Background: BAK and BAX permeabilize the mitochondrial membrane during apoptosis.
Results: Helices α2-α5 of BAK form the “BH3-in-groove homodimer” in the membrane, which oligomerizes by juxtaposing the carboxyl termini of α3 and α5, respectively.
Conclusion: A novel “α3:α3′, α5:α5′ oligomerization interface” exists in the BAK oligomeric pore.
Significance: These results support a model for BAX/BAK pore formation, which constitutes a key regulatory step in mitochondrial apoptosis.
Keywords: Apoptosis, Bax, Bcl-2, Cell Death, Electron Paramagnetic Resonance (EPR), Mitochondria, Bak, Oligomerization Interface, Spin Labeling
Abstract
The multidomain pro-apoptotic Bcl-2 family proteins BAK and BAX are believed to form large oligomeric pores in the mitochondrial outer membrane during apoptosis. Formation of these pores results in the release of apoptotic factors including cytochrome c from the intermembrane space into the cytoplasm, where they initiate the cascade of events that lead to cell death. Using the site-directed spin labeling method of electron paramagnetic resonance (EPR) spectroscopy, we have determined the conformational changes that occur in BAK when the protein targets to the membrane and forms pores. The data showed that helices α1 and α6 disengage from the rest of the domain, leaving helices α2-α5 as a folded unit. Helices α2-α5 were shown to form a dimeric structure, which is structurally homologous to the recently reported BAX “BH3-in-groove homodimer.” Furthermore, the EPR data and a chemical cross-linking study demonstrated the existence of a hitherto unknown interface between BAK BH3-in-groove homodimers in the oligomeric BAK. This novel interface involves the C termini of α3 and α5 helices. The results provide further insights into the organization of the BAK oligomeric pores by the BAK homodimers during mitochondrial apoptosis, enabling the proposal of a BAK-induced lipidic pore with the topography of a “worm hole.”
Introduction
Apoptosis, or programmed cell death, is an essential biological process in embryogenesis and in the maintenance of homeostasis in higher eukaryotic organisms (1). BAX2 (Bcl-2-associated X protein) or its homolog BAK (Bcl-2 antagonist/killer) acts as a key control point in the mitochondrial cell death pathways, which permeabilize the mitochondrial outer membrane (2, 3). BAX and BAK normally remain inactive in the cytosol and in the mitochondrial outer membrane, respectively (4, 5). Upon activation by various cell death signals, each protein oligomerizes via homo-dimerization (6–8), forming large pores in the outer membrane of mitochondria with an estimated diameter of ∼30–60 Å (9, 10) (see Fig. 1A).
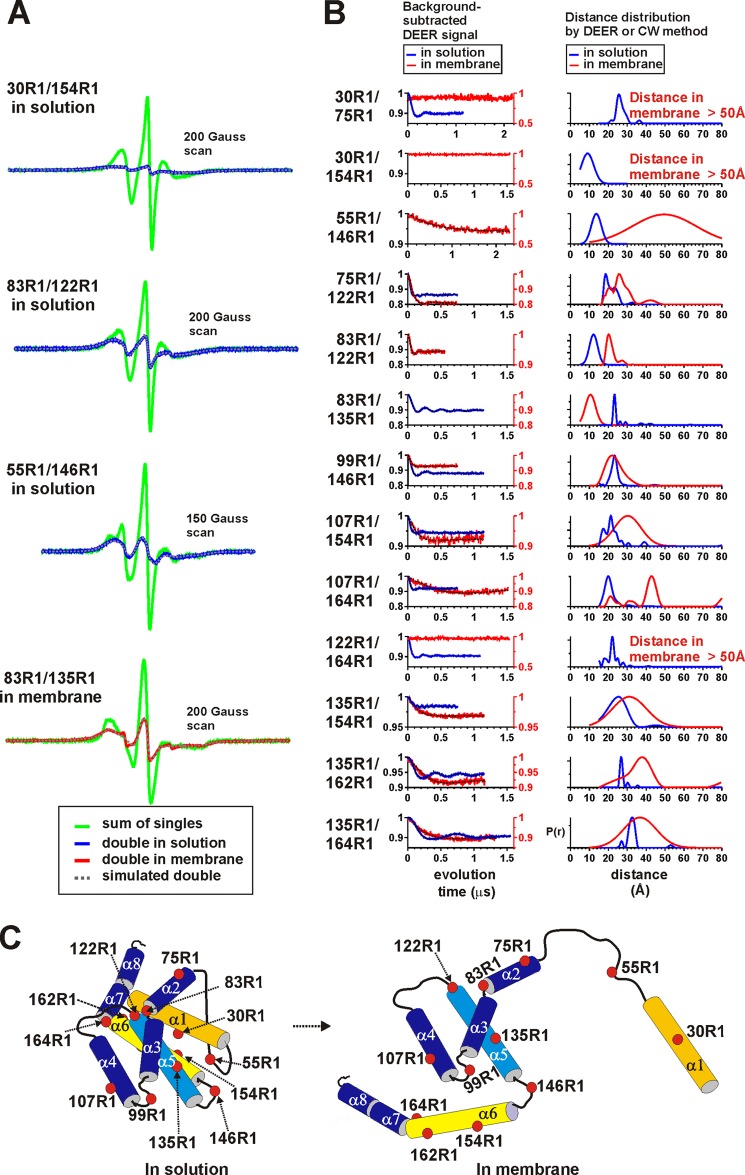
FIGURE 1.
Oligomeric pore formation by BAX (or BAK) and spin labeling of BAK cysteine mutant proteins and their membrane-permeabilizing activity. A, oligomeric pore formation by BAX (or BAK). BAX (or BAK) monomers (41) are hypothesized to form oligomeric pores in the mitochondrial outer membrane, first forming the BH3-in-groove homodimer (6) on the membrane surface, in which helices α2-α5 are highlighted in green and blue. B, sequence alignment of BAX and BAK proteins and the locations of site-directed spin labeling. The residue locations selected for cysteine substitution mutagenesis and spin labeling reaction or chemical cross-linking experiments in mouse BAK (mBAK, middle lines) are marked with colored dots above the amino acid sequences, which are aligned against human BAK and BAX proteins. The yellow and red dots represent the sites selected for spin labeling BAK singly and doubly, respectively. The cyan dots represent sites used for cross-linking experiments. Sequence similarities between the three proteins are rated with a star, colon, dot, and no sign (from the highest to lowest similarity). The α-helices and BH1–3 domains are indicated. This figure was adapted from Oh et al. (13). C, site-directed spin labeling (SDSL) reaction. The MTSSL reacts with the cysteine to form a spin-labeled cysteine residue designated as R1. D, spin labeling efficiency. The average percentage of spin labeling per cysteine residue is shown with the error ranges for each mutant from two experiments. The protein concentrations for certain mutants, e.g. 135R1/164R1, determined by the Bradford assay, might have been underestimated due to cysteine mutagenesis and/or spin labeling, resulting in % labeling efficiency and relative percent release (see Fig. 1E) values greater than 100%. E, the relative percent release of fluorescein isothiocyanate (FITC)-dextran (10 kDa) by the spin-labeled sBAK-ΔC-His proteins before correction for unlabeled cysteine mutant proteins. Liposome dye release experiments were carried out with the indicated spin-labeled sBAK/C154S-ΔC-His proteins (5 nm) in the presence of 25 nm N-terminally His-tagged p7/p15 Bid as described under “Experimental Procedures” (13). The error range is from duplicate experiments (except for 99R1/146R1). F, the relative percent release of FITC-dextran (10 kDa) by the spin-labeled sBAK-ΔC-His proteins after correction for unlabeled cysteine proteins. The contribution of the activity by the unlabeled proteins was corrected for each spin-labeled mutant protein as described under “Experimental Procedures.” G, the relative percent release of FITC-dextran (10 kDa) by the cysteine substitution sBAK-ΔC-His proteins without spin labeling. The dye release activity of the cysteine substitution sBAK-ΔC-His proteins was determined as described in E. All the ribbon diagrams were generated in PyMOL (42).
Recently, a symmetric homodimer x-ray structure of a BAX fragment consisting of helices α2-α5, known as “BH3-in-groove homodimer,” was reported by Czabotar et al. (6) (Fig. 1A). In this two-layered structure solved in the absence of membrane, helices α2 and α3 were arranged in an anti-parallel orientation forming the upper hydrophilic face and helices α4 and α5 were assembled in the lower layer presenting a hydrophobic surface (Fig. 1A), which is hypothesized to interact with the membrane and further oligomerize to form oligomeric pores (6). How the BH3-in-groove homodimers, i.e. the α2-α5 homodimer cores, are arranged in the pores is still unknown (6, 11).
In this current study, using the site-directed spin labeling method of electron paramagnetic resonance (EPR) spectroscopy (12), we measured the distances between 13 pairs of spin-labeled residues both in solution and in a membrane-inserted state in liposomes to map the conformational changes that occur in BAK upon membrane insertion (Fig. 1, B and C). The site-directed spin labeling method was also applied to residues in the α5-α6 helical hairpin in BAK to determine the organization of BAK molecules within the oligomeric pore in the membrane. These results support the formation of a BAK homodimer in the membrane that is structurally homologous to the BAX BH3-in-groove homodimer in crystals (6). Furthermore, the EPR and chemical cross-linking data reveal a novel interaction between the BAK BH3-in-groove homodimers in the oligomeric BAK, which involves the C termini of α3 and α5 helices (i.e. the C termini of α3 and α3′ and the C termini α5 and α5′). The data provide critical insights as to how BAK or BAX forms the homodimers and how they interact with each other in the membrane for higher order oligomerization and probable lipidic pore formation.
EXPERIMENTAL PROCEDURES
Expression and Purification of Recombinant Proteins
All single, double, and triple cysteine substitution BAK mutant proteins were prepared using the template plasmid pPosKJ-sBAKC154S-CHis engineered with the QuikChange® Site-directed mutagenesis kit (Stratagene) as described (13). All soluble forms of mouse BAK proteins contain residues 16–184 (helices α1-α8), with a C-terminal His6 tag and a C154S amino acid substitution mutation (designated as sBAK/C154S-ΔC-His) (13). N-terminally His-tagged p7/p15 Bid (designated as p7/p15 Bid) was prepared from the full-length p22 Bid protein by cleavage with caspase-8 (14, 15). All of the above proteins were treated with 18% (v/v) glycerol for storage at −80 °C.
Spin Labeling Reaction
The sBAK/C154S-ΔC-His proteins containing cysteine mutations were spin labeled with the methanethiosulfonate (MTSSL) or the perdeuterated MTSSL (MTSSL-d15) (Toronto Research Chemicals, Inc., Toronto, Canada) and unreacted spin labels were removed by gel filtration chromatography using a Supredex 75 column (GE Healthcare) as described (13). The spin-labeled proteins were concentrated to 1–10 mg/ml using mini-centrifugal concentrators. The protein concentration was determined by Bradford assay using a Bio-Rad Protein Assay solution (Bio-Rad) with BSA as a standard. The spin labeling efficiency (spin labeling percentage) was calculated from the molar ratio of the spin label and protein concentrations. To determine the spin label concentration, the spin labels were liberated from the protein by incubating the sample with 50 mm tris(2-carboxyethyl)phosphine for 30 min at room temperature (16). The spin label concentration was then quantitated by measuring the intensity of the EPR signal (either peak height or spectral area by double integration) of the samples using 10–100 μm 3-carboxyproxyl (Sigma) as a standard.
Preparation of Large Unilamellar Vesicles (LUVs)
LUVs that facilitate the targeting of His-tagged proteins were prepared with a mixture of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine, beef liver phosphatidylinositol, beef heart cardiolipin, cholesterol, and DOGS-NTA-Ni (all from Avanti Polar Lipids, Inc.) at a weight ratio of 36:22:9:8:20:5 in 20 mm Hepes, 150 mm KCl (pH 7.0) (buffer A) as described (16). The lipid composition of thus prepared LUVs resembles that of the mitochondrial outer membrane contact sites (17, 18). LUVs encapsulating fluorescein isothiocyanate-dextran 10 (FITC-dextran, 10 kDa, Invitrogen) were also prepared with the same lipid composition and stored in the presence of 18% (v/v) glycerol at −80 °C as described (14, 19–21). The liposomes were quantitated by phosphate assay (22).
Liposomal Release Assay
Liposome dye release experiments were carried out with the spin-labeled sBAK/C154S-ΔC-His proteins (5 nm) in the presence of 25 nm N-terminally His-tagged p7/p15 Bid after a 25-min incubation at 37 °C as described (16). To measure the dye-release activity of the cysteine substitution mutant BAK proteins, each corresponding spin-labeled protein was treated with 50 mm tris(2-carboxyethyl)phosphine for 30 min at room temperature (16) and diluted to a proper concentration for dye release assay. Alternatively, cysteine substitution mutant proteins (stored in 2 mm DTT) were directly used without tris(2-carboxyethyl)phosphine treatment.
The extent of release of the FITC-labeled dextran 10 from the LUVs was determined on a percentage basis as described (16). To account for the batch-to-batch variation in the extent of FITC-dextran release from the liposomes, the percent release of the FITC-dextran by the sBAK/C154S-ΔC-His protein (5 nm) was measured in the presence of 25 nm N-terminally His-tagged p7/p15 Bid as a reference for each liposomal preparation. The relative percent release of samples was then standardized against the reference. In all assays, the concentration of LUVs was 10 μg/ml of lipids or 0.125 nm as described (16). The relative percent release of each of the spin-labeled proteins was further corrected by subtracting the contribution of the unlabeled cysteine substitution BAK protein present in the spin-labeled samples as follows,
 |
where T is the relative percent release of the spin-labeled sample (i.e. mixture of spin labeled protein and unlabeled protein), C is the relative percent release of the cysteine substitution mutant protein (without spin labeling), and χ is the fraction of the spin-labeled protein in the sample (=spin labeling percentage/100). In this calculation, it was assumed that the dye-releasing activity of the mixture of a spin-labeled protein and an unlabeled one is additive and that the activity of each protein is approximately proportional to its protein concentration in the range of 0–5 nm.
Preparation of Membrane-inserted BAK Protein
Membrane-inserted sBAK-ΔC-His proteins were prepared in the presence of the equimolar activator protein p7/p15 Bid with a mixture of the spin-labeled sBAK-ΔC-His proteins and the unlabeled soluble BAK molecule (sBAK/C154S-ΔC-His) at various ratios such as 7:0, 3:4, and 1:6 (labeled:unlabeled BAK) as described (13), which were designated as 7:0, 3:4, or 1:6 mixture, respectively. Loosely bound sBAK-ΔC-His proteins and the Ni2+ ions bound to the DOGS-NTA-Ni in the proteoliposomes were removed by EDTA treatments and centrifugation as described (13). For the cross-linking experiment, a number of cysteine substitution mutant proteins of sBAK/C154S-ΔC-His were used to make the membrane-inserted BAK proteins following the same procedures as described here.
Continuous Wave (CW) EPR Spectroscopy
The CW deconvolution method (23) was used to determine the distance between the R1 residues in BAK that are apart less than 20 Å in solution or in membrane-inserted state. EPR spectra of the singly or doubly spin-labeled sBAK-ΔC-His proteins in 18% (v/v) glycerol solution were obtained on a Bruker EleXsys 580 spectrometer using a Bruker High Sensitivity resonator or a loop gap resonator (JAGMAR, Krakow, Poland) at 2 milliwatt incident microwave power using a field modulation of 1.0–1.5 Gauss at 100 kHz at room temperature. EPR spectra for membrane-inserted spin-labeled BAK samples were also obtained similarly. To measure the distance between two R1 residues in the BAK protein in solution, the line-broadening function and the distance distribution function were calculated using the spectrum of the doubly labeled sample and the spectral sum of the singles for each pair of spin labels by the deconvolution method (23) using a program developed by Altenbach (24). In this calculation, the percentage of interacting and non-interacting (due to contaminating singly labeled protein) spin populations are optimized to calculate the line-broadening function (24). In the membrane-inserted form (e.g. 83R1/135R1), first, a 1:6 (mol/mol) mixture of the doubly labeled protein and sBAK/C154S-ΔC-His was used to form oligomeric BAK pores in the membrane under the same conditions as described above. Then, a mixture of the corresponding singly labeled proteins (e.g. 83R1 and 135R1) and sBAK/C154S-ΔC-His at a ratio of 1:1:5 (mol/mol) was used to form pores similarly in the membrane. The spectra of these two samples were used for the deconvolution method (23) for distance estimation.
Pulse EPR Spectroscopy
The double electron electron resonance (DEER) method was used to determine the inter-spin distances in the range of ∼20–50 Å. DEER measurements were carried out at 80 K using the Bruker EleXys 580 system equipped with the flexline split-ring resonator ER 4118X-MS-3W1 using samples treated with 18% (v/v) glycerol. A four-pulse DEER sequence (25) was used for data acquisition using the PulseSPEL program provided by Bruker. The electron-electron double resonance π pulse length was set to 32 ns. A 2-step phase cycling was performed while recording the DEER signal. The total measurement time for each sample varied from several hours to days with signals averaging of up to 600 times. Data were analyzed primarily with DeerAnalysis2006 (26) to extract the distance information. DEFit program (27) or the Global Analysis program (28) were also used to check the analysis result by the DeerAnalysis2006.
Direct Spectral Simulation of Perdeuterated Spin Label R1-d15 at Residue 128 (128R1-d15) for Inter-spin Distance Estimation
Spectra simulation and fitting were performed as previously described (29–31) using a modified set of coordinate systems and corresponding rotation operators that impose C2 symmetry on the two dipolar coupled nitroxides. The fundamental coordinate system {x,y,z} was defined by the inter-electron vector, R{z}, and the C2 symmetry axis {y}. The orientation of the first nitroxide with respect to {x,y,z} was determined by three Euler angles (γ, β, α) and was located at +R/2 along the z axis. The second nitroxide was obtained by rotating the first by 180° about the y axis. The orientation of the magnetic field, H0, with respect to {x,y,z} was determined by Euler angles θ and φ. Following previously published work (30), the A-tensors for nitroxides 1 and 2 were given by,
 |
 |
where Ad is the diagonalized nitrogen hyperfine tensor for both nitroxides. Similar equations were applied to the g-tensors. The unique element of the dipolar coupling tensor was given by,
where r, the inter-electron distance, and all the other terms and symbols were as previously defined (30). Fitting was performed using Marquardt-Levenberg and simulated annealing algorithms developed in house. All code was developed in MATLAB R2012B (The Mathworks, Natick MA) and is available from the author (E.J.H.) upon request (28).
Homology Modeling of the Mouse BAK α2-α5 Homodimer Core
The homology model of the mouse BAK BH3-in-groove homodimer (α2-α5 homodimer core) was constructed using the crystal structures of the human BAX BH3-in-groove homodimer (Protein Data Bank code 4BDU) (6) and the human BAK solution structure (Protein Data Bank code 2IMS) (32) using COOT (33).
In Vitro Chemical Cross-linking by Disulfide Bond Formation
Oligomeric BAK formation was assessed by disulfide bond formation adapting the published procedures (34) as follows. The cysteine-substituted sBAK/C154S-ΔC-His protein samples in the membrane-inserted state were prepared as described above and diluted in buffer A (pH 7.0) to a final concentration of 15–20 μm. As a control for the solution state of BAK, an equimolar mixture of cysteine-substituted sBAK/C154S-ΔC-His protein and p7/p15 Bid was prepared in buffer A at a similar concentration for each mutant. For cross-linking, a volume of 8 μl BAK samples (in solution or in membrane) was added to 10 μl of 2× reaction buffer (40 mm Hepes/KOH, pH 7.5, 300 mm KCl, 200 mm sucrose, 5 mm MgCl2, 2 mm NaAsO2). The mixture was then treated with 2 μl of 10× CuPhe (copper(II)/(1,10-phenanthroline)3) redox catalyst solution on ice for 30 min, where the 10× CuPhe solution consisted of 150 mm copper sulfate (Sigma), 500 mm 1,10-phenanthroline (Sigma) in 20% (v/v) ethanol. The reaction was quenched by treating the reaction mixture with an equal volume (20 μl) of 2× quenching buffer (200 mm EDTA, 40 mm N-ethylmaleimide (Sigma), 130 mm Tris-HCl buffer, pH 6.8). As a control experiment (without cross-linking), identical steps were carried out with the same set of protein samples in the absence of CuPhe. All the resulting cross-linked or control samples were then diluted to a final concentration of ∼0.5 μm (∼10 ng/μl) BAK using the dilution buffer that was prepared by mixing 2× quenching buffer, 2× reaction buffer, buffer A, and the deionized water at a volume ratio of 10:5:4:1. A mixture of 10 μl of the resulting diluted BAK samples (∼100 ng) and an equal volume of 2× gel loading buffer (130 mm Tris-HCl, pH 6.8, 4% (w/v) SDS, 20% (v/v) glycerol, 0.05% (w/v) bromphenol blue with or without 200 mm DTT) was loaded onto 4–20% Precise Protein Gels (12-well, Pierce). The resulting gels were subjected to Western blot analysis with anti-BAK, NT antibody (Millipore) and anti-rabbit IgG (Goat), HRP-labeled (PerkinElmer Life Sciences) as the primary and secondary antibodies.
RESULTS
Most of the Spin-labeled BAK Proteins for Distance Measurement Retained Membrane-permeabilizing Activity
The 13 double cysteine substitution mutants of soluble BAK proteins (sBAK/C154S-ΔC-His) (Fig. 1B) displayed average spin labeling efficiency, ranging from ∼40% to almost 100% per thiol (Fig. 1D). All these samples demonstrated partial to full activity in liposome permeabilization as a mixture of spin-labeled and unlabeled proteins (Fig. 1E). To make corrections for the contribution by the unlabeled proteins, the activity of cysteine substitution mutant proteins were determined (Fig. 1G). The corrected relative percent release was calculated as described under “Experimental Procedures” (Fig. 1F). Spin-labeled BAK mutants 75R1/122R1, 83R1/122R1, 83R1/135R1, 99R1/146R1, and 135R1/162R1 displayed ∼50 to ∼85% activity relative to the fully active parent molecule with a C154S substitution (sBAK/C154S-ΔC-His, Fig. 1F). The rest of the doubly spin-labeled proteins retained full membrane permeabilizing activity (Fig. 1F). We thus determined the inter-nitroxide distances with these proteins in solution and in the membrane-inserted state by either the CW deconvolution method (for distance range of 6–20 Å) or the DEER method (for distance range of 20–50 Å) as summarized in Fig. 2 and Table 1.
FIGURE 2.
Distance between two spin labels in the doubly spin-labeled BAK mutants in solution and membrane-inserted states. A, CW EPR spectra of spin-labeled pairs in BAK in solution or in the membrane inserted state. The EPR spectra from the indicated doubly spin-labeled proteins, shown in blue (in solution) and red (in membrane) traces, were normalized to a unit area. The spectral sums of the normalized EPR spectra for the individual R1s in each pair are shown in green (for details, see “Continuous Wave (CW) EPR Spectroscopy” under “Experimental Procedures”). The simulated spectra calculated from the CW deconvolution method for the double mutants (23), showing large line broadening due to the close proximity of the spin labels, are in dotted gray traces. The corresponding distance distribution functions are shown in blue (in solution) and red (in membrane) traces in B (right column) along with the data obtained from the DEER experiments. B, DEER signals and distance distribution probability functions for the R1 pairs in BAK in solution or in the membrane-inserted state. The background-subtracted DEER signals are shown for the BAK mutants with spin labels attached at the indicated positions in solution (blue traces in the graphs on the left column) or in the membrane-inserted state (red traces in the graphs on the left column). Black dotted lines associated with each DEER signals represent the fitted DEER signals. The distance distribution probability functions, P(r), for the corresponding samples obtained either by the Tikhonov regularization or by Gaussian fit are also shown in blue and red traces, for solution and membrane-inserted states, respectively, in the right column. C, schematic representation of conformational changes occurring in BAK upon oligomeric pore formation in membrane. Red dots represent R1 residues. Helices α1 and α6 (and thus α7-α8 as well) move away from helices α2-α5, which appear to be bundled together. Helix α9 is not present in the soluble BAK.
TABLE 1.
Inter-spin distance in BAK in solution state and in the membrane-inserted state
Average distances between the indicated spin label pairs (first column) introduced in BAK mutants shown in Fig. 2 are summarized for the solution (third column) and membrane-inserted (fourth column) states in this table. The Cβ-Cβ distances for the original amino acid pairs are shown in the second column.
| Spin label pairs | Cβ-Cβ distance in solution structure | Inter-spin distance in solution (±S.D) by DEER or CW | Inter-spin distance in membrane (±S.D.) by DEER or CW |
|---|---|---|---|
| Å | Å | Å | |
| 30/75 (α1/α2) | 16.9 | 26.6 (± 2.9) | >50 |
| 30/154 (α1/α6) | 8.1 | 9.0 (± 6.5) (by CW) | >50 |
| 55/146 (α1–2 loop/α5–6 loop) | 9.9 | 13.7 (± 5.3) (by CW) | 49.6 (± 23.8) |
| 75/122 (α2/ α5 N-terminal) | 16.4 | 21.5 (± 3.4) | 27.1 (± 6.3) |
| 83/122 (α2–3 loop/α5 N-terminal) | 11.2 | 12.2 (± 4.4) (by CW) | 21.5 (± 2.6) |
| 83/135 (α2–3 loop/α5) | 15.3 | 26.4 (± 7.0) | 10.6 (± 5.4) (by CW) |
| 99/146 (α3–4 loop/α5–6 loop) | 15.2 | 23.3 (± 3.3) | 23.6 (± 5.5) |
| 107/154 (α4/α6) | 15.2 | 22.6 (± 6.6) | 30.5 (± 6.7) |
| 107/164 (α4/α6) | 21.3 | 20.9 (± 3.5) | 40.1 (± 12.8) |
| 122/164 (α5 N-terminal/α6 N-terminal) | 13.2 | 22.6 (± 5.1) | > 50 |
| 135/154 (α5/α6) | 16.7 | 25.2 (± 5.8) | 30.7 (± 12.5) |
| 135/162 (α5/α6) | 21.8 | 27.7 (± 2.2) | 36.9 (± 10.5) |
| 135/164 (α5/α6) | 23.6 | 33.0 (± 5.0) | 38.9 (± 10.5) |
Inter-nitroxide Spin Label Distances in BAK in Solution State
Spin-labeled residues 30R1 and 154R1, located on two neighboring helices α1 and α6, respectively, in the solution structure (Figs. 1B and 2C), were separated by 9.0 Å distance (top panel in Fig. 2A, right panel in Fig. 2B), close to the predicted Cβ-Cβ distance (i.e. 8.1 Å) (Table 1). Similarly, 83R1 and 122R1, which are located in the α2–3 loop and at the amino terminus of the α5 helix, respectively, were apart by 12.2 Å, close to the predicted Cβ-Cβ distance (11.2 Å) (Fig. 2, A and B, Table 1). The measured inter-spin distances for other pairs by either the CW or DEER methods (Fig. 2, A and B) were all greater than the known Cβ-Cβ distances by 4–10 Å (Table 1), which can be attributed to location-specific rotameric structures of the spin label in the R1 side chain (35, 36).
Intramolecular Inter-nitroxide Distances in Membrane-inserted BAK Reveal Conformational Changes in BAK upon Membrane Insertion
In the presence of 6-fold excess of unlabeled BAK protein (sBAK/C154S-ΔC-His) (Fig. 3A, left panel), we were able to successfully detect the DEER modulation for pairs of nitroxide spin labels within a monomer that constitutes the BAK oligomeric pore in the membrane (Figs. 2B and 3B, Table 1). In control experiments with mixtures of two singly labeled proteins and the unlabeled protein at a molar ratio of 1:1:5 (Fig. 3A, right panel), the DEER modulations were not observed (Fig. 3B, green traces), demonstrating that the above condition (i.e. in the presence of 6-fold excess unlabeled proteins) ensured the detection of the intramolecular spin-spin interactions specifically. The short inter-spin distance for 83R1/135R1 in the membrane was determined by the CW method under similar conditions (Fig. 2A) (see also Table 1). The data summarized in Fig. 2 and Table 1 collectively revealed the following conformational changes.
FIGURE 3.
Detection of the intramolecular spin-spin interactions by the DEER (or CW) experiment. A, rationale for the intra-molecular spin-spin interaction by DEER measurement. Left panel, in the presence of 6-fold excess of unlabeled BAK (sBAK/C154S-ΔC-His) protein (black dots), doubly labeled BAK protein (red dot) forms pores in the membrane by the activation with p7/p15 Bid. Only the intramolecular spin-spin interactions between the two nitroxide spin labels (XR1-YR1) will be observed by the DEER approach because the distance between the doubly labeled BAK proteins increase beyond the detection limit. Right panel, a mixture of two singly spin-labeled BAK proteins (green dots) is used in the presence of unlabeled BAK (sBAK/C154S-ΔC-His) at the indicated ratio to form pores. DEER modulations will not be observed in the mixture of two singly labeled proteins due to an increase in the inter-spin distance by dilution with the unlabeled proteins. A CW experiment can also be performed this way. B, DEER data for the doubly spin-labeled proteins and a mixture of two singly labeled proteins in the presence of excess unlabeled proteins. The DEER modulation curves for the indicated membrane-inserted BAK samples prepared with the doubly labeled BAK or the mixture of two singly labeled proteins are shown in red and green traces, respectively. Their two-dimensional (left panels) or three-dimensional (right panels) background signals fitted are shown in black solid lines (for the doubly labeled sample) or black dotted lines (for the mixture of two singly labeled BAK).
Disengagement of Helices α1 and α6
The distances between 30R1 on helix α1 and two other locations on helices α2 (75R1) and α6 (154R1) were measured in solution and in the membrane-inserted state of BAK (Fig. 2). The distance for 30R1/75R1 increased from ∼27 Å to a distance beyond the detection limit (50 Å) (Fig. 2B and Table 1). Similarly, the distance for 30R1/154R1 also increased from 9.0 Å to a distance beyond the detection limit (50 Å) (Fig. 2, A and B, and Table 1). These indicated that helix α1 is removed from both helices α2 and α6 in the membrane-inserted state (Fig. 2C) as was qualitatively observed with antibodies in BAX by others (37). Consistent with this interpretation, two residues, 55R1 and 146R1, which are located in the loops interconnecting helices α1 and α2 (designated as α1–2) and helices α5 and α6 (α5–6), respectively, became separated from each other by an average distance of ∼50 Å in the membrane-inserted state (Fig. 2B, Table 1).
Residues 122R1 and 164R1 that are located, respectively, near the amino terminus of helix α5 and the carboxyl terminus of α6 were apart by 22.6 Å in the solution state (Fig. 2B, Table 1). In the membrane, the 122R1/164R1 distance increased to a distance beyond detection (>50 Å) (Fig. 2B), indicating that the α5-α6 helical hairpin structure is disrupted upon membrane insertion (Fig. 2C). Consistent with this, the distances for spin label pairs, 135R1 (α5)/154R1 (α6), 135R1 (α5)/162R1 (α6), and 135R1 (α5)/164R1 (α6), all increased upon membrane insertion of BAK (Fig. 2). In addition, the distances from 107R1 on helix α4 to two α6 residues 154R1 and 164R1 also increased in the membrane (Fig. 2B, Table 1), indicating that α6 also moves away from helix α4 in the membrane (Fig. 2C). The above results together indicated that helices α1 and α6 disengage from each other and also from the rest of the protein domains upon membrane insertion (Fig. 2C).
Reorganization of the α2-α5 Core Folding Unit
The distances between several spin labels located within helices α2-α5 of BAK were determined in the membrane-inserted state to determine whether the BAK molecule is unfolded globally upon membrane insertion (Fig. 2B, Table 1). Unexpectedly, the average distance between 99R1 and 146R1 that are located in the α3–4 and α5–6 loops, respectively, remained almost the same before and after membrane insertion (Fig. 2B), which were 23.3 (±3.3) Å and 23.6 (±5.5) Å, respectively (Table 1). The small increase in the standard deviation of the distance distribution function upon membrane insertion indicated a slight increase in the flexibility of the two loops in membrane. These indicated that α3–4 and α5–6 loops remain close to each other even after BAK inserts into the membrane (Fig. 2C). Additionally, the distance between residue 75R1 on helix α2 and residue 122R1 at the amino terminus of helix α5 increased by less than 6 Å from 21.5 to 27.1 Å upon membrane insertion (Fig. 2, B and C, Table 1). Noting that these two BAK mutants, 99R1/146R1 and 75R1/122R1, were 60–85% as active as the parent C154S BAK molecule (Fig. 1F), the results above suggested that helices α2-α5 are folded together in the membrane-inserted state (Fig. 2C).
The inter-spin distances from 83R1 to two other positions, 122R1 at the N terminus of α5 and 135R1 that is ∼3 helical turns away from it, further supported the above finding. Residue 83R1 is located in the short α2–3 loop (Figs. 1B and 2C). The inter-nitroxide distance for the 83R1/122R1 pair increased from 12.2 to 21.5 Å upon membrane insertion of BAK (Fig. 2, A and B, Table 1). In contrast, the distance for 83R1/135R1 decreased from 26.4 Å to 10.6 Å upon membrane insertion (Fig. 2, A and B, Table 1). This indicated that 83R1 moves away from the N terminus of α5 (i.e. 122R1) but toward the C terminus of α5 (i.e. 135R1) upon membrane insertion of BAK as schematically shown in Fig. 2C.
Distance Measurements Suggest That Helix α5 Is Juxtaposed in an Anti-parallel Orientation in the BAK Homodimer
Proximity of α5 Residues in Oligomeric BAK Pore by CW Experiments
Previous chemical cross-linking studies suggested an anti-parallel juxtaposition of the α2-α3 extended helices to form the “BH3:BH3 interface,” forming a symmetric BAK dimer (8). For the reorganized BAK monomer (Fig. 2C) to form such a symmetric dimer, other domains including α5 should also be brought close to each other as in the BAX BH3-in-groove homodimer (6) (see Fig. 1A). To test this, we carried out scanning spin dilution experiments using the single cysteine substitution mutant proteins in helix α5 (residues 122–145) of BAK in the membrane-inserted state (Fig. 4). Among the tested residues, 124R1, 128R1, 142R1, and 143R1 clearly showed a spin dilution effect (hd/h0 >1.25) (Fig. 5A). The approximate inter-spin distances were successfully calculated for 124R1, 142R1, and 143R1 with the CW deconvolution method (23) by approximating the spectra of the spin-diluted samples (3:4 mixture) as the unbroadened EPR signal (Fig. 5A). The distances were 15.0, 18.0, and 12.7 Å for 124R1, 142R1, and 143R1, respectively (Fig. 5B).
FIGURE 4.
Sites of site-directed spin labeling for inter-residue distance measurement between two neighboring BAK proteins in membrane-inserted state. A, sites of spin labeling in BAK. The α-carbon atoms of the 18 residues selected for cysteine substitution mutagenesis and spin labeling reaction are shown in black spheres in a ribbon diagram of the solution structure of sBAK/C154S-ΔC-His. The ribbon diagrams were generated in PyMOL (42) using the coordinates of the mouse BAK obtained by homology modeling after the human counterpart (13). Residues 180–184 are not shown. B, spin labeling efficiency. The percentage of spin labeling per cysteine residue (i.e. per thiol) in each protein was determined (see “Experimental Procedures”). The average values of two measurements are shown with the error ranges indicated. C, the relative percent release of fluorescein isothiocyanate (FITC)-dextran (10 kDa) by the spin-labeled sBAK-ΔC-His proteins after correction for the unlabeled proteins. Liposome dye release experiments were carried out with the indicated spin-labeled sBAK-ΔC-His proteins (5 nm) in the presence of 25 nm N-terminally His-tagged p7/p15 Bid using liposomes (10 μg/ml) encapsulating FITC-dextran (10 kDa) as described (13). The same experiments were carried out with unlabeled cysteine substitution mutants. The corrected activity values for the spin-labeled mutants were calculated as described under “Experimental Procedures.” The average values of two measurements are shown with the error ranges indicated. The protein concentrations for certain mutants, e.g. 135R1, 137R1, 149R1, and 163R1, determined by the Bradford assay, might have been underestimated due to cysteine mutagenesis and/or spin labeling, resulting in % labeling efficiency (Fig. 4B) and/or relative percent release values (Fig. 4C) greater than 100%.
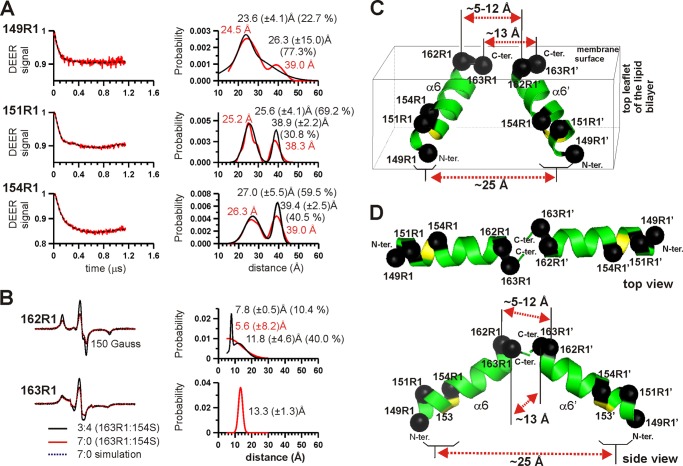
FIGURE 5.
Proximity of helix α5 residues in membrane-inserted BAK oligomers. A, spin dilution experiment for helix α5 residues. The membrane-inserted BAK samples were prepared with the indicated spin-labeled proteins in the presence (3:4) or absence (7:0) of sBAK/C154S-ΔC-His at the indicated ratio as described under “Experimental Procedures.” The area-normalized EPR spectra of the spin-diluted samples (3:4 mixture, black trace) of the indicated membrane-inserted BAK are superimposed to those of the undiluted samples (7:0 mixture, red trace). The ratios of the amplitude of the central line for the 7:0 mixture (h0) to that for 3:4 mixture (hd) are indicated. The red arrows for 128R1 indicate the splitting of the EPR lines in the absence of the spin dilution (7:0 mixture) due to the strong spin-spin interactions between two 128R1 residues in close proximity in the membrane-inserted state of BAK (see also Fig. 5C). The hyperfine extrema of the spectrum for the spin-diluted sample (3:4 mixture) are denoted by “im,” which indicate a severely restricted tumbling motion (i.e. immobile) in this spin label. B, distances of 124R1, 142R1, and 143R1 in membrane-inserted BAK to their nearest neighbors estimated by the CW deconvolution method. The CW deconvolution method (23) was applied to the spectra for the indicated mutants (red traces, left panel), resulting in simulated fits (black dotted lines on the left panel) superimposed to the spectra for 7:0 mixture of the indicated residues. The corresponding distance distribution functions are shown on the right panel. C, direct spectral simulation of perdeuterated spin label R1-d15 at residue 128 (128R1-d15) for inter-spin distance estimation. The EPR spectra of the BAK Cys-128 mutant labeled with a perdeuterated spin label (R1-d15, top panel) were obtained in the presence or absence of sBAK/C154S-ΔC-His in membrane at the indicated ratios (dotted traces) at −30 °C. The spectrum for the 1:6 mixture was used to calculate the line width and the principal elements of the g and A tensors of the spin label, which gave the corresponding fit (middle panel, red trace). The spectrum from the 7:0 mixture was fitted theoretically with these spectral parameters for the two C2-symmetry-related nitroxides that are oriented relative to each other as defined by the Euler angles α, β, and γ as described under “Experimental Procedures” (29–31). This resulted in eight sets of symmetry-equivalent Euler angles that fit the data best, all yielding an identical inter-spin distance of 14.3 Å. Only one set of Euler angles is shown here. D, DEER data for membrane-inserted BAK in the absence of spin dilution. DEER experiments were carried out without spin dilution with the indicated membrane-inserted BAK proteins. The DEER data (left column) were analyzed by the DeerAnalysis program using Tikhonov regularization, resulting in distance distribution functions on the right panel (red traces), which could be best fitted with two Gaussian models (blue traces, right panel) with the average inter-spin distance <r> and the standard deviation for the shorter distances indicated. The dotted lines superimposed to the DEER signals are the calculated DEER signal for the distance fit. E, angular clustering of certain residues in helix α5. Side chains of residues Ser-122, Gly-124, Ala-128, Arg-135, Gln-142, and Arg-143, which show spin-spin interactions for the corresponding R1 residues, are clustered on one side of helix α5. Ser-122 is in the loop just upstream of α5 N terminus. F, anti-parallel arrangement of two α5 helices in the BAK homodimer. The Cα carbon atoms of the three residues indicated are shown in spheres on α5 helices along with the inter-spin distances for the symmetry-related spin label pairs in the BAK homology model of BAX BH3-in-groove homodimer model (6).
BAK 128R1 showed a strong spin-spin interaction in the membrane-inserted state in the absence of extra sBAK/C154S-ΔC-His (7:0 mixture) (red trace in Fig. 5A). This is evidenced by the splitting in the EPR spectrum of BAK 128R1 indicated by the red arrows in Fig. 5A (also see Fig. 5C, bottom panel). Note that this splitting was not observed in the presence of excess unlabeled protein sBAK/C154S-ΔC-His (3:4 mixture) (black trace in Fig. 5A), in which 3 parts of the spin-labeled BAK 128R1 is mixed with 4 parts of the unlabeled sBAK/C154S-ΔC-His for pore formation in the membrane. These results indicate that residue 128R1 is at or near the homo-dimerization/oligomerization interface, consistent with the BAK homology model of the BH3-in-groove homodimer that was built after the BAX BH3-in-groove homodimer (6) (Fig. 5F).
To determine the inter-nitroxide distance between two neighboring 128R1 residues accurately, a perdeuterated spin label, MTSSL-d15, was used to get the 128R1-d15 spectra with and without spin dilution (Fig. 5C). The inter-spin distance was estimated by direct spectral simulation of the EPR spectra (29–31), resulting in the distance r of 14.3 Å (Fig. 5C, bottom panel). This is close to the Cβ-Cβ distance of 11.0 Å for the two nearby Ala-128 residues in the mouse BAK homology model of the BAX BH3-in-groove homodimer (6), indicating that BAK forms a homodimer structure similar to BAX (Fig. 5F).
Proximity of α5 Residues in Oligomeric BAK Pore by DEER Experiments
DEER measurements with the BAK oligomers formed with the singly spin-labeled BAK proteins also gave additional distances between neighboring α5 residues that are apart greater than 20 Å (Fig. 5D). Residues 122R1 and 135R1 gave very clear DEER modulation curves (Fig. 5D). The distances for the nearest spin labels for 122R1 and 135R1 were 30.9 and 32.0 Å, respectively. These values were greater than their corresponding Cβ-Cβ distances in the homology model of BAK BH3-in-groove homodimer, which were 24.4 and 22.0 Å, respectively (Fig. 5F). This discrepancy is likely to have originated from the R1 side chain orientation that has a ∼7.5 Å length (24).
Anti-parallel Arrangement of α5 Helices
Of note, residues 122R1 and 135R1, which are near the N terminus (residue 123) and near the center of helix α5 (Fig. 5E), gave very similar distances (Fig. 5D). We also note that residues 124R1, 128R1, 135R1, 142R1, and 143R1 are angularly clustered on one side of helix α5 (Fig. 5E). Interestingly, residues 124R1, 128R1, and 142R1 are all within ∼20 Å distance from their nearest neighbors (i.e. 124R1′, 128R1′, and 142R1′, Fig. 5B), whereas the intercalating residue 135R1 is much farther removed from its nearest neighbor (135R1′) at ∼32 Å distance (Fig. 5D). This indicates that two neighboring α5 helices are brought close to each other near the amino (i.e. near 124R1 and 128R1) and the carboxyl termini (e.g. near 142R1 and 143R1) of the helix, but not near the middle of it (e.g. 135R1). This indicates anti-parallel arrangement of two neighboring α5 helices as in the case of the BAX BH3-in-groove homodimer structure (6) (Fig. 5F), suggesting that the arrangement of the α5 helices in BAK homodimer in the membrane is very similar to that of BAX BH3-in-groove homodimer x-ray crystal structure. The results also exclude the possibility of domain-swapped dimer formation by BAK, unlike in the case of BCL-XL (38) or BAX (6).
In addition, the close proximity of residues 142R1 and 143R1 to their respective nearest neighbors (Fig. 5, B and E) indicates that two neighboring symmetric BH3-in-groove homodimers are also interacting with each other near the carboxyl termini of the helix 5 in the oligomeric structure (Fig. 5F). High spin labeling efficiency and high activity in membrane permeabilization observed for BAK 122R1 (13), 135R1 and 143R1 strongly support this interpretation (Fig. 4, B and C).
Distance Measurements between α6 Residues Indicate That the Two Neighboring α6 Helices Are in a Trapezoidal Arrangement
Residues 149R1, 151R1, and 154R1 in the membrane-inserted BAK were separated from their closest neighbors at a distance of 23.6, 25.6, and 27.0 Å, respectively, by the DEER measurements (Fig. 6A). These R1 residues in the α6 helix had high spin labeling efficiencies (>80%) and intact membrane-permeabilizing activities (Fig. 4). Residue 162R1 located near the C terminus of α6, however, was much closer to its nearest neighbor 162R1′ with the inter-spin distance of 5–12 Å with a broad distance distribution in a one or two Gaussian fit (Fig. 6B). Similarly, another C-terminal residue 163R1 was also in close proximity to its neighboring residue (163R1′) (Fig. 6B). Given the adsorption of helix α6 to the membrane surface,3 consistent with its amphipathic nature, the distance constraints above indicate that two neighboring α6 helices are in one of the two possible trapezoidal arrangements with their C termini brought close to each other; in pseudo parallel (Fig. 6C) or pseudo-anti-parallel orientation on the membrane surface (Fig. 6D).
FIGURE 6.
Proximity of helix α6 residues in membrane-inserted BAK oligomers. A, DEER data for residues 149R1, 151R1, and 154R1 in the absence of spin dilution. DEER experiments were carried out for the indicated proteins without spin dilution. The DEER data (left column) were analyzed by the DeerAnalysis program using Tikhonov regularization, resulting in the distance distribution functions on the right panel (red traces). The black dotted lines superimposed to the DEER signals (red traces, left panel) are the calculated DEER signal for the distance fit. Results by two Gaussian models are superimposed (black traces, right panel) with the average inter-spin distance, the standard deviation and the percent of the spin pairs for the distances indicated. B, proximity of helix α6 residues 162R1 and 163R1 in BAK oligomers. The area-normalized EPR spectra of the spin-diluted samples (3:4 mixture, black trace) of the indicated membrane-inserted BAK are superimposed to those of the undiluted samples (7:0 mixture, red trace). The spectra for 162R1 and 163R1 were acquired at −30 and 22 °C, respectively. The data could be best-fitted with one or two Gaussian distance distributions (right panel). Residue 162R1, with a spin labeling efficiency of 74.2 (±0.6)%, had the interacting spin pairs close to the expected value of 55% (52.3% interacting spins for the single Gaussian fit (red trace) and 50.4% (=10.4 + 40.0) for the double Gaussian fit (black trace). Similarly, residue 163R1 (58.6 (±1.1)% labeling efficiency), resulted in ∼35% interacting spin pairs (34% expected) in the data fitting. C and D, pseudo parallel or pseudo anti-parallel arrangement of two neighboring α6 helices in the BAK oligomer. Two tilted α6 helices are arranged side by side (C) or in a head-to-head orientation (D) with their C termini in close proximity. The average inter-spin distances (dotted arrows) are shown for the indicated pairs of residues (Cα carbon atoms in black spheres).
Additional Distance Measurements and Cross-linking Studies Further Reveal the Organization of the BAK BH3-in-groove Homodimers in the Oligomeric BAK
Proximity of the Carboxyl Termini of Helix 3 in the Oligomeric BAK
Based on (i) possible adsorption of the BAK “BH3-in-groove dimer” to membrane surface (6), (ii) the close proximity of C-terminal residues of α5 helices, e.g. 142R1 and 143R1 (Fig. 5B), and (iii) the possible pseudo-parallel arrangement of α6 helices on the membrane surface (39) (Fig. 6C), we reasoned that the C termini of α3 helices might also be brought close to each other between two neighboring BAK homodimers in the oligomeric pore (Fig. 7A). To test this, we carried out a CW spin dilution experiment using 96R1 that is located at the C terminus of α3 of BAK in the membrane-inserted state (Fig. 7, A and B). The average inter-spin distance estimated from the data were ∼9 Å (Fig. 7B). The Cβ-Cβ distance between the two Glu-96 residues in the BAK homology model of the BH3-in-groove homodimer was ∼38 Å (see Fig. 7A). This is beyond the detection limit of the CW spin dilution experiment (∼25 Å). Thus, the strong spin-spin interaction observed in 96R1 must be due to the inter-homodimer interactions (Fig. 7B), confirming the juxtaposition of the two C termini of α3 helices between two neighboring BAK homodimers (Fig. 7A).
FIGURE 7.
BAK forms symmetric BH3-in-groove homodimers that associate by α6:α6 interactions to generate higher order oligomers in membrane, juxtaposing the C termini of helices α3 and α5, respectively, between two neighboring homodimers. A, association of the BH3-in-groove homodimers in the membrane is schematically shown. The Cα-carbon atoms of the residue locations chosen for cysteine substitution mutation are shown in colored spheres (red and black for Cys-69 and Cys-111, and green for Cys-96, Cys-143, and Cys-162, respectively). The α-helices and amino acid locations are indicated with primed and unprimed numbers for the two polypeptide chains in two gray shades, respectively. The double headed arrows indicate cross-linkable cysteines. B, spin dilution experiment for 96R1, a C-terminal residue of helix α3. The spin dilution experiment was carried out with membrane-inserted BAK 96R1 samples as indicated. The best fitted one-Gaussian distance distribution (assuming 57.3% interacting and 42.7% non-interacting spins) resulted in the average distance and S.D. indicated. BAK 96R1 had a spin labeling efficiency of 84.9 (±1.0)% and a specific relative percent release activity of 96.0 (±6.1)% (see Fig. 4). C and D, rationale for the detection of various BAK homodimers and homooligomers by disulfide bond formation. The letter M represents a monomer of BAK. The letter C represents cysteine residues substituted at the indicated locations of BAK. D2 represents dimers of BAK with two disulfide bonds formed between the monomers, resulting in a slightly reduced electrophoretic mobility than other dimers. M2n represents oligomers of even-numbered BAKs formed by the polymerization of BAK homodimers. This figure was adapted from Dewson et al. (7). E, dye releasing activity of the cysteine substitution BAK mutant proteins. Liposome dye release experiments were carried out with the indicated cysteine-substituted sBAK-ΔC-His proteins (5 nm) in the presence of 25 nm N-terminally His-tagged p7/p15 Bid as described under “Experimental Procedures.” The average values of two experiments are shown with the error ranges. F–I, cross-linking of BAK in membrane-inserted or in solution state. The indicated BAK single, double, and triple cysteine mutants in solution state (G and I) or in the membrane-inserted state (F and H) were cross-linked by copper(II)/phenanthroline complex (Cu(Phe)3) (34) and were analyzed by Western blotting analysis against BAK after PAGE under a reducing (data not shown) or a non-reducing condition (F–I). In lanes 5 and 6 in F and G, 69C+111C represents a mixture of equal quantity of two single cysteine substitution mutant proteins either in solution (G) or in the membrane-inserted state (F). D1, D2, and 3x, 4x, etc., represent BAK single disulfide-bonded dimer, double disulfide-bonded dimer, trimer, tetramer, etc., respectively.
Chemical Cross-linking Studies Also Show That the C Termini of Two Nearby α3, α5, or α6 Helices Are in Close Proximity to Each Other, Respectively, in the Oligomeric BAK in the Membrane
To probe the possible oligomerization interface(s) involving the C termini of α3, α5, and α6 helices, which were suggested by the distance measurements above, we adopted the procedures by Dewson et al. (8) (Fig. 7, C and D).
In the homology model of mouse BAK BH3-in-groove homodimer, residue Leu-69 (Met-71 in human BAK, Fig. 1B) on helix α2 of one BAK molecule is in close proximity to residue Lys-111 (Lys-113 in human BAK) on helix α4 of another BAK monomer (Fig. 7A). The two residues, Leu-69 and Lys-111, are apart at a large distance from their respective symmetry-related partners, Leu-69′ and Lys-111′, respectively (6, 8). When these two residues were substituted with cysteine residues in BAK (Cys-69/Cys-111), and then oxidized by copper(II)/phenanthroline (Cu(Phe)3), they readily formed a dimer in the membrane-inserted state (Fig. 7, C, D2, and F, lanes 7 and 8) unlike in the case of individual mutants (Fig. 7F, lanes 1–4, D1) or in the case of a 1:1 mixture of the two (Fig. 7, F, lane 6, and G). Typically, D2 had a slightly reduced electrophoretic mobility than D1 (data not shown), demonstrating that dimerization of the BAK L69C/K111C double mutant was due to the formation of two disulfide bonds by the cross-linking between two pairs of Cys-69 and Cys-111 that are in close proximity as predicted by the BAK homology model (Fig. 7A).
When an additional mutation Cys-162 was present in the α6 helix of the L69C/K111C BAK mutant, it resulted in BAK oligomers made of predominantly even-numbered BAK monomers upon oxidation (Fig. 7F, lanes 9 and 10), which were susceptible to the reducing agent3 and were not readily formed in solution state (Fig. 7G, lanes 9 and 10). The data showed that an additional disulfide bond was formed between the dimers by the introduction of Cys-162. Thus, these data showed that the oligomerization interface between the dimers involved helix α6 in mouse BAK as reported for human BAK (7). Furthermore, when mutations such as Cys-96 or Cys-143 were introduced to the L69C/K111C BAK mutant, similar results were observed (Fig. 7, H and I), indicating that Cys-96 and Cys-143 were also at the oligomerization interface. The BAK mutant proteins with Cys-96, Cys-143, or Cys-162 substitution formed dimers readily in the membrane upon oxidation (data not shown). The liposomal release assay showed that all the single, double, and triple cysteine substitution BAK mutant proteins retained their membrane-permeabilizing activity similar to its parent molecule (sBAK/C154S-ΔC-His) (Fig. 7E). Thus, the EPR data above and these cross-linking data here all together support that, in addition to α6 helices, the C termini of α3 helices and the C termini of α5 helices are, respectively, brought into close proximity between two neighboring BH3-in-groove homodimers in the active oligomeric BAK pore (Fig. 7A).
DISCUSSION
In this current study, we have shown that the intra- and inter-molecular distances in the oligomeric BAK in the membrane are consistent with the BH3-in-groove homodimer structure (6). Furthermore, additional distance constraints between the BAK homodimers provide clues as to how the BAK homodimers might oligomerize to form pores (Fig. 8).
FIGURE 8.
Theoretical models of the BAK α3:α3′, α5:α5′ oligomerization interface on a flat surface and in a pore. A, the best geometry of BAK dimers on a flat membrane surface satisfying the distance constraints for the α3:α3′, α5:α5′ oligomerization interface. Two BAK homodimers, each consisting of two α2-α5 polypeptides in two shades of gray, are brought to each other, satisfying the distance constraints for the 96R1–96R1 and 143R1–143R1 spin pairs at the α3:α3′, α5:α5′ oligomerization interface with the assumption that the R1-R1 distances are the same as the Cα-Cα distances for the pairs. In each homodimer shown in a top view (upper panel) and a side view (bottom panel), the C2-symmetry axes are perpendicular to a horizontal line (line a) on the membrane surface (coplanar with the page). In the best geometry simulated, the angle between line a and the lines that connect the two symmetry-related 143Cα atoms in each homodimer (b lines) was ∼15° with the distance between the two C2-symmetry axes of ∼45 Å. B, a theoretical model of a hexamer of BAK homodimers. Six BAK BH3-in-groove homodimers (i.e. six α2-α5 homodimers in side view of A) are arranged in a hexagonal geometry, forming a pore (only two homodimers are shown). In this geometry, BAK molecules are arranged in a way that the horizontal projections of the b lines of the six homodimers form a hexagon with the edge length of 50 Å, where the indicated distance between the C2-axes would be ∼43 Å (top view). The radius of the aqueous pore formed by the hydrophilic surface (helices α2-α3) of the BAK homodimers will be ∼25 Å due to the thickness of the homodimer (∼18 Å). A vertical section of a putative lipidic pore, which would interact with the hydrophobic surface of the hexameric arrangement of the BAK dimers, is schematically shown (side view, bottom).
Our data clearly demonstrated the existence of an additional interface between the membrane-inserted BAK dimers in addition to the “α6:α6 interface” (7) (Figs. 5–7). This new interface was formed by the carboxyl termini of the α3 and α5 helices between two neighboring BH3-in-groove homodimers (α2-α5 homodimers), thus will be referred to as “α3:α3′, α5:α5′ oligomerization interface” (Fig. 8A). In this interface, residues 96R1 and 143R1 were brought close to their respective neighbors at an average distance of ∼9 and ∼13 Å, respectively, as shown in Figs. 7B and 5B. To simplify the modeling of oligomerized homodimers, we examined possible arrangements of the two BAK BH3-in-groove homodimers in a linear oligomer on a flat surface assuming that the R1-R1 distance is the same as the Cα-Cα distances for the above pairs (96R1–96R1 and 143R1–143R1). By adjusting the inter-homodimer distance and the rotational angle of the homodimer relative to the inter-homodimer vector, the error between the calculated distances and the measured distances for 96R1–96R1 and 143R1–143R1 pairs was minimized (Fig. 8A). This resulted in a best solution with the inter-homodimer distance of ∼45 Å and a ∼15° angle between the inter-dimer vector (C2-axis to C2-axis, i.e. Fig. 8A, line a) and the vector connecting the two symmetry-related 143 Cα atoms (Fig. 8A, line b). Now if the surface is curved tangential to line a, a model of BAK oligomeric pores of various sizes might be approximated (Fig. 8B).
SDS-PAGE/Western blot analysis of the BAK oligomers formed by the disulfide-mediated cross-linking of the L69C/K111C/H162C BAK mutant showed that BAK octamers (4 BAK homodimers) were clearly present and perhaps decamers (5 BAK homodimers) as well (Fig. 7F, lane 10). In the case of L69C/K111C/H162C and L69C/K111C/R143C BAK mutants, unresolved oligomers larger than 250 kDa (∼12-mer of BAK, i.e. 6 homodimers) were formed (Fig. 7H). These results are consistent with the in vivo cross-linking experiments for human BAK protein by Dewson et al. (7).
If six BAK homodimers are arranged in a hexamer with an edge of ∼50 Å, a pore with a radius of ∼25 Å can be constructed considering the thickness of the BAK homodimer (Fig. 8B). In this arrangement, the distance between two neighboring C2-axes will be ∼43 Å (Fig. 8B), close to the value on a flat surface (Fig. 8A). If the BAK oligomers induce the formation of lipidic pores as suggested by others (6, 19, 40), the hexamer of BAK homodimers could be arranged within the lipidic pore with the hydrophobic surface of the homodimers interacting with the curved membrane surface (Fig. 8B, bottom). From these considerations, it is theoretically possible to obtain a BAK oligomeric pore with a diameter of ∼50 Å, consistent with the reported range of ∼30–60 Å (9, 10). Further studies are needed, however, to define the exact orientation of the BAK homodimers relative to the membrane normal.
Dewson et al. (7) proposed the existence of the α6:α6 interface between the BAK dimers in human BAK oligomeric structure. Recent data by Ma et al. (39) showed that, in the BAK oligomer, the α6 helices with a single cysteine amino substitution mutation in human BAK were chemically cross-linked to each other best at certain periodic residue locations such as 157 (155 in mouse) and 164 (162 in mouse), suggesting a parallel arrangement of the α6 helices. The cross-linking by cysteine residue at 164 in their report corresponds to Cys-162–Cys-162 cross-linking in mouse BAK in this study (Fig. 7), which was also supported by the direct distance measurements (Fig. 6). These results lend credence to the biological relevance of the proteoliposomal system used in this study. Additionally, Dewson et al. (7) reported that the core α2–α5 helices of BAK are sufficient for dimerization but that the α6–α8 helices are essential for apoptotic function of BAK (39). The BAK protein construct we used in this study (sBAK/C154S-ΔC-His) have all the above helices except helix α9, further supporting the biological relevance of the system.
If the α6:α6 interface is formed by the pseudo-parallel arrangement of two nearby surfaced-adsorbed α6 helices as implied by the results from this study and also as suggested by Ma et al. (39), the C2-symmetry that exists within the BH3-in-groove homodimer (or α2-α5 homodimer) would not apply to helices α6-α9 (Fig. 7A). This asymmetric association of the α6 helices in combination with the α3:α3′, α5:α5′ oligomerization interface between the BAK homodimers might result in a curved oligomer on the flat membrane surface. This could then eventually lead to a circular oligomer by forming the last α6:α6 interface and the α3:α3′, α5:α5′ oligomerization interface between the initiating BAK homodimer and the terminating one. This, in turn, could deform the membrane surface in a concerted motion, or gradually during oligomerization, resulting in the pore formation, probably the lipidic pore (Fig. 8B). In contrast, if the α6:α6 interface is formed by a pseudo-anti-parallel arrangement as shown in Fig. 6D or by an arrangement that retains the C2-symmetry of the entire α2-α6 domains within a homodimer, it would result in a linear polymerization of the BAK homodimers. This will leave the α7-α9 domains on both sides of the linear polymer of the α2-α5 homodimers on the membrane surface. It is hard to envision how such a linear structure could close to form a pore.
Due to the presence of the α3:α3′, α5:α5′ oligomerization interface between BAK homodimers in the membrane, there is a possibility of simultaneous intra- and inter-homodimer spin-spin interactions even when a singly spin labeled BAK forms the oligomeric pores. For example, if a spin-spin pair, A(1)-A(1)′, in homodimer 1 is brought close to a neighboring homodimer 2 with identical pairs, i.e. A(2)-A(2)′, it can result in additional inter-homodimer interactions between spin pairs such as A(1)-A(2), A(1)′-A(2)′, A(1)-A(2)′, and A(1)′-A(2), which might be revealed by the DEER experiment. We have a hint of such possibility in the DEER data for 135R1, displaying a second distance population (Fig. 5D). Lack of knowledge in the dynamic nature of the interface(s) and the rotameric conformations of the spin-labeled residue at this location, however, make it difficult to assign these distances to specific inter-dimer interactions.
In conclusion, we have demonstrated that helices α2-α5 of BAK form a homodimeric structure in the membrane similar to the BAX BH3-in-groove homodimer x-ray crystal structure (6). We have also demonstrated the existence of molecular contacts, α3:α3′, α5:α5′ oligomerization interface in the membrane, enabling the formation of a BAK- lipidic pore. These results shed light into the mechanism of oligomerization of the BAK (or BAX), a critical step in the mitochondrial apoptotic pathway.
Acknowledgments
We thank Robert Galvin, Pawan Singh, Kyungro Lee, Kelly Foss, Shinyoub Lee, Minji Park, Steffi Lee, Matthew Park, Puja Singh, Ryung-Suk Kim, and Jindrich Symersky for technical support. We thank Drs. David Mueller, Carl Correll, and Binal Shah for sharing equipment for the EPR sample preparation and help in fluorometry; Drs. Kenneth Neet and Christina Valeria Iancu for comments on the manuscript; and Drs. Wayne Hubbell, Christian Altenbach, Likai Song, and Yeon-Kyun Shin for help in EPR data analyses.
This work was supported, in whole or in part, by National Institutes of Health Grant 5 R01 GM097508, Scientist Development Grant 0835026G from the American Heart Association, the EPR Center at the Rosalind Franklin University of Medicine and Science (RFUMS), and the Start-up fund from RFUMS.
S. Aluvila, T. Mandal, E. Hustedt, P. Fajer, J. Y. Choe, and K. J. Oh, unpublished data.
- BAX
- Bcl-2-associated X protein
- BAK
- Bcl-2 antagonist/killer
- Bcl-2
- B-cell lymphoma-2
- BH
- Bcl-2 homology
- BH3
- Bcl-2 homology domain 3
- CuPhe
- copper(II)/(1,10-phenanthroline)3
- CW
- continuous wave
- DEER
- double electron electron resonance
- DOGS-NTA-Ni
- 1,2-dioleoyl-sn-glycero-3-{[N-5-amino-1-carboxylpentyl)iminodiacetic acid]succinyl} (nickel salt)
- EPR
- electron paramagnetic resonance
- LUV
- large unilamellar vesicle
- MTSSL
- (1-oxyl-2,2,5,5,-tetramethylpyrroline-3-methyl)methanethiosulfonate spin label
- sBAK
- soluble BAK.
REFERENCES
- 1. Danial N. N., Korsmeyer S. J. (2004) Cell death. Critical control points. Cell 116, 205–219 [DOI] [PubMed] [Google Scholar]
- 2. Llambi F., Moldoveanu T., Tait S. W., Bouchier-Hayes L., Temirov J., McCormick L. L., Dillon C. P., Green D. R. (2011) A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol. Cell 44, 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wei M. C., Zong W. X., Cheng E. H., Lindsten T., Panoutsakopoulou V., Ross A. J., Roth K. A., MacGregor G. R., Thompson C. B., Korsmeyer S. J. (2001) Proapoptotic BAX and BAK. A requisite gateway to mitochondrial dysfunction and death. Science 292, 727–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wolter K. G., Hsu Y. T., Smith C. L., Nechushtan A., Xi X. G., Youle R. J. (1997) Movement of Bax from the cytosol to mitochondria during apoptosis. J. Cell Biol. 139, 1281–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cheng E. H., Sheiko T. V., Fisher J. K., Craigen W. J., Korsmeyer S. J. (2003) VDAC2 inhibits BAK activation and mitochondrial apoptosis. Science 301, 513–517 [DOI] [PubMed] [Google Scholar]
- 6. Czabotar P. E., Westphal D., Dewson G., Ma S., Hockings C., Fairlie W. D., Lee E. F., Yao S., Robin A. Y., Smith B. J., Huang D. C., Kluck R. M., Adams J. M., Colman P. M. (2013) Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell 152, 519–531 [DOI] [PubMed] [Google Scholar]
- 7. Dewson G., Kratina T., Czabotar P., Day C. L., Adams J. M., Kluck R. M. (2009) Bak activation for apoptosis involves oligomerization of dimers via their α6 helices. Mol. Cell 36, 696–703 [DOI] [PubMed] [Google Scholar]
- 8. Dewson G., Kratina T., Sim H. W., Puthalakath H., Adams J. M., Colman P. M., Kluck R. M. (2008) To trigger apoptosis, Bak exposes its BH3 domain and homodimerizes via BH3:groove interactions. Mol. Cell 30, 369–380 [DOI] [PubMed] [Google Scholar]
- 9. Saito M., Korsmeyer S. J., Schlesinger P. H. (2000) BAX-dependent transport of cytochrome c reconstituted in pure liposomes. Nat. Cell Biol. 2, 553–555 [DOI] [PubMed] [Google Scholar]
- 10. Martinez-Caballero S., Dejean L. M., Kinnally M. S., Oh K. J., Mannella C. A., Kinnally K. W. (2009) Assembly of the mitochondrial apoptosis-induced channel, MAC. J. Biol. Chem. 284, 12235–12245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dewson G., Kluck R. M. (2009) Mechanisms by which Bak and Bax permeabilise mitochondria during apoptosis. J. Cell Sci. 122, 2801–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hubbell W. L., Lopez C. J., Altenbach C., Yang Z. (2013) Technological advances in site-directed spin labeling of proteins. Curr. Opin. Struct. Biol. 23, 725–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oh K. J., Singh P., Lee K., Foss K., Lee S., Park M., Lee S., Aluvila S., Park M., Singh P., Kim R. S., Symersky J., Walters D. E. (2010) Conformational changes in BAK, a pore-forming proapoptotic Bcl-2 family member, upon membrane insertion and direct evidence for the existence of BH3-BH3 contact interface in BAK homo-oligomers. J. Biol. Chem. 285, 28924–28937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oh K. J., Barbuto S., Meyer N., Kim R. S., Collier R. J., Korsmeyer S. J. (2005) Conformational changes in BID, a pro-apoptotic BCL-2 family member, upon membrane binding. A site-directed spin labeling study. J. Biol. Chem. 280, 753–767 [DOI] [PubMed] [Google Scholar]
- 15. Luo X., Budihardjo I., Zou H., Slaughter C., Wang X. (1998) Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 94, 481–490 [DOI] [PubMed] [Google Scholar]
- 16. Oh K. J., Barbuto S., Pitter K., Morash J., Walensky L. D., Korsmeyer S. J. (2006) A membrane-targeted BID BCL-2 homology 3 peptide is sufficient for high potency activation of BAX in vitro. J. Biol. Chem. 281, 36999–37008 [DOI] [PubMed] [Google Scholar]
- 17. Lutter M., Fang M., Luo X., Nishijima M., Xie X., Wang X. (2000) Cardiolipin provides specificity for targeting of tBid to mitochondria. Nat Cell Biol. 2, 754–761 [DOI] [PubMed] [Google Scholar]
- 18. Ardail D., Privat J. P., Egret-Charlier M., Levrat C., Lerme F., Louisot P. (1990) Mitochondrial contact sites. Lipid composition and dynamics. J. Biol. Chem. 265, 18797–18802 [PubMed] [Google Scholar]
- 19. Terrones O., Antonsson B., Yamaguchi H., Wang H. G., Liu J., Lee R. M., Herrmann A., Basañez G. (2004) Lipidic pore formation by the concerted action of proapoptotic BAX and tBID. J. Biol. Chem. 279, 30081–30091 [DOI] [PubMed] [Google Scholar]
- 20. Kuwana T., Bouchier-Hayes L., Chipuk J. E., Bonzon C., Sullivan B. A., Green D. R., Newmeyer D. D. (2005) BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol. Cell 17, 525–535 [DOI] [PubMed] [Google Scholar]
- 21. Kuwana T., Mackey M. R., Perkins G., Ellisman M. H., Latterich M., Schneiter R., Green D. R., Newmeyer D. D. (2002) Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell 111, 331–342 [DOI] [PubMed] [Google Scholar]
- 22. Böttcher C. J. F., Van gent C. M., Pries C. (1961) A rapid and sensitive sub-micro phosphorus determination. Anal. Chim. Acta 24, 203–204 [Google Scholar]
- 23. Rabenstein M. D., Shin Y. K. (1995) Determination of the distance between two spin labels attached to a macromolecule. Proc. Natl. Acad. Sci. U.S.A. 92, 8239–8243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Altenbach C., Oh K. J., Trabanino R. J., Hideg K., Hubbell W. L. (2001) Estimation of inter-residue distances in spin labeled proteins at physiological temperatures. Experimental strategies and practical limitations. Biochemistry 40, 15471–15482 [DOI] [PubMed] [Google Scholar]
- 25. Pannier M., Veit S., Godt A., Jeschke G., Spiess H. W. (2000) Dead-time free measurement of dipole-dipole interactions between electron spins. J. Magn. Reson. 142, 331–340 [DOI] [PubMed] [Google Scholar]
- 26. Jeschke G., Chechik V., Ionita P., Godt A., Zimmermann H., Banham J., Timmel C. R., Hilger D., Jung H. (2006) DeerAnalysis2006 - a comprehensive software package for analyzing pulsed ELDOR data. Appl. Magn. Res. 30, 473–498 [Google Scholar]
- 27. Sen K. I., Logan T. M., Fajer P. G. (2007) Protein dynamics and monomer-monomer interactions in AntR activation by electron paramagnetic resonance and double electron-electron resonance. Biochemistry 46, 11639–11649 [DOI] [PubMed] [Google Scholar]
- 28. Brandon S., Beth A. H., Hustedt E. J. (2012) The global analysis of DEER data. J. Magn. Reson. 218, 93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hustedt E. J., Beth A. H. (1999) Nitroxide spin-spin interactions. Applications to protein structure and dynamics. Annu. Rev. Biophys. Biomol. Struct. 28, 129–153 [DOI] [PubMed] [Google Scholar]
- 30. Hustedt E. J., Smirnov A. I., Laub C. F., Cobb C. E., Beth A. H. (1997) Molecular distances from dipolar coupled spin-labels. The global analysis of multifrequency continuous wave electron paramagnetic resonance data. Biophys. J. 72, 1861–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hustedt E. J., Stein R. A., Sethaphong L., Brandon S., Zhou Z., Desensi S. C. (2006) Dipolar coupling between nitroxide spin labels. The development and application of a tether-in-a-cone model. Biophys. J. 90, 340–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moldoveanu T., Liu Q., Tocilj A., Watson M., Shore G., Gehring K. (2006) The x-ray structure of a BAK homodimer reveals an inhibitory zinc binding site. Mol. Cell 24, 677–688 [DOI] [PubMed] [Google Scholar]
- 33. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Careaga C. L., Falke J. J. (1992) Thermal motions of surface α-helices in the d-galactose chemosensory receptor. Detection by disulfide trapping. J. Mol. Biol. 226, 1219–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Langen R., Oh K. J., Cascio D., Hubbell W. L. (2000) Crystal structures of spin labeled T4 lysozyme mutants. Implications for the interpretation of EPR spectra in terms of structure. Biochemistry 39, 8396–8405 [DOI] [PubMed] [Google Scholar]
- 36. Fleissner M. R., Bridges M. D., Brooks E. K., Cascio D., Kálai T., Hideg K., Hubbell W. L. (2011) Structure and dynamics of a conformationally constrained nitroxide side chain and applications in EPR spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 108, 16241–16246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim H., Tu H. C., Ren D., Takeuchi O., Jeffers J. R., Zambetti G. P., Hsieh J. J., Cheng E. H. (2009) Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol. Cell 36, 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. O'Neill J. W., Manion M. K., Maguire B., Hockenbery D. M. (2006) BCL-XL dimerization by three-dimensional domain swapping. J. Mol. Biol. 356, 367–381 [DOI] [PubMed] [Google Scholar]
- 39. Ma S., Hockings C., Anwari K., Kratina T., Fennell S., Lazarou M., Ryan M. T., Kluck R. M., Dewson G. (2013) Assembly of the Bak Apoptotic Pore, A critical role for the BAK protein α6 helix in the multimerization of homodimers during apoptosis. J. Biol. Chem. 288, 26027–26038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schafer B., Quispe J., Choudhary V., Chipuk J. E., Ajero T. G., Du H., Schneiter R., Kuwana T. (2009) Mitochondrial outer membrane proteins assist Bid in Bax-mediated lipidic pore formation. Mol. Biol. Cell 20, 2276–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Suzuki M., Youle R. J., Tjandra N. (2000) Structure of Bax. Coregulation of dimer formation and intracellular localization. Cell 103, 645–654 [DOI] [PubMed] [Google Scholar]
- 42. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]