Background: Delaying DNA synthesis kinetics of HIV-1 reverse transcriptase by lowering dNTP concentrations elevates biochemical RNA template switch.
Results: Degradation of SAMHD1 by Vpx, which increases dNTP levels, reduces HIV-1 template switching in macrophages, but not in CD4+ T cells.
Conclusion: Altering cellular dNTP levels directly affects in vivo HIV-1 template switching.
Significance: Cellular dNTP abundance controlled by SAMHD1 contributes to HIV-1 template switching in macrophages.
Keywords: HIV-1, Homologous Recombination, Macrophages, Nucleotide, Reverse Transcription, SAMHD1, Vpx
Abstract
Template switching can occur during the reverse transcription of HIV-1. Deoxynucleotide triphosphate (dNTP) concentrations have been biochemically shown to impact HIV-1 reverse transcriptase (RT)-mediated strand transfer. Lowering the dNTP concentrations promotes RT pausing and RNA template degradation by RNase H activity of the RT, subsequently leading to strand transfer. Terminally differentiated/nondividing macrophages, which serve as a key HIV-1 reservoir, contain extremely low dNTP concentrations (20–50 nm), which results from the cellular dNTP hydrolyzing sterile α motif and histidine aspartic domain containing protein 1 (SAMHD1) protein, when compared with activated CD4+ T cells (2–5 μm). In this study, we first observed that HIV-1 template switching efficiency was nearly doubled in human primary macrophages when compared with activated CD4+ T cells. Second, SAMHD1 degradation by viral protein X (Vpx), which elevates cellular dNTP concentrations, decreased HIV-1 template switching efficiency in macrophages to the levels comparable with CD4+ T cells. Third, differentiated SAMHD1 shRNA THP-1 cells have a 2-fold increase in HIV-1 template switching efficiency. Fourth, SAMHD1 degradation by Vpx did not alter HIV-1 template switching efficiency in activated CD4+ T cells. Finally, the HIV-1 V148I RT mutant that is defective in dNTP binding and has DNA synthesis delay promoted RT stand transfer when compared with wild type RT, particularly at low dNTP concentrations. Here, we report that SAMHD1 regulation of the dNTP concentrations influences HIV-1 template switching efficiency, particularly in macrophages.
Introduction
Retroviruses contain two copies of a single-stranded RNA genome. During the reverse transcription process, the genetic segments encoded in each of the two RNA copies harbored in a single viral particle can be exchanged. Moreover, in HIV-1 this recombination event occurs 20% of the time during viral replication (1, 2). First, superinfection of a cell with two genetically distinct virions can generate new viral progenies containing heterodimeric RNA genomes from the two initial parental virions. Next, during the reverse transcription step upon infection of a new target cell, recombination of the heterodimeric genomes can occur. This can lead to the production of recombinant viral progenies containing a mixture of both heterodimeric genome sequences. Importantly, this can ultimately accelerate viral genetic diversification and viral escape from immunological or drug pressure. Moreover, HIV-1 can rapidly develop multiple drug-resistant mutations by recombining with pre-existing mutations resistant to each of individual antiviral drugs (3, 4).
Several mechanistic factors affecting the HIV-1 recombination event have been identified. Viral recombination between homologous regions of two viral RNA copies requires both DNA synthesis and RNA template degradation, which are catalyzed by DNA polymerase and RNase H activities of RT,2 respectively (5, 6). Upon pausing of RT during the DNA synthesis, the RNase H activity degrades the RNA templates. This creates ssDNA regions that can initiate the strand transfer and homologous recombination by the primer annealing to the homologous region of the other RNA genome harbored in the same viral particle (7, 8). In this “strand invasion” mechanism model, factors that cause RT pausing directly affect recombination and strand transfer frequency. Basically, frequent HIV RT pausing leads to higher strand transfer events (1, 9–11). The secondary structures of RNA templates, such as stem loops, are also known to induce RT pausing and enhance strand transfer (1, 12, 13). Another factor that influences strand transfer is the ability of RT to bind the template/primer. The dipeptide insertion mutation at the finger domain of RT alters the interaction with template/primer and affects biochemical strand transfer of RT (14).
In vitro strand transfer assays have also revealed that when dNTP concentrations are titrated lower, there is a linear increase in strand transfer products (15). Kinetic delays made by low dNTP concentration can contribute to frequent RT pausing, which can promote RNA template degradation and strand invasion (5, 15). Thus it has been hypothesized that HIV-1 may undergo more strand transfer events during viral replication in cell types containing lower dNTP concentrations (5, 15). HIV-1 infects both activated/dividing CD4+ T cells and terminally differentiated/nondividing monocyte-derived macrophages (MDMs) (16–18). Importantly, we reported that human primary macrophages harbor 20–50 nm cellular dNTP concentrations, whereas activated CD4+ T cells harbor 2–5 μm (16, 17, 19).
Recently, a cellular protein called sterile α motif and histidine aspartic domain containing protein 1 (SAMHD1) was discovered as a new anti-lentivirus host restriction protein. SAMHD1 restricts the reverse transcription of lentiviruses by lowering cellular dNTP concentrations by using its dNTP triphosphohydrolase activity to hydrolyze dNTPs to deoxynucleosides (dN) and triphosphates (20–22). Indeed, SAMHD1 restricts lentivirus replication particularly in nondividing cells by reducing the cellular dNTP concentrations below the level of the substrate necessary for efficient reverse transcription of viral DNA synthesis (19, 23, 24). HIV-2 and simian immunodeficiency virus sooty mangabey (SIVsm) encode a viral protein X (Vpx), which targets SAMHD1 for proteasomal degradation (22, 25). Although HIV-1 does not express Vpx, we can introduce Vpx by transducing the cells with virus-like particles (VLP) containing Vpx to modulate dNTP levels through SAMHD1 degradation. The degradation of SAMHD1 by Vpx does not require the de novo expression of Vpx in the infecting cells because VLP containing Vpx can sufficiently accelerate the reverse transcription kinetics of the co-infecting viruses in trans (26, 27). This elevates cellular dNTP concentration and ultimately rescues the virus from the restricted viral DNA synthesis in macrophages (19, 28). This Vpx-mediated rescue of HIV-1 was also observed in resting CD4+ T cells and dendritic cells (23, 29). Furthermore, Vpx can enhance reverse transcription of other lentiviruses such as SIV and feline immunodeficiency virus (30, 31). Furthermore, we also reported that SAMHD1 restricts the replication of various DNA viruses that infect macrophages, such as vaccinia virus and herpesvirus (32).
Although biochemical evidences support that lowering dNTP concentrations promotes RT-mediated template switch and recombination, there have been conflicting studies regarding the recombination frequency in CD4+ T cells and macrophage. Levy et al. (33) reported higher recombination in MDMs when compared with CD4+ T cells, whereas Chen et al. (34) reported there were no significant differences in the recombination frequency between these two different cell types.
In this study, we investigated the effect of the SAMHD1-Vpx network, which regulates the in vivo cellular dNTP concentration availability, on HIV-1 template switching frequency. We employed not only multiple primary cells and cell line models, but also an HIV-1 vector system engineered to measure viral template switching efficiency. This system expresses the entire HIV-1 genome except env and nef. The nef gene was replaced with a bicistronic reporter expressing heat-stable antigen (HSA) for detecting transduction efficiency and a nonfluorescent GFP mutant for monitoring viral template switching events (6, 35). Here, we report that SAMHD1 degradation by Vpx+ VLP affected HIV-1 template switching frequency in MDMs due to an increase in dNTP levels.
MATERIALS AND METHODS
Cell Cultures and Reagents
CD4+ T cells and monocytes were isolated from buffy coats (New York Blood Bank), and activated CD4+ T cells and MDMs were prepared as described previously (23, 24, 36). Scramble or SAMHD1 shRNA expressing THP-1 cells were kindly provided from Dr. N. Landau (New York University) and prepared as described previously (37). Human lung fibroblast (HLF) cells (ATCC) were cultured as described previously (38, 39). Cells were transduced with Vpx−/Vpx+ VLP for 18 h before Western blot analysis using anti-SAMHD1, anti-β-actin, or anti-GAPDH antibodies (Abcam). HSA CD24 antibody was purchased from BD Biosciences. 25 μg of proteins were loaded for Western blot analysis.
dNTP Extraction and Assay
This assay protocol has been published previously (16). Cells were treated with Vpx−/Vpx+ VLP for 18 h before dNTPs were extracted.
HIV-1 Vector Construction and Preparation
The NL4-3-based pHIG plasmid was a gift from Dr. Louis Mansky (35). It contains the entire HIV-1 genome except for env and nef genes. An HSA reporter flanked by an internal ribosomal binding site (IRES) sequence, followed by a GFP reporter that replaced the Nef gene, will be referred to in this study as the WT vector. This IRES-GFP portion in pHIG was replaced with the IRES-GFFP (a kind gift from Dr. Vinay Pathak from the NCI, National Institutes of Health) recombinant sequence, which was previously used to measure HIV-1 template switching (6). This vector we refer to as GFFP. pHIG and GFFP plasmids were transfected independently into 293FT cells along with vesicular stomatitis virus G using polyethylenimine (Sigma) for 6 h before fresh media was replaced. Viral supernatant for pHIG was collected 24 h after transfection. For GFFP vector production, 6 h after transfection, 10 μm azidothymidine was added to the fresh media to prevent vector from reinfecting 293FT. GFFP viral vectors were collected at 24 h and purified through a 20% sucrose cushion to remove azidothymidine (6, 40). VLP vector production was done as described previously (24, 27). V148I RT mutation was created by site-directed mutagenesis using the primers previously described (38).
Transduction Efficiency and Flow Cytometry
Viral vectors produced for the experiment were normalized for the same transduction efficiency based on the ELISA p27 (Advanced Bioscience Laboratories, Inc.) assay for VLP and p24 assay (Abcam) for the pHIG and GFFP. VLP were transduced at 4.5 × 104 pg of p27 per 300,000 cells. 300,000 CD4+ T cells were transduced through spinoculation at 3000 rpm for 10 min with 1.7 × 105 pg of p24 under Vpx−/Vpx+ VLP treatment. Cells were washed after spinoculation. 300,000 MDMs were transduced with 5.8 × 105 and 1.6 × 105 pg of p24, respectively, under Vpx−/Vpx+ VLP treatment. THP-1 cells were transduced with 2.5 × 105 and 8.3 × 104 pg of p24 with shRNA scramble and shRNA SAMHD1, respectively. HLFs transduced with V148I mutant had 2.5 × 105 and 8.3 × 104 pg of p24, respectively, under Vpx−/Vpx+ VLP treatment, whereas the WT control used the same amount of p24 as in CD4+ T cells. The transduced cells were collected for flow cytometry analysis between 2 and 6 days after transduction as described below. All flow cytometry analysis was done using Auto Macs except for THP-1, which used Accuri. The percentages of live HSA+/− and GFP+/− populations were quantified for each viral vector at specific time points using FlowJo software. Results were done in triplicates for at least three different donors.
RESULTS
HIV-1 Template Switching Frequency in Human Primary Monocyte-derived Macrophages (MDMs) and Activated CD4+ T Cells
First, to investigate the effect of the SAMHD1-Vpx network on HIV-1 template switching frequency, we utilized the pHIG NL4-3 HIV-1 vector system, which has been established for investigating the viral mutation rate (35). This construct was used as our WT control vector. In addition, to investigate the HIV template switching efficiency, we replaced the enhanced GFP (eGFP) gene of pHIG vector with a nonfluorescent “GFFP” mutant construct that contains a 225-bp duplicated “F” region that is responsible for the loss of the protein fluorescence (Fig. 1A) (6). This duplication results in a GFP protein that is not functional, (GFP−), in cells by flow analysis. If template switching occurs at the duplicated F region of the GFFP gene, this results in the loss of one of the duplicated F segments; this may reconstitute a functional eGFP protein, which is read as GFP+ cells by flow analysis as described previously (Fig. 1A) (6, 35, 41). Cell surface expression of HSA was used to determine the total number of transduced (HSA+) versus nontransduced (HSA−) cells by flow analysis. All HIV-1 vectors used in this study were pseudotyped with vesicular stomatitis virus G, and the vectors were normalized by p24 ELISA.
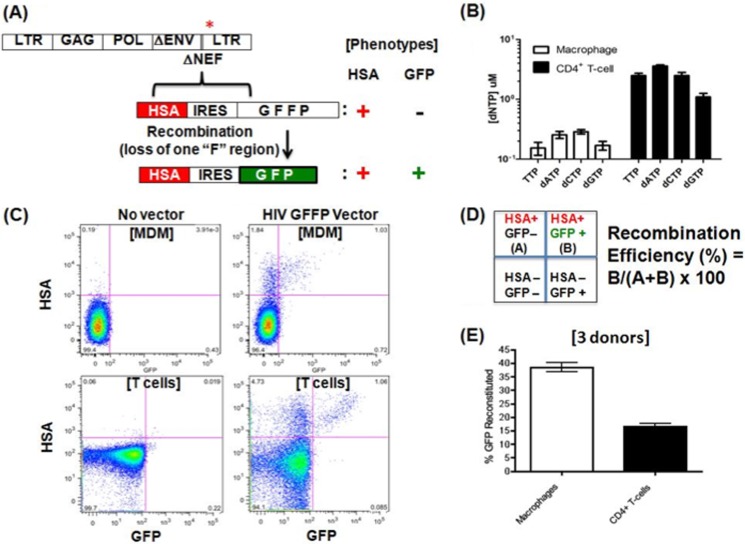
FIGURE 1.
Template switching frequency in macrophage and activated CD4+ T cells. A, HIV-1 vector system for measuring viral template switching efficiency. The previously constructed NL4-3-based HIV-1 vector was modified by replacing the nef gene with a dual expression cassette expressing HSA and flanked by an IRES, followed by a modified/non-fluorescent GFP (GFFP), which encodes an internally duplicated 225-bp GFP sequence (6, 35). Upon template switching the functional/fluorescent GFP is reconstituted, generating cells with HSA+/GFP+ readouts. B, dNTP concentrations of MDMs were 0.15–0.28 μm, and dNTP concentrations of human primary activated CD4+ T cells were 1–3.5 μm with error bars representing an experiment from three donors. C, flow cytometry results from CD4+ T cells and MDMs transfected with the GFFP vector (no vector control cells at right). Activated CD4+ T cells and MDMs were collected at days 2 and 6 after transduction, respectively, when no further increase of the HSA+ population was observed. D, quadrants of the analyzed cell populations in the flow plots and template switching efficiency calculation using reconstituted GFP+/total HSA+ population. A, HSA+/GFP−; B, HSA+/GFP+. E, template switching efficiency comparison of activated CD4+ T cells and MDMs. Error bars represent experiment from three donors.
MDMs and activated CD4+ T cells were prepared from three different donors. We first confirmed the dNTP concentration difference between these two cell types by the HIV RT primer extension-based dNTP assay (16). Indeed, MDMs from these donors display much lower dNTP concentrations than activated CD4+ T cells (Fig. 1B). We then transduced these two cell types with the GFFP vector, and cells were collected and stained with anti-HSA antibody for flow analysis at days 2 and 6 after transduction for CD4+ T cells and MDMs, respectively. Flow cytometry plots (Fig. 1C) show expression of HSA and functional GFP for MDMs and CD4+ T cells. Importantly, GFP expression in cells only occurs after template switching of the nonfluorescent GFFP mutant. As illustrated in Fig. 1D, the upper left quadrant shows HSA+ GFP− cells, whereas the upper right quadrant displays the HSA+ GFP+ cell population. The template switching efficiency in each of these cells types was determined by a ratio between number of cells (B) that express both HSA and reconstituted GFP (HSA+/GFP+) to the total number of transduced HSA+ cells, which include both HSA+/GFP− (A) and HSA+/GFP+ (B) cell populations: (B/(A+B) × 100) (Fig. 1D). Note that under our experimental condition, the HIV-1 wild type GFP vector control, in which the numbers of GFP+ and HSA+ cells are at a 1:1 ratio, suggest equal detection power of both GFP and HSA markers in this system (data not shown). Upon the determination of the template switching efficiency with three donors (Fig. 1E), HIV-1 has nearly 2-fold higher template switching frequency in MDMs when compared with CD4+ T cells. This 2-fold difference in HIV-1 template switching is consistent with the observation previously reported by using a different recombination assay system (33).
Effect of SAMHD1 and Vpx on HIV-1 Template Switching in MDMs
When MDMs are treated with VLP containing Vpx protein, Vpx induces E3-ubiqutin-mediated proteasomal degradation of SAMHD1, which is followed by the rapid increase of cellular dNTP concentrations in MDMs (22, 24, 27, 42). Therefore, we tested whether the cellular dNTP levels controlled by SAMHD1 and Vpx affect HIV-1 template switching efficiency in MDMs. First, MDMs were treated with Vpx− or Vpx+ VLP, and we confirmed the efficient degradation of SAMHD1 by Western analysis from three donors (Fig. 2A). Second, we conducted the dNTP assay with MDMs (from the same donors) treated with Vpx− or Vpx+ VLP to confirm that Vpx VLP treatment elevated dNTP concentrations across all four nucleotides (Fig. 2B). Indeed the Vpx-induced SAMHD1 degradation elevated dNTP concentrations of macrophages close to that of CD4+ T cells (near 1 μm). Next, we pretreated MDMs from the same donors with Vpx− or Vpx+ VLP and then transduced the MDMs with the GFFP HIV vectors. The template switching frequency was determined by flow analysis, as shown in Fig. 2C. In Fig. 2D, quantification of the flow plots from three different donors for MDMs showing no VLP and Vpx− VLP displayed a similar high template switching efficiency (35–40%), whereas the Vpx+ VLP MDMs showed a reduced template switching efficiency, which is close to that of the activated CD4+ T cells. Collectively, these data support that SAMHD1 affects HIV-1 template switching in MDMs.
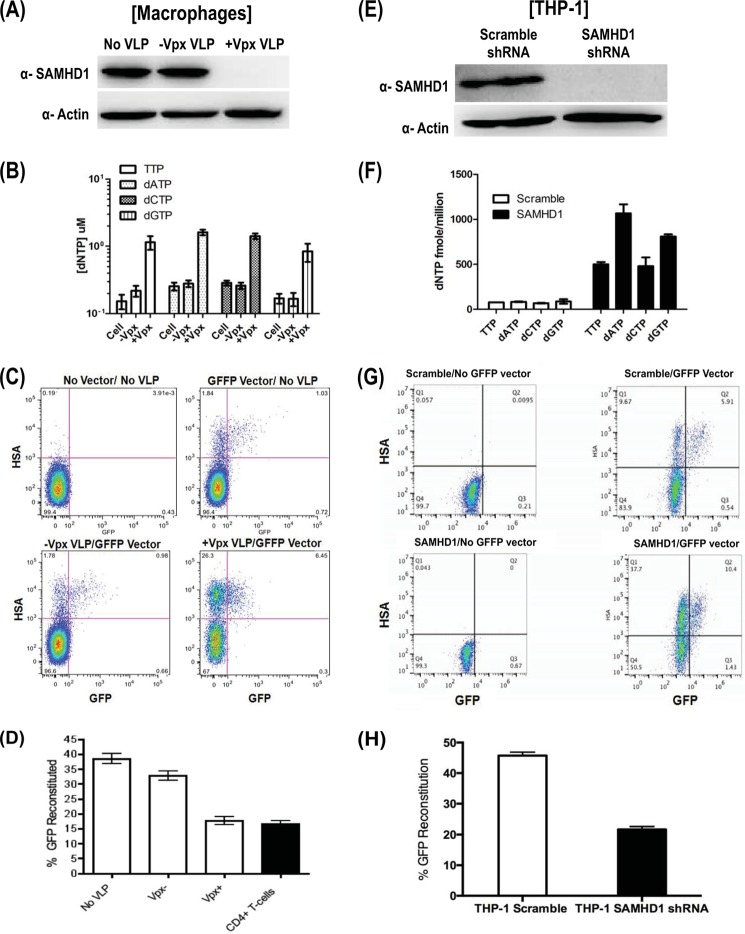
FIGURE 2.
Effects of Vpx-mediated SAMHD1 degradation on HIV-1 template switching in MDMs. A, MDMs from three donors were treated with the Vpx− or Vpx+ VLP (no VLP control) for 18 h, and the SAMHD1 level and actin control were examined by Western blot analysis. B, dNTP level of MDMs shows Vpx−/+ VLP 0.16–0.28 and 0.85–1.6 μm, respectively. Error bars represent experiment from three donors. C, MDMs pretreated with Vpx− or Vpx+ VLP (no-VLP control) were transduced with GFFP vector, and the cells were harvested at day 6 after transduction for flow analysis. D, quantification of flow data from panel C with error bars representing experiment from three donors. Template switching efficiency determined from the reconstituted functional GFP+ population is described in the legend for Fig. 1D. The template switching efficiency of activated CD4+ T cells in the black bar is for comparison. E, Western blot analysis of SAMHD1 in differentiated THP-1 macrophage cell lines expressing shRNA against SAMHD1 or control scrambled shRNA. F, dNTP levels range from 499–1067 fmol/million and 68–87 fmol/million for scramble shRNA and SAMHD1 shRNA, respectively. Error bars represent duplicate experiment. G, flow analysis of the scramble and SAMHD1 shRNA expressing THP-1 transduced with GFFP vector. H, the template switching efficiencies of these two cell lines were determined in triplicates and compared as described in the legend for Fig. 1D.
Effect of SAMHD1 Knockdown on HIV-1 Template Switching in THP-1 Cells
Next, we further confirmed the role of SAMHD1 in HIV-1 template switching by using a monocytic cell line, THP-1, that expresses either shRNA against SAMHD1 or a scrambled control shRNA (19, 31, 42). After differentiating the scrambled or SAMHD1 shRNA expressing THP-1 cell lines into macrophages with phorbol 12-myristate 13-acetate, we ensured that the SAMHD1 knockdown was efficient via Western blot analysis. As shown in Fig. 2E, shRNA against SAMHD1 had efficient knockdown protein level, whereas the scrambled shRNA control had detectable SAMHD1 expression. In addition, Fig. 2F shows that THP-1 cells with shRNA against SAMHD1 exhibited elevated dNTP levels when compared with the scrambled THP-1 control cells. Finally, when the THP-1 cells were transduced with the GFFP vector seen in the flow plot (Fig. 2G), the GFP reconstitution was diminished by one-half when compared with the scrambled THP-1 control when the flow plot was quantified from the triplicate experiments shown in Fig. 2H. These data further support our hypothesis that SAMHD1 affects HIV-1 template switching in macrophages.
Effects of SAMHD1 Degradation on HIV-1 Templates Switching in Primary Activated CD4+ T Cells
Although SAMHD1 expression in activated/dividing CD4+ T cells has been reported (23), the cellular dNTP levels are maintained at high levels, likely due to cell cycle-related dNTP biosynthesis. Next, we tested whether the Vpx-mediated SAMHD1 degradation also affects HIV-1 template switching in activated CD4+ T cells. First, we examined whether Vpx+ VLP can degrade SAMHD1 in activated CD4+ T cells. As shown in Fig. 3A, the treatment of activated CD4+ T cells by Vpx+ induced SAMHD1 degradation (when compared with no VLP and Vpx− VLP-treated cells). Also, the dNTP assay (Fig. 3B) showed that Vpx+ VLP treatment elevated the dNTP concentrations of activated CD4+ T cells (three donors) from 1–3.5 to 7–18 μm. As shown in Fig. 3, C and D, the cells transduced with the GFFP vector with Vpx+ VLP treatment (Fig. 3D) displayed the same template switching efficiency with the CD4+ T cells treated with Vpx− VLP or no VLP by flow analysis. This was expected because the dNTP concentrations of activated CD4+ T cell are already above the steady state Km level of HIV-1 RT. HIV-1 RT is already fully capable of processive DNA synthesis with little pause required for template switching. Basically, the higher dNTP increased above the Km value of HIV-1 RT does not significantly improve the kinetics and processive DNA synthesis of wild type HIV-1 RT. This likely resulted in no significant alteration of HIV-1 template switching upon the SAMHD1 degradation in activated CD4+ T cells.
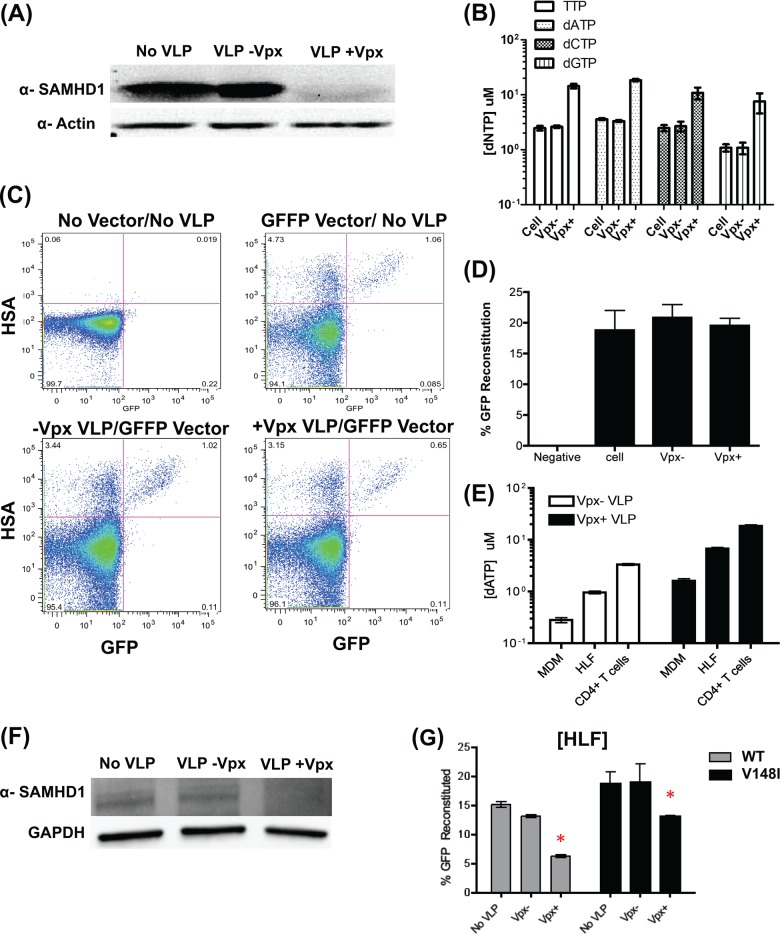
FIGURE 3.
Effect of Vpx-mediated SAMHD1 degradation in activated CD4+ T cells and RT V148I dNTP binding mutant on viral template switching in human lung fibroblast. Activated CD4+ T cells were treated with the Vpx- or Vpx+ VLP (no VLP control) for 18 h, and the levels of SAMHD1 via Western blot are shown in panel A. B, dNTP of CD4+ T cells with error bars representing experiment from three donors shows levels of 1–3.5 μm with Vpx− VLP to 7–18 μm with Vpx+ VLP. C, The same donor CD4+ T cells pretreated for 18 h with Vpx− or Vpx+ VLP before the cells were transduced with GFFP vectors. The cells were collected at 48 h after transduction for flow cytometry. Activated CD4+ T cells with no vector/no VLP as well as transduced with GFFP vector without the VLP pretreatment were used as control. D, the template switching efficiency of activated CD4+ T cells was determined by the reconstituted GFP+ population with error bars representing experiment from three donors. E, HLF dNTP assay in duplicate experiment showed that Vpx−/+ VLP have an average of 0.960 and 6.74 μm/million, respectively. dATP concentrations for MDMs and CD4+ T cells are shown for comparison. F, Western blot analysis of HLFs treated with VLP and no VLP treatment after 18 h. G, HLFs were pretreated with Vpx− or Vpx+ VLP for 18 h or no VLP treatment as control and were transduced with GFFP vector containing wild type (WT) or V148I RT mutant. The cells were harvested at 48 h after transduction, and the template switching efficiency of the cells was determined by flow cytometry in triplicates. Error bars represent experiment from triplicate experiments. *, V148I vector showed a 2-fold higher template switching efficiency than WT vector when the cells were pretreated with Vpx+ VLP.
Effect of SAMHD1 on Template Switching of HIV-1 Vector Harboring a dNTP Binding V148I RT Mutant
Finally, we investigated the relationship between HIV-1 RT kinetics and SAMHD1 in terms of HIV-1 template switching. We employed the HIV-1 V148I RT mutant, which has reduced dNTP binding affinity and thus displays diminished processive DNA synthesis (15, 38). This causes the RT to pause more frequently, which increases RNA template degradation and increases strand transfer when compared with wild type HIV-1 RT, particularly at unsaturated dNTP concentrations (i.e. below the Km value of HIV-1 RT) (15, 43, 44). First, we introduced the V148I RT mutation into the pol gene of the GFFP vector. The V148I HIV-1 vector transduced MDMs poorly due to the reduced dNTP binding affinity and the low dNTP concentrations. However, this mutant vector is able to transduce primary HLFs, which harbors an average of 0.960 μm dATP per million cells in Fig. 3E, which is 3.4-fold higher than MDMs and 3.5-fold lower than CD4+ T cells. As shown in Fig. 3G, both WT and V148I RT mutant GFFP vectors show reduced template switching efficiency upon Vpx+ VLP treatment. This is due to SAMHD1 degradation, which is shown in Fig. 3F. This is consistent with the observation in MDMs. Interestingly, the V148I vector showed a 2-fold higher template switching efficiency than WT vector when the cells were pretreated with Vpx+ VLP (see * in Fig. 3G). These data support our hypothesis that delayed RT kinetics by the V148I mutation enhances viral template switching efficiency due to low dNTP abundance.
DISCUSSION
Biochemical studies have shown that low dNTP concentrations are known to interfere with the RT-mediated processive DNA synthesis and increase pausing, which enhances dynamic copy choice strand transfer events (5, 11, 15, 45). Slow polymerization during reverse transcription allows RT to exert its RNase H activity to degrade viral RNA template for strand invasion to occur (1, 11, 15). SAMHD1 has been shown to restrict HIV-1 replication in MDMs due to its dNTP hydrolase activity, which reduces the substrate availability for RT (19, 24). The results from this study show that SAMHD1 activity also indirectly increases template switching frequency by decreasing viral replication kinetics due to the reduction of dNTPs (24).
Our results show that SAMHD1 degradation by Vpx elevates dNTP concentrations of MDMs to the micromolar range, which lowers HIV-1 template switching efficiency in MDMs. At this dNTP concentration, the Km values for RT where dNTP availability is no longer the rate-limiting step of viral DNA synthesis result in less frequent pausing and strand transfer (5, 15, 46). Our V148I dNTP binding-defective mutation resulted in a higher template switching frequency, which compares well with previous findings that dNTP binding-defective mutations such as Q151N and M184I also show increased template switching events (6, 15). This further supports the dynamic copy choice strand transfer mechanism.
Our results show that macrophage has higher template switching frequency than CD4+ T cells, which correlates with the findings of Levy et al. (33). Although the results are different from those of Chen et al. (34), we find that these varied results may be due to the differing experimental design. The recombination system used by Chen et al. (34) has a heterologous virion and a specific theoretical measurable maximum recombination frequency calculation where the numbers are based on genetic random assortment probabilities to calculate recombination frequency.
Lastly, because HIV-1 does not encode Vpx, unlike HIV-2/SIVsm, it is a testable hypothesis that HIV-1 may undergo more frequent genetic recombination than HIV-2 and SIVsm, particularly in macrophages. In conclusion, the virological data presented in this study support that SAMHD1 protein, which modulates cellular dNTP abundance, contributes to HIV-1 template switching efficiency particularly in macrophages.
This work was supported, in whole or in part, by National Institutes of Health Grants AI077401 and AI049781 (to B. K.). This work was also supported by the Emory Center for AIDS Research.
This paper is dedicated in memory of our dear friend and colleague Shilpa Aggarwal. We will remember the times spent together with you and miss you always.
- RT
- reverse transcriptase
- dNTP
- deoxynucleotide triphosphate
- MDM
- monocyte-derived macrophage
- SAMHD1
- sterile α motif and histidine aspartic domain containing protein 1
- SIVsm
- simian immunodeficiency virus sooty mangabey
- Vpx
- viral protein X
- VLP
- virus-like particles(s)
- HLF
- human lung fibroblast
- HSA
- heat-stable antigen
- IRES
- internal ribosomal entry site.
REFERENCES
- 1. Basu V. P., Song M., Gao L., Rigby S. T., Hanson M. N., Bambara R. A. (2008) Strand transfer events during HIV-1 reverse transcription. Virus Res. 134, 19–38 [DOI] [PubMed] [Google Scholar]
- 2. Onafuwa-Nuga A., Telesnitsky A. (2009) The remarkable frequency of human immunodeficiency virus type 1 genetic recombination. Microbiol. Mol. Biol. Rev. 73, 451–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gu Z., Gao Q., Faust E. A., Wainberg M. A. (1995) Possible involvement of cell fusion and viral recombination in generation of human immunodeficiency virus variants that display dual resistance to AZT and 3TC. J. Gen. Virol. 76, 2601–2605 [DOI] [PubMed] [Google Scholar]
- 4. Moutouh L., Corbeil J., Richman D. D. (1996) Recombination leads to the rapid emergence of HIV-1 dually resistant mutants under selective drug pressure. Proc. Natl. Acad. Sci. U.S.A. 93, 6106–6111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gao L., Hanson M. N., Balakrishnan M., Boyer P. L., Roques B. P., Hughes S. H., Kim B., Bambara R. A. (2008) Apparent defects in processive DNA synthesis, strand transfer, and primer elongation of Met-184 mutants of HIV-1 reverse transcriptase derive solely from a dNTP utilization defect. J. Biol. Chem. 283, 9196–9205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nikolenko G. N., Svarovskaia E. S., Delviks K. A., Pathak V. K. (2004) Antiretroviral drug resistance mutations in human immunodeficiency virus type 1 reverse transcriptase increase template-switching frequency. J. Virol. 78, 8761–8770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rigby S. T., Van Nostrand K. P., Rose A. E., Gorelick R. J., Mathews D. H., Bambara R. A. (2009) Factors that determine the efficiency of HIV-1 strand transfer initiated at a specific site. J. Mol. Biol. 394, 694–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rigby S. T., Rose A. E., Hanson M. N., Bambara R. A. (2009) Mechanism analysis indicates that recombination events in HIV-1 initiate and complete over short distances, explaining why recombination frequencies are similar in different sections of the genome. J. Mol. Biol. 388, 30–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wisniewski M., Balakrishnan M., Palaniappan C., Fay P. J., Bambara R. A. (2000) Unique progressive cleavage mechanism of HIV reverse transcriptase RNase H. Proc. Natl. Acad. Sci. U.S.A. 97, 11978–11983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wisniewski M., Balakrishnan M., Palaniappan C., Fay P. J., Bambara R. A. (2000) The sequential mechanism of HIV reverse transcriptase RNase H. J. Biol. Chem. 275, 37664–37671 [DOI] [PubMed] [Google Scholar]
- 11. Gao L., Balakrishnan M., Roques B. P., Bambara R. A. (2007) Insights into the multiple roles of pausing in HIV-1 reverse transcriptase-promoted strand transfers. J. Biol. Chem. 282, 6222–6231 [DOI] [PubMed] [Google Scholar]
- 12. Kim J. K., Palaniappan C., Wu W., Fay P. J., Bambara R. A. (1997) Evidence for a unique mechanism of strand transfer from the transactivation response region of HIV-1. J. Biol. Chem. 272, 16769–16777 [DOI] [PubMed] [Google Scholar]
- 13. Shen W., Gao L., Balakrishnan M., Bambara R. A. (2009) A recombination hot spot in HIV-1 contains guanosine runs that can form a G-quartet structure and promote strand transfer in vitro. J. Biol. Chem. 284, 33883–33893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nguyen L. A., Daddacha W., Rigby S., Bambara R. A., Kim B. (2012) Altered strand transfer activity of a multiple-drug-resistant human immunodeficiency virus type 1 reverse transcriptase mutant with a dipeptide fingers domain insertion. J. Mol. Biol. 415, 248–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Operario D. J., Balakrishnan M., Bambara R. A., Kim B. (2006) Reduced dNTP interaction of human immunodeficiency virus type 1 reverse transcriptase promotes strand transfer. J. Biol. Chem. 281, 32113–32121 [DOI] [PubMed] [Google Scholar]
- 16. Diamond T. L., Roshal M., Jamburuthugoda V. K., Reynolds H. M., Merriam A. R., Lee K. Y., Balakrishnan M., Bambara R. A., Planelles V., Dewhurst S., Kim B. (2004) Macrophage tropism of HIV-1 depends on efficient cellular dNTP utilization by reverse transcriptase. J. Biol. Chem. 279, 51545–51553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kennedy E. M., Gavegnano C., Nguyen L., Slater R., Lucas A., Fromentin E., Schinazi R. F., Kim B. (2010) Ribonucleoside triphosphates as substrate of human immunodeficiency virus type 1 reverse transcriptase in human macrophages. J. Biol. Chem. 285, 39380–39391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Klatzmann D., Barré-Sinoussi F., Nugeyre M. T., Danquet C., Vilmer E., Griscelli C., Brun-Veziret F., Rouzioux C., Gluckman J. C., Chermann J. C., et al. (1984) Selective tropism of lymphadenopathy associated virus (LAV) for helper-inducer T lymphocytes. Science 225, 59–63 [DOI] [PubMed] [Google Scholar]
- 19. Lahouassa H., Daddacha W., Hofmann H., Ayinde D., Logue E. C., Dragin L., Bloch N., Maudet C., Bertrand M., Gramberg T., Pancino G., Priet S., Canard B., Laguette N., Benkirane M., Transy C., Landau N. R., Kim B., Margottin-Goguet F. (2012) SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13, 223–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goldstone D. C., Ennis-Adeniran V., Hedden J. J., Groom H. C., Rice G. I., Christodoulou E., Walker P. A., Kelly G., Haire L. F., Yap M. W., de Carvalho L. P., Stoye J. P., Crow Y. J., Taylor I. A., Webb M. (2011) HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382 [DOI] [PubMed] [Google Scholar]
- 21. Powell R. D., Holland P. J., Hollis T., Perrino F. W. (2011) Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J. Biol. Chem. 286, 43596–43600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Laguette N., Sobhian B., Casartelli N., Ringeard M., Chable-Bessia C., Ségéral E., Yatim A., Emiliani S., Schwartz O., Benkirane M. (2011) SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Baldauf H. M., Pan X., Erikson E., Schmidt S., Daddacha W., Burggraf M., Schenkova K., Ambiel I., Wabnitz G., Gramberg T., Panitz S., Flory E., Landau N. R., Sertel S., Rutsch F., Lasitschka F., Kim B., König R., Fackler O. T., Keppler O. T. (2012) SAMHD1 restricts HIV-1 infection in resting CD4+ T cells. Nat. Med. 18, 1682–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim B., Nguyen L. A., Daddacha W., Hollenbaugh J. A. (2012) Tight interplay among SAMHD1 protein level, cellular dNTP levels, and HIV-1 proviral DNA synthesis kinetics in human primary monocyte-derived macrophages. J. Biol. Chem. 287, 21570–21574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hrecka K., Hao C., Gierszewska M., Swanson S. K., Kesik-Brodacka M., Srivastava S., Florens L., Washburn M. P., Skowronski J. (2011) Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sunseri N., O'Brien M., Bhardwaj N., Landau N. R. (2011) Human immunodeficiency virus type 1 modified to package Simian immunodeficiency virus Vpx efficiently infects macrophages and dendritic cells. J. Virol. 85, 6263–6274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Berger G., Durand S., Goujon C., Nguyen X. N., Cordeil S., Darlix J. L., Cimarelli A. (2011) A simple, versatile and efficient method to genetically modify human monocyte-derived dendritic cells with HIV-1-derived lentiviral vectors. Nat. Protoc. 6, 806–816 [DOI] [PubMed] [Google Scholar]
- 28. Goujon C., Arfi V., Pertel T., Luban J., Lienard J., Rigal D., Darlix J. L., Cimarelli A. (2008) Characterization of simian immunodeficiency virus SIVSM/human immunodeficiency virus type 2 Vpx function in human myeloid cells. J. Virol. 82, 12335–12345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. St Gelais C., de Silva S., Amie S. M., Coleman C. M., Hoy H., Hollenbaugh J. A., Kim B., Wu L. (2012) SAMHD1 restricts HIV-1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4+ T-lymphocytes cannot be upregulated by interferons. Retrovirology 9, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goujon C., Rivière L., Jarrosson-Wuilleme L., Bernaud J., Rigal D., Darlix J. L., Cimarelli A. (2007) SIVSM/HIV-2 Vpx proteins promote retroviral escape from a proteasome-dependent restriction pathway present in human dendritic cells. Retrovirology 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Behrendt R., Schumann T., Gerbaulet A., Nguyen L. A., Schubert N., Alexopoulou D., Berka U., Lienenklaus S., Peschke K., Gibbert K., Wittmann S., Lindemann D., Weiss S., Dahl A., Naumann R., Dittmer U., Kim B., Mueller W., Gramberg T., Roers A. (2013) Mouse SAMHD1 has antiretroviral activity and suppresses a spontaneous cell-intrinsic antiviral response. Cell Rep. 4, 689–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hollenbaugh J. A., Gee P., Baker J., Daly M. B., Amie S. M., Tate J., Kasai N., Kanemura Y., Kim D. H., Ward B. M., Koyanagi Y., Kim B. (2013). Host factor SAMHD1 restricts DNA viruses in non-dividing myeloid cells. PLoS Pathog. 9, e1003481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Levy D. N., Aldrovandi G. M., Kutsch O., Shaw G. M. (2004) Dynamics of HIV-1 recombination in its natural target cells. Proc. Natl. Acad. Sci. U.S.A. 101, 4204–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen J., Rhodes T. D., Hu W. S. (2005) Comparison of the genetic recombination rates of human immunodeficiency virus type 1 in macrophages and T cells. J. Virol. 79, 9337–9340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dapp M. J., Clouser C. L., Patterson S., Mansky L. M. (2009) 5-Azacytidine can induce lethal mutagenesis in human immunodeficiency virus type 1. J. Virol. 83, 11950–11958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chugh P., Bradel-Tretheway B., Monteiro-Filho C. M., Planelles V., Maggirwar S. B., Dewhurst S., Kim B. (2008) Akt inhibitors as an HIV-1 infected macrophage-specific anti-viral therapy. Retrovirology 5, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gramberg T., Kahle T., Bloch N., Wittmann S., Müllers E., Daddacha W., Hofmann H., Kim B., Lindemann D., Landau N. R. (2013) Restriction of diverse retroviruses by SAMHD1. Retrovirology 10, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jamburuthugoda V. K., Chugh P., Kim B. (2006) Modification of human immunodeficiency virus type 1 reverse transcriptase to target cells with elevated cellular dNTP concentrations. J. Biol. Chem. 281, 13388–13395 [DOI] [PubMed] [Google Scholar]
- 39. Van Cor-Hosmer S. K., Daddacha W., Kim B. (2010) Mechanistic interplay among the M184I HIV-1 reverse transcriptase mutant, the central polypurine tract, cellular dNTP concentrations and drug sensitivity. Virology 406, 253–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Baekelandt V., Eggermont K., Michiels M., Nuttin B., Debyser Z. (2003) Optimized lentiviral vector production and purification procedure prevents immune response after transduction of mouse brain. Gene Ther. 10, 1933–1940 [DOI] [PubMed] [Google Scholar]
- 41. Svarovskaia E. S., Delviks K. A., Hwang C. K., Pathak V. K. (2000) Structural determinants of murine leukemia virus reverse transcriptase that affect the frequency of template switching. J. Virol. 74, 7171–7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hofmann H., Logue E. C., Bloch N., Daddacha W., Polsky S. B., Schultz M. L., Kim B., Landau N. R. (2012) The Vpx lentiviral accessory protein targets SAMHD1 for degradation in the nucleus. J. Virol. 86, 12552–12560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Diamond T. L., Souroullas G., Weiss K. K., Lee K. Y., Bambara R. A., Dewhurst S., Kim B. (2003) Mechanistic understanding of an altered fidelity simian immunodeficiency virus reverse transcriptase mutation, V148I, identified in a pig-tailed macaque. J. Biol. Chem. 278, 29913–29924 [DOI] [PubMed] [Google Scholar]
- 44. Jamburuthugoda V. K., Santos-Velazquez J. M., Skasko M., Operario D. J., Purohit V., Chugh P., Szymanski E. A., Wedekind J. E., Bambara R. A., Kim B. (2008) Reduced dNTP binding affinity of 3TC-resistant M184I HIV-1 reverse transcriptase variants responsible for viral infection failure in macrophage. J. Biol. Chem. 283, 9206–9216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. DeStefano J. J., Mallaber L. M., Fay P. J., Bambara R. A. (1993) Determinants of the RNase H cleavage specificity of human immunodeficiency virus reverse transcriptase. Nucleic Acids Res. 21, 4330–4338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kennedy E. M., Amie S. M., Bambara R. A., Kim B. (2012) Frequent incorporation of ribonucleotides during HIV-1 reverse transcription and their attenuated repair in macrophages. J. Biol. Chem. 287, 14280–14288 [DOI] [PMC free article] [PubMed] [Google Scholar]