strain and were complemented when the Rv1747 gene was replaced. This was particularly striking for iniB and iniA where there was approximately three times as much transcript present in the mutant strain compared with the WT.
Figure 1.
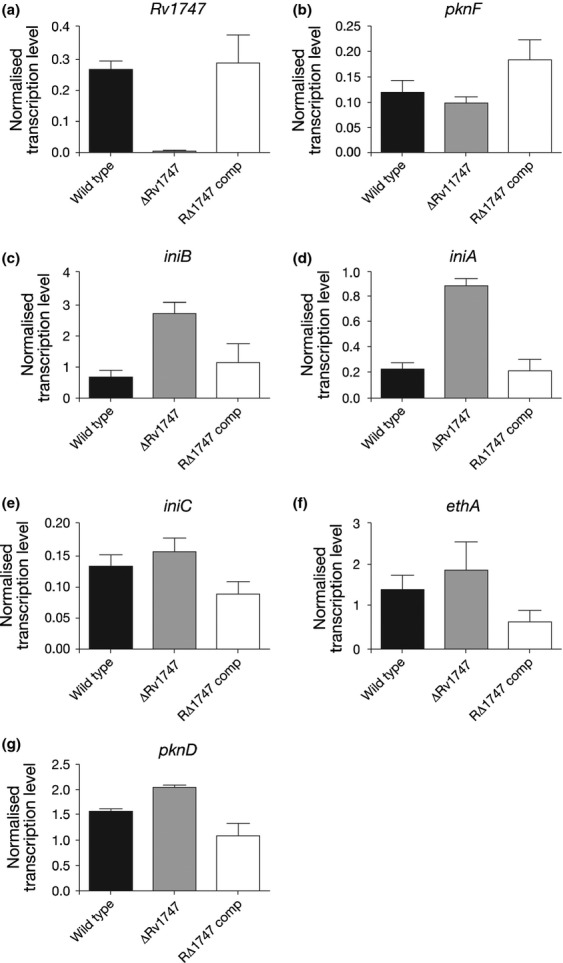
qRT-PCR to confirm the effect of Rv1747 deletion and complementation on the relative transcription levels of selected genes. Each panel shows the normalised transcription level of each gene investigated in WT,ΔRv1747 and complement strains. Data plotted are the mean of three biological replicates, and the error bars show the standard deviations. Data were normalised to sigA expression.
The results of qRT-PCR for the WT, ΔpknF and the pknF-complemented strain (Fig. 2) confirmed that expression of pknF in the ΔpknF mutant was undetectable and transcription was restored to almost WT level in the complementing strain. Expression of Rv1747 was the same in all three strains; thus, the transcriptional changes seen in the ΔpknF mutant are not due to changes in Rv1747 expression. The transcriptional profiles of iniB,iniA and iniC (Fig. 2c–e) follow the same pattern as shown by the microarray data. The iniC gene did not appear in the gene lists generated by microarrays in the pknF mutant or complement strain, but Fig. 2e clearly shows that the level of iniC transcript did not change in the ΔpknF mutant strain but was slightly decreased in the pknF-complemented strain.
Figure 2.
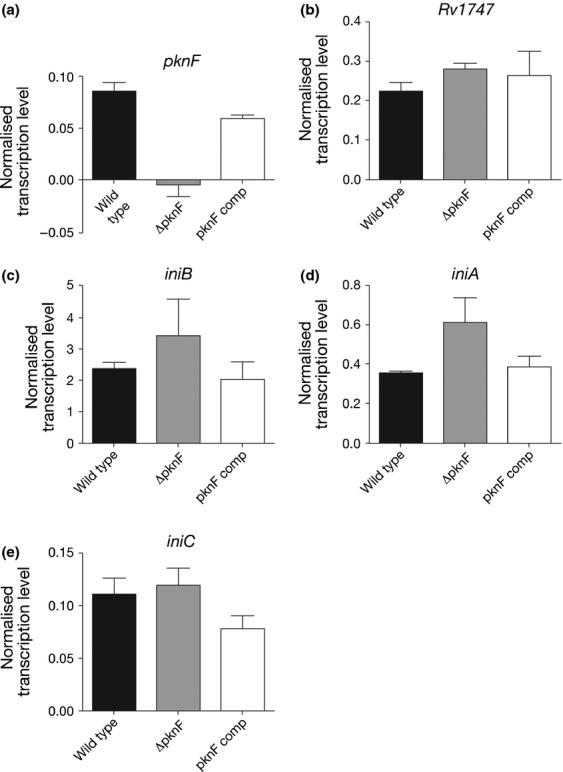
qRT-PCR to confirm the effect of pknF deletion and complementation on the relative transcription levels of selected genes. Each panel shows the normalised transcription level of each gene investigated in WT,ΔpknF and complement strains. Data plotted are the mean of three biological replicates, and the error bars show the standard deviations. Data were normalised to sigA expression.
The cell wall of the ΔpknF and ΔRv1747 mutants appeared normal by drug sensitivity assays, electron microscopy and lipoarabinomannan content
Many of the genes which were differentially regulated between WT and ΔpknF and ΔRv1747 mutants are involved in cell wall processes. We therefore tested whether the mutants were any more susceptible to cell wall and other stresses, viz. isoniazid, ethambutol, streptomycin, ciprofloxacin, ofloxacin, hydrogen peroxide, S-nitrosoglutathione, ethidium bromide, mitomycin C and sodium dodecyl sulphate, using a microplate Alamar Blue assay. However, no differences were observed in MICs between the mutants and the WT (Fig. S1).
Moreover, the transcription of the pknF-Rv1747 operon was not altered by isoniazid or ethambutanol (or indeed by a number of other stresses: gentamicin, streptomycin, hydrogen peroxide, t-butyl hydrogen peroxide, diamide, mitomycin C, S-nitrosoglutathione, ofloxacin, plumbagin, sodium nitroprusside, pH 7.2, pH 5.5 or in stationary phase; Fig. S2).
Transmission electron microscopy was performed to examine whether there were any differences in the cell wall structure and composition between the WT, ΔpknF,ΔRv1747 and the respective complementing strains. No discernible differences in cell wall structure were evident between the strains (Fig. S3).
The possibility that Rv1747-transported lipoarabinomannan was tested by analysing this molecule using two anti-lipoarabinomannan antibodies, one primarily recognising not only PIM6 but also ManLAM capped with three mannosyl residues (Mab F183-24) and one recognising the Ara6 structure in lipoarabinomannan (Mab F30-5). There were however no differences in the levels of ManLAM between cell suspensions of the WT, ΔRv1747 and Rv1747 complementing strains (Fig. S4).
Discussion
The substrate of the Rv1747 transporter is presently unknown. Moreover, other than the attenuation of growth of a ΔRv1747 mutant in mice and macrophages, there are presently no further mutant phenotypes known, growth being normal in vitro (Curry et al., 2005). We have used transcriptional microarray analysis to see whether this would throw light on the nature of Rv1747 transport function. This has demonstrated significant changes in transcriptional profiles between the ΔRv1747 and ΔpknF mutants and the WT. Interestingly, the genes most up-regulated in both of the mutant strains were in the iniBAC operon, identified as isoniazid-inducible (Alland et al., 1998). The iniA gene was also induced by ethambutol, another M. tuberculosis therapeutic agent that also targets the cell wall but with a different mechanism of action. Using M. bovis BCG, the promoter of the iniBAC operon was shown to be specifically induced by a broad range of inhibitors to cell wall biosynthesis including antibiotics that inhibited the synthesis of peptidoglycan, arabinogalactans, mycolic acids and fatty acids (Alland et al., 2000). iniA is also essential for the activity of an efflux pump, which confers resistance to isoniazid and ethambutol, although IniA does not directly transport isoniazid from the cell (Colangeli et al., 2005). All these findings would be compatible with the Rv1747 transporter exporting a component of the cell wall necessary for growth of the bacillus in vivo. The sensitivity of the ΔRv1747 or ΔpknF mutants was not however altered towards isoniazid or indeed any other of the agents tested. Neither was the transcription of the pknF-Rv1747 operon altered by isoniazid or ethambutanol. Moreover, there were no observable changes in morphology of the cell wall as seen by electron microscopy. Thus, Rv1747 does not appear to export any component of the cell wall that is involved in formation of an observable structure.
Other significant changes in expression of cell wall-associated genes were also found in the microarray study. Thus, ethA and ino1 were up-regulated in the ΔRv1747 strain. EthA functions to activate the prodrugs ethionamide, thiacetazone and isoxyl, which all use different mechanisms to inhibit mycolic acid synthesis (Dover et al., 2007). Ino1 is involved in the PI biosynthetic pathway; this phospholipid is also a component of the cell envelope. The list of the top ten down-regulated genes included two genes annotated as being conserved integral membrane proteins, namely Rv1999c and Rv2265, and one gene, Rv1382, annotated as a probable export or membrane protein. Up-regulation of all these genes may be acting as a compensatory mechanism for the loss of the function of Rv1747.
As PknF positively regulates Rv1747 function (Spivey et al., 2011), it may be expected that a ΔpknF mutant would have a similar phenotype to a ΔRv1747 mutant. Significantly, the iniB and iniA were also up-regulated in the ΔpknF mutant, correlating with the demonstration that PknF positively modulates the function of Rv1747 (Spivey et al., 2011). There were fewer genes whose expression level changed upon pknF deletion. This may be because of cross-talk between the 11 M. tuberculosis STPKs, which are able to cross-talk and recognise the same substrate in vitro (and probably also in vivo; Greenstein et al., 2005; Grundner et al., 2005; Molle & Kremer, 2010; Prisic et al., 2010). There is also a high degree of cross-reactivity between inhibitors of PknB and PknF (Lougheed et al., 2011).
The role of the STPK PknF that controls Rv1747 function has previously been examined in a pknF antisense strain (Deol et al., 2005). These authors described changes in cell morphology, including aberrant septum formation and reported a 16-fold increase in the uptake of d-glucose in the antisense strain. In contrast, in the present study, we did not find any morphological changes in the ΔpknF mutant.
Pitarque et al. (2008) have proposed that an unidentified transporter may be required to translocate the lipoglycans lipoarabinomannan and lipomannan, to the cell surface as the virulence of M. tuberculosis depends upon the export of these immunomodulatory molecules to the cell surface, as shown for the translocation of PDIMs (Sulzenbacher et al., 2006). The involvement in PDIM transport of an ABC transporter, DrrABC, together with the RND family protein MmpL7 (Cox et al., 1999; Camacho et al., 2001), which is a potential substrate of the STPK PknD (Pérez et al., 2006), is a possible paradigm for Rv1747 and PknF. Thus, the translocation of virulence-critical molecules such as lipoglycans could be a plausible explanation for the attenuation of the ΔRv1747 mutant in mice. However, our limited analysis with two anti-lipoarabinomannan antibodies failed to find any difference in this molecule in the ΔRv1747 mutant.
In this study, we have demonstrated that there are significant increases in gene expression in the efflux pump-related iniBAC genes in the ΔRv1747 and ΔpknF deletion strains. As this operon is inducible by a broad range of inhibitors of cell wall biosynthesis, we conclude that the loss of the Rv1747 transporter system affects cell wall biosynthesis and results in the accumulation of intermediates that may play a role in cell wall processes or biosynthesis, causing an induction of the iniBAC operon expression and implicates Rv1747 in exporting a component of the cell wall, which is required for virulence.
Acknowledgments
We thank Kathyrn Lougheed for help in setting up the Alamar Blue assays, Ben Appelmelk and Jeroen Geurtsen (VU University Medical Centre Amsterdam) for the gift of anti-lipoarabinomannan antibodies and Arnold Driessen (University of Groningen) for helpful discussion on ABC transporter function. We acknowledge BμG@S (the Bacterial Microarray Group at St. George's, University of London) and particularly Philip Butcher, Jason Hinds, Kate Gould and Denise Waldron for the supply of the M. tuberculosis microarrays and advice. This work was supported by an MRC studentship to V.L.S. and by the Medical Research Council (Grant numbers U117585867 and U117584228).
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Growth inhibition assays assessing the susceptibility of WT H37Rv, ΔpknF and ΔRv1747 strains to a range of drug and stress reagents.
β-Galactosidase assays on the pknF promoterlacZ strain and pEJ414 control strain in Mycobacterium tuberculosis after a panel of treatments.
Transmission electron micrographs of Mycobacterium tuberculosis comparing cell wall structure in (a) WT H37Rv, ΔRv1747 and Rv1747 complement strains, and (b) WT H37Rv, ΔpknF and pknF complement strains.
Mycobacterium tuberculosis whole cell ELISAs comparing the levels of ManLAM in H37Rv WT, ΔRv1747 and Rv1747 complement strains.
Supplementary Materials and methods
References
- Alland D, Kramnik I, Weisbrod TR, Otsubo L, Cerny R, Miller LP, Jacobs WR, Jr, Bloom BR. Identification of differentially expressed mRNA in prokaryotic organisms by customized amplification libraries (DECAL): the effect of isoniazid on gene expression in Mycobacterium tuberculosis. P Natl Acad Sci USA. 1998;95:13227–13232. doi: 10.1073/pnas.95.22.13227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alland D, Steyn AJ, Weisbrod T, Aldrich K, Jacobs WR., Jr Characterization of the Mycobacterium tuberculosis iniBAC promoter, a promoter that responds to cell wall biosynthesis inhibition. J Bacteriol. 2000;182:1802–1811. doi: 10.1128/jb.182.7.1802-1811.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braibant M, Gilot P, Content J. The ATP binding cassette (ABC) transport systems of Mycobacterium tuberculosis. FEMS Microbiol Rev. 2000;24:449–467. doi: 10.1111/j.1574-6976.2000.tb00550.x. [DOI] [PubMed] [Google Scholar]
- Camacho LR, Constant P, Raynaud C, Laneelle MA, Triccas JA, Gicquel B, Daffe M, Guilhot C. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Evidence that this lipid is involved in the cell wall permeability barrier. J Biol Chem. 2001;276:19845–19854. doi: 10.1074/jbc.M100662200. [DOI] [PubMed] [Google Scholar]
- Colangeli R, Helb D, Sridharan S, et al. The Mycobacterium tuberculosis iniA gene is essential for activity of an efflux pump that confers drug tolerance to both isoniazid and ethambutol. Mol Microbiol. 2005;55:1829–1840. doi: 10.1111/j.1365-2958.2005.04510.x. [DOI] [PubMed] [Google Scholar]
- Cox JS, Chen B, McNeil M, Jacobs WR., Jr Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature. 1999;402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- Curry JM, Whalan R, Hunt DM, Gohil K, Strom M, Rickman L, Colston MJ, Smerdon SJ, Buxton RS. An ABC transporter containing a forkhead-associated domain interacts with a serine–threonine protein kinase and is required for growth of Mycobacterium tuberculosis in mice. Infect Immun. 2005;73:4471–4477. doi: 10.1128/IAI.73.8.4471-4477.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassa E. Phylogenetic and functional classification of ABC (ATP-binding cassette) systems. In: Holland IB, Cole SPC, Kuchler K, Higgins CF, editors. ABC Proteins; From Bacteria to Man. Academic Press, London, UK; 2003. pp. 3–35. [Google Scholar]
- Davidson AL, Dassa E, Orelle C, Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev. 2008;72:317–364. doi: 10.1128/MMBR.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson RJ, Hollenstein K, Locher KP. Uptake or extrusion: crystal structures of full ABC transporters suggest a common mechanism. Mol Microbiol. 2007;65:250–257. doi: 10.1111/j.1365-2958.2007.05792.x. [DOI] [PubMed] [Google Scholar]
- Deol P, Vohra R, Saini AK, Singh A, Chandra H, Chopra P, Das TK, Tyagi AK, Singh Y. Role of Mycobacterium tuberculosis Ser/Thr Kinase PknF: implications in glucose transport and cell division. J Bacteriol. 2005;187:3415–3420. doi: 10.1128/JB.187.10.3415-3420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover LG, Alahari A, Gratraud P, Gomes JM, Bhowruth V, Reynolds RC, Besra GS, Kremer L. EthA, a common activator of thiocarbamide-containing drugs acting on different mycobacterial targets. Antimicrob Agents Chemother. 2007;51:1055–1063. doi: 10.1128/AAC.01063-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher D, Jackson SP. The FHA domain. FEBS Lett. 2002;513:58–66. doi: 10.1016/s0014-5793(01)03294-x. [DOI] [PubMed] [Google Scholar]
- Greenstein AE, Grundner C, Echols N, Gay LM, Lombana TN, Miecskowski CA, Pullen KE, Sung PY, Alber T. Structure/function studies of Ser/Thr and Tyr protein phosphorylation in Mycobacterium tuberculosis. J Mol Microbiol Biotechnol. 2005;9:167–181. doi: 10.1159/000089645. [DOI] [PubMed] [Google Scholar]
- Grundner C, Gay LM, Alber T. Mycobacterium tuberculosis serine/threonine kinases PknB, PknD, PknE, and PknF phosphorylate multiple FHA domains. Protein Sci. 2005;14:1918–1921. doi: 10.1110/ps.051413405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds J, Mahenthiralingam E, Kempsell KE, Duncan K, Stokes RW, Parish T, Stoker NG. Enhanced gene replacement in mycobacteria. Microbiology. 1999;145:519–527. doi: 10.1099/13500872-145-3-519. [DOI] [PubMed] [Google Scholar]
- Holland IB, Cole S, Kuchler K, Higgins C. ABC Proteins: From Bacteria to Man. London, UK: Academic Press; 2003. [Google Scholar]
- Hunt DM, Saldanha JW, Brennan JF, Benjamin P, Strom M, Cole JA, Spreadbury CL, Buxton RS. Single nucleotide polymorphisms that cause structural changes in the cyclic AMP receptor protein transcriptional regulator of the tuberculosis vaccine strain Mycobacterium bovis BCG alter global gene expression without attenuating growth. Infect Immun. 2008;76:2227–2234. doi: 10.1128/IAI.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D, Kunimoto DY. Secretion of human interleukin 2 by recombinant Mycobacterium bovis BCG. Infect Immun. 1995;63:799–803. doi: 10.1128/iai.63.3.799-803.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lougheed KE, Osborne SA, Saxty B, et al. Effective inhibitors of the essential kinase PknB and their potential as anti-mycobacterial agents. Tuberculosis (Edinb) 2011;91:277–286. doi: 10.1016/j.tube.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molle V, Kremer L. Division and cell envelope regulation by Ser/Thr phosphorylation: Mycobacterium shows the way. Mol Microbiol. 2010;75:1064–1077. doi: 10.1111/j.1365-2958.2009.07041.x. [DOI] [PubMed] [Google Scholar]
- Molle V, Soulat D, Jault JM, Grangeasse C, Cozzone AJ, Prost JF. Two FHA domains on an ABC transporter, Rv1747, mediate its phosphorylation by PknF, a Ser/Thr protein kinase from Mycobacterium tuberculosis. FEMS Microbiol Lett. 2004;234:215–223. doi: 10.1016/j.femsle.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Oatway WH, Steenken W. The pathogenesis and fate of tubercle produced by dissociated variants of tubercle bacilli. J Infect Dis. 1936;59:306–325. [Google Scholar]
- Papavinasasundaram KG, Anderson C, Brooks PC, Thomas NA, Movahedzadeh F, Jenner PJ, Colston MJ, Davis EO. Slow induction of RecA by DNA damage in Mycobacterium tuberculosis. Microbiology. 2001;147:3271–3279. doi: 10.1099/00221287-147-12-3271. [DOI] [PubMed] [Google Scholar]
- Pérez J, Garcia R, Bach H, de Waard JH, Jacobs WR, Jr, Av-Gay Y, Bubis J, Takiff HE. Mycobacterium tuberculosis transporter MmpL7 is a potential substrate for kinase PknD. Biochem Biophys Res Commun. 2006;348:6–12. doi: 10.1016/j.bbrc.2006.06.164. [DOI] [PubMed] [Google Scholar]
- Pitarque S, Larrouy-Maumus G, Payré B, Jackson M, Puzo G, Nigou J. The immunomodulatory lipoglycans, lipoarabinomannan and lipomannan, are exposed at the mycobacterial cell surface. Tuberculosis (Edinb) 2008;88:560–565. doi: 10.1016/j.tube.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prisic S, Dankwa S, Schwartz D, Chou MF, Locasale JW, Kang CM, Bemis G, Church GM, Steen H, Husson RN. Extensive phosphorylation with overlapping specificity by Mycobacterium tuberculosis serine/threonine protein kinases. P Natl Acad Sci USA. 2010;107:7521–7526. doi: 10.1073/pnas.0913482107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickman L, Scott C, Hunt DM, Hutchinson T, Menéndez MC, Whalan R, Hinds J, Colston MJ, Green J, Buxton RS. A member of the cAMP receptor protein family of transcription regulators in Mycobacterium tuberculosis is required for virulence in mice and controls transcription of the rpfA gene coding for a resuscitation promoting factor. Mol Microbiol. 2005;56:1274–1286. doi: 10.1111/j.1365-2958.2005.04609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin W, Hofnung M, Dassa E. Getting in or out: early segregation between importers and exporters in the evolution of ATP-binding cassette (ABC) transporters. J Mol Evol. 1999;48:22–41. doi: 10.1007/pl00006442. [DOI] [PubMed] [Google Scholar]
- Spivey VL, Molle V, Whalan RH, Rodgers A, Leiba J, Stach L, Walker KB, Smerdon SJ, Buxton RS. Forkhead-associated (FHA) domain containing ABC transporter Rv1747 is positively regulated by Ser/Thr phosphorylation in Mycobacterium tuberculosis. J Biol Chem. 2011;286:26198–261209. doi: 10.1074/jbc.M111.246132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer B, Sander P, Sedlacek L, Ellrott K, Boettger EC. Instability and site-specific excision of integration-proficient mycobacteriophage L5 plasmids: development of stably maintained integrative vectors. Int J Med Microbiol. 2001;290:669–675. doi: 10.1016/S1438-4221(01)80004-7. [DOI] [PubMed] [Google Scholar]
- Sulzenbacher G, Canaan S, Bordat Y, et al. LppX is a lipoprotein required for the translocation of phthiocerol dimycocerosates to the surface of Mycobacterium tuberculosis. EMBO J. 2006;25:1436–1444. doi: 10.1038/sj.emboj.7601048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth inhibition assays assessing the susceptibility of WT H37Rv, ΔpknF and ΔRv1747 strains to a range of drug and stress reagents.
β-Galactosidase assays on the pknF promoterlacZ strain and pEJ414 control strain in Mycobacterium tuberculosis after a panel of treatments.
Transmission electron micrographs of Mycobacterium tuberculosis comparing cell wall structure in (a) WT H37Rv, ΔRv1747 and Rv1747 complement strains, and (b) WT H37Rv, ΔpknF and pknF complement strains.
Mycobacterium tuberculosis whole cell ELISAs comparing the levels of ManLAM in H37Rv WT, ΔRv1747 and Rv1747 complement strains.
Supplementary Materials and methods


