Background: Mechanisms filamentous fungi use to sense cellulose in the environment remain unclear.
Results: Neurospora crassa cellodextrin transporters, CDT-1 and CDT-2, are required to induce the expression and secretion of cellulases, using a mechanism distinct from that in Trichoderma reesei.
Conclusion: CDT-1 and/or CDT-2 may act as transporting receptors (transceptors).
Significance: CDT-1- and/or CDT-2-mediated cellodextrin signaling may be conserved in industrially useful fungi.
Keywords: Bioenergy, Biofuel, Cellulase, Fungi, Genetics, Cellodextrin Transporter, Neurospora crassa, Transceptor
Abstract
Neurospora crassa colonizes burnt grasslands and metabolizes both cellulose and hemicellulose from plant cell walls. When switched from a favored carbon source to cellulose, N. crassa dramatically up-regulates expression and secretion of genes encoding lignocellulolytic enzymes. However, the means by which N. crassa and other filamentous fungi sense the presence of cellulose in the environment remains unclear. Previously, we have shown that a N. crassa mutant carrying deletions of three β-glucosidase enzymes (Δ3βG) lacks β-glucosidase activity, but efficiently induces cellulase gene expression and cellulolytic activity in the presence of cellobiose as the sole carbon source. These observations indicate that cellobiose, or a modified version of cellobiose, functions as an inducer of lignocellulolytic gene expression and activity in N. crassa. Here, we show that in N. crassa, two cellodextrin transporters, CDT-1 and CDT-2, contribute to cellulose sensing. A N. crassa mutant carrying deletions for both transporters is unable to induce cellulase gene expression in response to crystalline cellulose. Furthermore, a mutant lacking genes encoding both the β-glucosidase enzymes and cellodextrin transporters (Δ3βGΔ2T) does not induce cellulase gene expression in response to cellobiose. Point mutations that severely reduce cellobiose transport by either CDT-1 or CDT-2 when expressed individually do not greatly impact cellobiose induction of cellulase gene expression. These data suggest that the N. crassa cellodextrin transporters act as “transceptors” with dual functions - cellodextrin transport and receptor signaling that results in downstream activation of cellulolytic gene expression. Similar mechanisms of transceptor activity likely occur in related ascomycetes used for industrial cellulase production.
Introduction
Liquid biofuels produced from lignocellulosic biomass are an environmentally clean and renewable source of energy that could displace a significant fraction of the current demand for petroleum (1–4). However, the costs associated with conversion of insoluble polysaccharides in plant cell walls to easily fermentable sugars represent significant barriers to the production of cost-competitive biofuels (4–6). Filamentous fungi have the capacity to secrete large amounts of lignocellulosic enzymes that release fermentable sugars from plant cell walls, and this ability has been exploited by industry to produce cellulases in quantities exceeding 100 g/liter of culture (7).
The best inducers of plant cell wall-degrading enzyme expression by filamentous fungi are insoluble substrates that include cellulose, hemicellulose, or mixtures of plant polymers. Since these naturally inducing substances cannot enter fungal cells, it is generally believed that oligosaccharides released from the polymers and their derivatives function as the actual molecules that trigger enzyme induction (8). Early work in Trichoderma reesei using [U-14C]cellobiose confirmed the presence of a cellobiose uptake system (9). This uptake system was specific for β-linked diglucosides, could be inhibited by glucose and uptake was stimulated following treatment with sophorose. Despite characterization of this cellobiose uptake system, no specific transporter or transporters were described to enable the diglucoside uptake. More recently, studies in T. reesei identified four different transporters involved in cellulase production in response to a media containing lactose or cellobiose. In the first study, deletion of one transporter (Trire2:3405) was sufficient to prevent both lactose uptake and cellulase induction in T. reesei (10). However a second study using a different T. reesei strain found that deletion of the genes encoding either Trire2:77517 or Trire2:79202 showed a decrease in both lactose uptake and cellulase induction (11). While both studies identified different transporters involved in cellulase induction via lactose, the mechanisms of cellulase induction by sophorose (another soluble inducer of cellulases in T. reesei) and by crystalline cellulose (Avicel) remained unknown. In a third study using sophorose, cellobiose, or crystalline cellulose as inducers, a dual cellobiose/glucose transporter named Stp1 (Trire2:47710) was shown to be involved in the T. reesei carbon catabolite repression response (12). Intriguingly, although deletion of Trire2:3405 abolished lactose uptake (10), the deletion strain still imported sophorose and cellobiose. However, deletion of Trire2:3405 (Crt1) abolished the accompanying induction of cellulases (12). These data suggest that, similar to nutrient sensing in Saccharomyces cerevisiae (13, 14), T. reesei uses multiple transporters that may function in signal transduction (so called transceptors) to induce cellulase gene expression.
The filamentous fungus N. crassa, has been used as a model laboratory organism for nearly 100 years (15). In nature, N. crassa is only associated with plant material, primarily killed by exposure to fire (16–19). N. crassa is an effective degrader of lignocellulose (20). When grown on crystalline cellulose, N. crassa increases the transcription of seven major facilitator superfamily (MFS)3 sugar transporters (21). Two of these transporters, CDT-1 and CDT-2, were capable of transporting cellodextrins when expressed in Saccharomyces cerevisiae (22). In this study, we examine the hypothesis that CDT-1 and CDT-2 function as transporters and also are required for signaling for cellulase induction by crystalline cellulose. We show that the presence of at least one of the transporters CDT-1 or CDT-2 is required for cellulase gene induction and that deletion of both cdt-1 and cdt-2 results in a N. crassa strain that is unable to sense the presence of crystalline cellulose. Furthermore, by using strains lacking the genes encoding three β-glucosidase enzymes (Δ3βG) (23), we show that additional absence of both cdt-1 and cdt-2 results in a strain of N. crassa that is unable to sense the presence of cellobiose.
EXPERIMENTAL PROCEDURES
Deletion Strains
Strains obtained from the Fungal Genetics Stock Center (FGSC) (24) include the WT (FGSC 2489), and deletion strains for the two cellodextrin transporters: NCU00801 (FGSC 16575) and NCU08114 (FGSC 17868 and 17869). The Δ3βG deletion strain was described previously (23). Multiple deletion strains were made by performing sequential crosses performed on Westergaard media (25). The genotype of each multiple deletion strain was confirmed using a gene-specific primer and a common primer for the hygromycin phosphotransferase (hph) resistance cassette. The primer for hph was 5′-CGA CAG ACG TCG CGG TGA GTT CAG-3′. Reverse primers were NCU00801: 5′-TAG GGT TGT AGA CAC CTG C-3′; NCU08114: 5′-GAC GAC CAG AAC TAG GTA GG-3′; NCU00130: 5′-TAG TGT ACA AAC CCC AAG C-3′; NCU004952: 5′-AAC ACA CAC ACA CAC ACT GG-3′; and NCU08755: 5′-ACA GTG GAG GTG AGA AAG G-3′.
Shake Flask Studies
Conidia from deletion strains were inoculated at a concentration equal to 106 conidia per milliliter in 50 ml of Vogel's salts (26) with 2% w/v sucrose, 2% Avicel PH 101 (Sigma), or 2% cellobiose (Sigma) in a 250 ml Erlenmeyer flask and grown under constant light at 200 rpm. Images were taken at 48, 72, and 96 h.
Transcriptional Studies
Conidia from strains were inoculated at a concentration equal to 106 conidia per milliliter in 50 ml Vogel's salts (26) with 2% w/v sucrose in a 250 ml Erlenmeyer flask and grown under constant light at 200 rpm for 16 h. Biomass was then spun at 1,000 × g for 10 min and washed in Vogel's salts twice to remove any excess sucrose. Biomass was then added to a new flask containing 50 ml of Vogel's salts supplemented with 1% w/v sucrose, 1% w/v Avicel PH 101 (Sigma) or cellobiose (Sigma) at the indicated concentration. Cultures were induced for 4 h under constant light at 200 rpm (23). The culture biomass was then harvested by filtration over a Whatman glass microfiber filter (GF/F) on a Büchner funnel and washed with 50 ml of Vogel's salts. The biomass was flash frozen in liquid nitrogen and stored at −80 °C. Three independent biological replicates (flasks) were evaluated for each time point.
RNA Isolation
Total RNA from frozen samples was isolated using Zirconia/silica beads (0.5 mm diameter; Biospec) and a Mini-Beadbeater-96 (Biospec) with 1 ml TRIzol reagent (Invitrogen) according to the manufacturer's instructions. The total RNA was further purified by digestion with TURBO DNA-free (Ambion) and an RNeasy kit (Qiagen). RNA concentration and integrity was checked by Nanodrop and agarose gel electrophoresis, respectively.
RT-PCR
Quantitative RT-PCR was performed using the EXPRESS One-Step SYBR GreenER Kit (Invitrogen) and the StepOnePlus Real-Time PCR System (Applied Biosystems). Reactions were performed in triplicate with a total reaction volume of 10 μl including 300 nm each forward and reverse primers and 75 ng of template RNA. Data analysis was performed using the StepOne Software (Applied Biosystems) with the Relative Quantitation/Comparative CT (ΔΔCT) setting. Data were normalized to the endogenous control actin (NCU04173) with expression on sucrose as the reference sample.
RT-PCR Primers
The primers for actin (NCU04173) were forward 5′-TGA TCT TAC CGA CTA CCT-3′ and reverse 5′-CAG AGC TTC TCC TTG ATG-3′. The primers for cbh-1 (NCU07340) were forward 5′-ATC TGG GAA GCG AAC AAA G-3′ and reverse 5′-TAG CGG TCG TCG GAA TAG-3′. The primers for gh5-1 (NCU00762) were forward 5′-GAG TTC ACA TTC CCT GAC A-3′ and reverse 5′-CGA AGC CAA CAC GGA AGA-3′. RT-PCR primers were previously identified and optimized in Tian et al. (21) and Dementhon et al. (27).
Glucose and Cellobiose Concentration during Growth on Crystalline Cellulose
Conidia from the wild type strain were inoculated in duplicate at a concentration equal to 106 conidia per milliliter in 50 ml of Vogel's salts (26) with 2% w/v Avicel PH 101 (Sigma) in a 250 ml Erlenmeyer flask and grown under constant light at 200 rpm for 7 days. Samples were taken daily and spun at 1,000 × g for 10 min to remove biomass and excess Avicel. The concentration of glucose and cellobiose were measured on a DIONEX ICS-3000 HPLC (Dionex Corporation) using a CarboPac PA20 Analytical Column (3 × 150 mm) and a CarboPac PA20 guard column (3 × 30 mm) at 30 °C. Following injection of 25 μl of diluted samples, elution was performed with 100 mm KOH (isocratic) at 0.4 ml/min. Sugars were detected using pulsed amperometric detection, Four-Potential Carbohydrate Waveform and Peaks were analyzed using the Chromeleon software package.
GFP-tagged Transporters
Template gDNA from N. crassa WT strain (FGSC 2489) was extracted according to the method of Lee et al. (51). Strains tagged with GFP were created by adapting the tools described in Honda and Selker (28). The 10xGly::GFP::hph::LoxP fragment was subcloned from FJ457011 (28) into pRS426 using XhoI (NEB) and KpnI (NEB). The upstream region was amplified by PCR (∼1000 bp upstream and coding region) using primers containing XhoI (NEB) sites and ligated using T4 DNA Ligase (NEB). The downstream region was then amplified by PCR (∼1000 bp downstream of the coding region) using primers containing KpnI (NEB) sites and ligated using T4 DNA Ligase (NEB). The final vectors were sequenced using several primers to ensure that the proper sequence was obtained. The primers used to clone the upstream region are as follows NCU08114: 5′-GGG CCC CCC CTC GAG GTC GAC GGT ATC GAT AAG CTT CAA GTT TCG GTA CAC-3′ and 5′-GCC TCC GCC TCC GCC GCC TCC GCC TCG AGA AGC AAC AGA CTT GCC CTC ATG-3′; NCU00801: 5′-GGG CCC CCC CTC GAG GTC GAC GGT ATC GAT ACA TCG TCC CGC CAT CCC GG-3′ and 5′-GCC TCC GCC TCC GCC GCC TCC GCC TCG AGA AGC AAC GAT AGC TTC GGA CA-3′. The primers used to clone the downstream region as as follows: NCU08114: 5′-CGA AGT TAT GGA TCC GAG CTC GGT ACC GTG TCT GTT TGA GAT TG-3′ and 5′-GGG CTG CAG GAA TTC GAT ATC AAG CTG ACG ACC AGA ACT AGG TAG G-3′; NCU00801: 5′-CGA AGT TAT GGA TCC GAG CTC GGT ACC ACA GGC GAC AAG GAA G-3′ and 5′-CTA GTG GAT CCC CCG GGC TGC AGG AAT TCG ATA TCA AGC TCG TTG GAG CTG TCC CC-3′. The vectors were then transformed into the Ku70 deletion strain as described below.
Mutagenesis of CDT-1 and CDT-2 and Strain Construction
Alanine mutants were produced in previously described 2μ yeast expression plasmids (22). These plasmids contained the CDT-1 or CDT-2 open reading frame fused to GFP and the S. cerevisiae PGK1 promoter and CYC transcriptional terminator within the pRS426 backbone. The QuikChange (Agilent Technologies) protocol was used to introduce alanine mutations, and results were confirmed by sequencing the plasmids. All plasmids were transformed into the S. cerevisiae strain YPH449 (MATa ura3-52 lys2–801_amber ade2-101_ochre trp1-Δ63 his3-Δ200 leu2-Δ1), which contained a previously described (22) plasmid containing the GH1–1 open reading frame and the S. cerevisiae PGK1 promoter within the pRS424 backbone.
Determination of Strain Growth Rates
Colonies corresponding to independent transformants of S. cerevisiae strain YPH449 containing GH1–1 and a given alanine mutant of CDT-1 or CDT-2 were resuspended in 400 μl of YNP (with ammonium sulfate) plus 2% cellobiose and CSM (-uracil, -leucine) at a starting OD (600 nm) of ∼0.2. Growth curves were recorded using a Bioscreen C (Oy Growth Curves Ab Ltd), and growth rates determined from the exponential phase of the curve.
GFP-tagged Point Mutants
GFP-tagged point mutants were created using a QuikChange II XL Site-directed Mutagenesis Kit (Stratagene) according to the manufacturer's instructions. The primers used to create the point mutations are as follows NCU00801 D307A: 5′-ACC GAC GGT GTC GCC AAG GTC TGG TGG-3′ and 5′-CCA CCA GAC CTT GGC GAC ACC GTC GGT-3′; NCU00801 E476A: 5′-GTG TTA TTC CCA CCG CGG CTC TCG AGA CCA-3′ and 5′-GTG GTC TCG AGA GCC GCG GTG GGA ATA ACA A-3′; NCU08114 S224A: 5′-GTG GGT TCC CGA GGC TCC CCG TTT CCT 3′ and 5′-AGG AAA CGG GGA GCC TCG GGA ACC CAC-3′; NCU08114 E417A: 5′-CGG CTA CGC CAT CGC AAT CCT CCC TTA CC-3′ and 5′-GGT AAG GGA GGA TTG CGA TGG CGT AGC CG-3′. The final vectors were sequenced using several primers to ensure that the proper sequence was obtained and were then transformed into the N. crassa Ku70 deletion strain (29, 30) as described below.
Transformation of N. crassa Strains
1 μg of plasmid DNA was transformed into the Δmus-52 strain (FGSC 9719) by electroporation as described (31). The constructs were targeted to the native locus by homologous recombination and integration of the constructs at the native locus in heterokaryotic transformants was confirmed by GFP fluorescence and PCR with primers designed to the region upstream and downstream of the integration cassette. To recover homokaryotic strains, and repair the mus-52 gene, transformants were crossed with the Δ3βG strain (23) and hygromycin B (Sigma-Aldrich) resistant/GFP-positive/PEST (Glufosinate-ammonium; Sigma-Aldrich) sensitive progenies were screened for the absence of NCU00130, NCU04952, and NCU08755 as described previously (23).
Cellobiose Uptake Assays
Conidia from each strain were inoculated at a concentration equal to 106 conidia per milliliter in 3 ml of Vogel's salts with 2% w/v sucrose in a 24 well plate and grown under constant light at 200 rpm for 40 h. Biomass was transferred to 5 ml of Vogel's salts containing no carbon and was incubated at 450 rpm for 5 min. Biomass was again transferred to 3 ml of Vogel's salts containing 2% w/v cellobiose (Sigma-Aldrich) and was incubated under constant light at 200 rpm for 24 h to induce transporter expression. Following induction, biomass was washed three times with Vogel's salts containing no carbon to remove any excess cellobiose and transferred to a new 24-well plate with 3 ml of cellobiose uptake assay solution (5 mm MES pH 6.0, 100 mm NaCl, 100 μm cellobiose) and vortexed at 900 rpm for 10 s. 300 μl samples were taken at 2.5, 10, 20, and 30 min intervals. Samples were centrifuged at 14,000 rpm at 4 °C for 10 min. 200 μl of each sample were transferred to an HPLC vial containing 800 μl of 0.1 m NaOH. The concentration of remaining cellobiose was measured on a DIONEX ICS-3000 HPLC (Dionex Corporation) as described above. The remaining biomass was washed with water, dried for 24 h at 95 °C, and weighed with a Sartorius SE2 ultra-micro analytic balance.
Fluorescence Microscopy of GFP-tagged Transporters
Mycelia for microscopy were prepared in the same way as biomass was prepared for cellobiose uptake assays, without the final Vogel's salts washes. Confocal microscopy was performed on a Leica SD6000 microscope equipped with a Yokogawa CSU-X1 spinning disk head and 488-nm lasers controlled by Metamorph software (Molecular Devices). All images were acquired using a 100 × 1.4 NA oil immersion objective.
RESULTS
Induction of Cellulase Gene Transcripts Is Blocked in Cellulose-induced Cultures of N. crassa Lacking Two Cellodextrin Transporter Genes
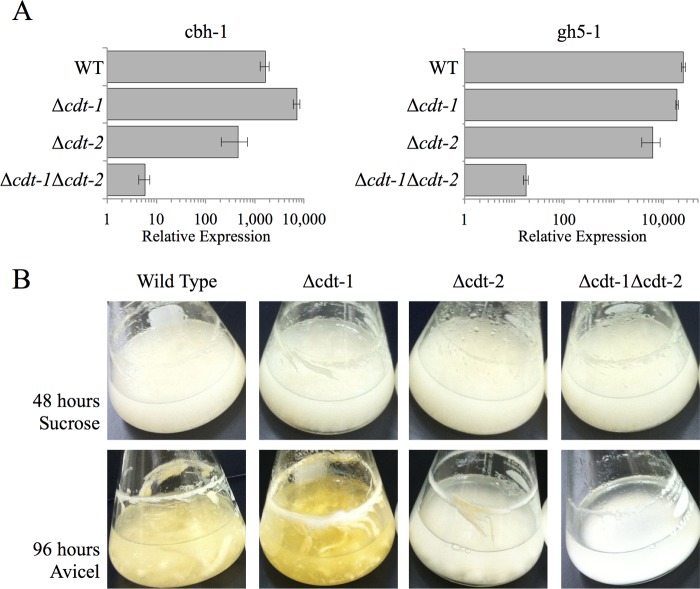
A previous systems analysis identified several predicted transporters that increase in expression level when N. crassa is grown on crystalline cellulose (21). When expressed in S. cerevisiae, two of these transporters were capable of efficiently transporting cellooligosaccharides (22). We hypothesized that these up-regulated transporters are involved in sensing of crystalline cellulose and contribute to the induction and secretion of active cellulases. To examine this hypothesis, we initially tested whether the expression of two major cellulase genes (cbh-1, NCU07340; gh5-1, NCU00762) was induced in strains carrying deletions of the eight previously identified transporters (21) (“Experimental Procedures”). Because strains carrying single deletions were unable to produce a significant defect in induction (Fig. 1A), a mutant strain was constructed that lacked both cdt-1 and cdt-2. Unlike the single mutants, this strain (Δcdt-1 Δcdt-2) showed a significant defect in the induction of both cbh-1 and gh5-1 following a 4-h transfer of N. crassa to 1% w/v Avicel (Fig. 1A). In addition to the induction phenotype, we also examined the growth phenotype of strains carrying both single deletions and double deletions of cdt-1 and/or cdt-2 on both sucrose and Avicel as compared with wild type (Fig. 1B). All strains were able to grow equally well on sucrose. However, a minor growth defect was seen in strain lacking cdt-2 after 96 h growth on 2% w/v Avicel. A severe growth defect was observed when a strain lacking both cdt-1 and cdt-2 was cultivated on Avicel. These data indicate that cdt-1 and cdt-2 are involved in both the induction of cellulases as well as the growth of N. crassa on crystalline cellulose. Deletion of both genes was sufficient to prevent N. crassa from sensing the presence of crystalline cellulose in its environment.
FIGURE 1.
Cellulase gene expression levels in WT and cellodextrin transporter deletion strains after induction with Avicel. A, gene expression of the cellulases cbh-1 and gh5-1 after 4 h induction with 1% Avicel in WT, Δcdt-1, Δcdt-2, and Δcdt-1Δcdt-2 strains. Gene expression levels of cbh-1 and gh5-1 were normalized to 1 when induced with 1% sucrose. Expression levels of actin (NCU04173) were used as an endogenous control in all samples. Each strain was grown in triplicate, and error bars indicate 1 S.D. B, growth phenotype of WT, Δcdt-1, Δcdt-2, and Δcdt-1Δcdt-2 strains when grown in sucrose or Avicel. Strains were grown for 48 h on 2% sucrose or 96 h on 2% Avicel.
Deletion of cdt-1 and cdt-2 in a Strain Lacking Genes Encoding Three β-Glucosidase Enzymes (Δ3βG) Results in a Strain Unable to Respond to Cellobiose
Previously, we showed that a N. crassa strain lacking genes encoding three β-glucosidase enzymes (two extracellular and one intracellular; Δ3βG) responds to cellobiose in the same way that wild type N. crassa responds to crystalline cellulose (23). We therefore hypothesized that a strain lacking the genes encoding the three β-glucosidase enzymes as well as cdt-1 and cdt-2 (Δ3βGΔcdt-1Δcdt-2) would be unable to induce cellulases in response to a physiologically relevant concentration of cellobiose. To examine this hypothesis, we first defined the physiologically relevant concentration of cellobiose required for cellulase induction in N. crassa.
We first measured the concentration of soluble glucose and cellobiose from cultures growing over 7 days on 2% w/v Avicel (Fig. 2A). The glucose concentration reached a maximum of ∼70 μm on the second day and proceeded to decrease until day 6 when it reached its minimum concentration of ∼20 μm. On the other hand, the cellobiose concentration was much lower, reaching a maximum of about 1 μm over the 7 day time course. In a second experiment, we used the Δ3βG background strain to examine the induction of two major cellulase genes (cbh-1, NCU07340; gh5-1, NCU00762) in response to decreasing concentrations of cellobiose. We confirmed that 0.2% w/v (5.8 mm) cellobiose produced maximal induction (23), but were also able to detect induction of cbh-1 and gh5-1 above that seen by starvation (derepression) with concentrations of cellobiose as low as 10 nm (Fig. 2B). Taken together, the results of these two experiments indicate that 1 μm cellobiose should serve as a physiologically relevant concentration for examining cellulase gene induction in strains lacking cdt-1 and/or cdt-2 in the Δ3βG background strain.
FIGURE 2.
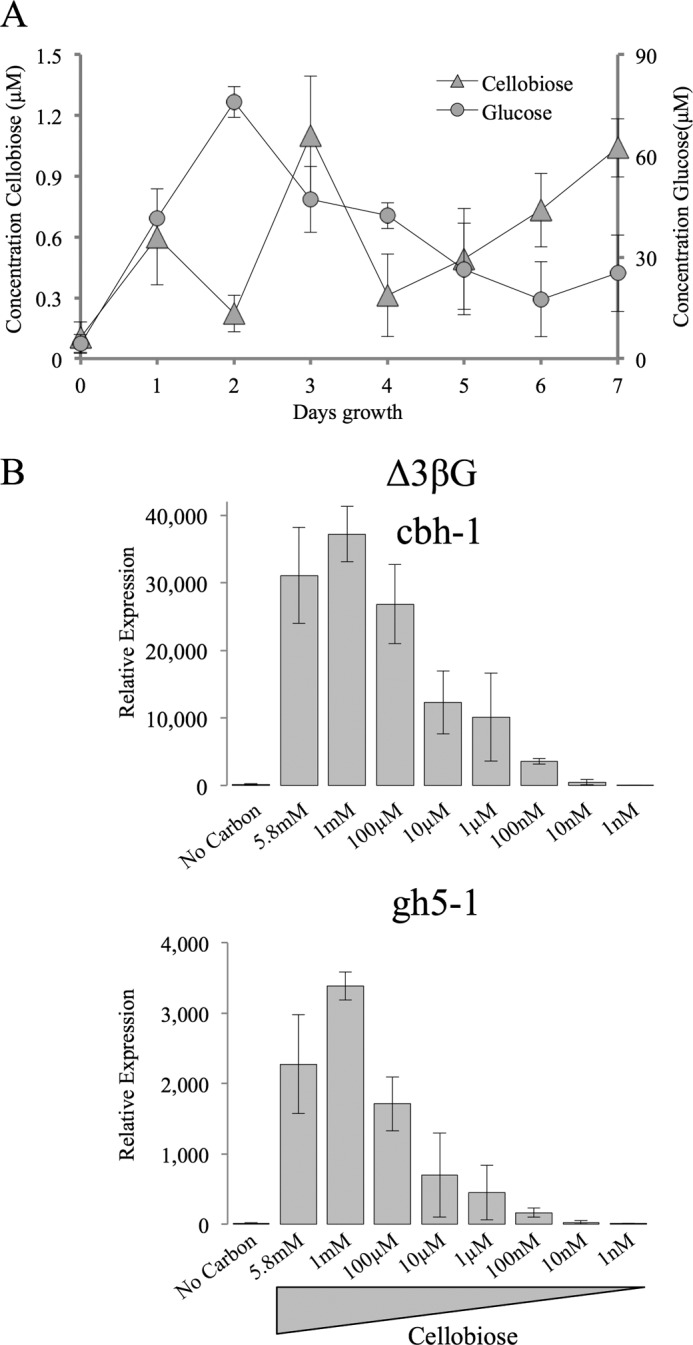
Concentration of glucose and cellobiose in WT cultures and the effect of cellobiose concentration on cellulase gene induction in the Δ3βG strain. A, concentration of glucose and cellobiose in WT cultures grown on Avicel. The concentration of glucose and cellobiose were measured every 24 h from cultures growing on 2% Avicel. Samples were run as biological triplicates and error bars indicate 1 S.D. Time point 0 represents the concentration of glucose and cellobiose before inoculation. B, gene expression of the cellulases cbh-1 and gh5-1 in the Δ3βG strain after 4 h induction with varying concentrations of cellobiose. Gene expression levels of cbh-1 and gh5-1 were normalized to 1 when induced with 1% sucrose. Expression levels of actin (NCU04173) were used as an endogenous control in all samples. Each strain was grown in triplicate, and error bars indicate 1 S.D.
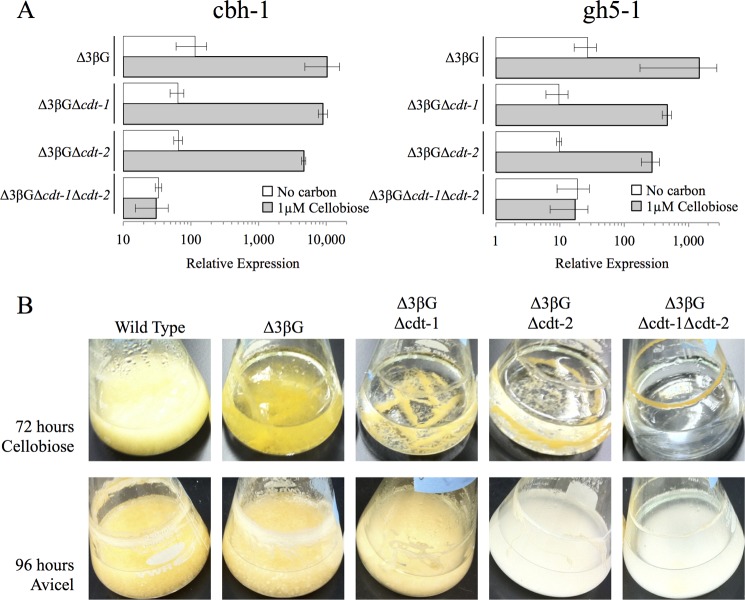
In a 4-h transfer experiment using 1 μm cellobiose, we found that strains lacking either cdt-1 or cdt-2 (in the Δ3βG background strain) showed an induction profile for the two major cellulase genes (cbh-1, NCU07340; gh5-1, NCU00762) similar to that observed in the Δ3βG background strain alone (Fig. 3A). However, when both cdt-1 and cdt-2 were deleted (Δ3βGΔcdt-1Δcdt-2), the induction profile for both cbh-1 and gh5-1 matched the starvation/derepression profile (Fig. 3A). These data imply that this strain is unable to sense the presence of cellobiose. In addition to cellulase gene induction, we examined the growth of strains carrying both the single deletion and double deletions of cdt-1 and/or cdt-2 (Fig. 3B) in the Δ3βG background strain. The Δ3βG strain grows slowly on cellobiose, likely due to the expression of minor predicted β-glucosidase enzymes (23, 32). Although additional deletion of either cdt-1 or cdt-2 did not exacerbate this growth defect, a Δ3βG strain carrying deletions of both cdt-1 and cdt-2 (Δ3βGΔcdt-1Δcdt-2) showed almost no growth after 72 h on 2% cellobiose (Fig. 3B). Taken together, these results reveal that either cdt-1 or cdt-2 are required for the induction of cellulose gene expression when N. crassa is exposed to cellobiose as well as for its growth on cellobiose or Avicel.
FIGURE 3.
Cellulase gene expression levels in Δ3βG and cellodextrin transporter deletion strains after induction with cellobiose. A, gene expression of the cellulases cbh-1 and gh5-1 after 4 h induction with 1 μm cellobiose or no carbon (starvation) in the Δ3βG, Δ3βGΔcdt-1, Δ3βGΔcdt-2, and Δ3βGΔcdt-1Δcdt-2 strains. Gene expression levels of cbh-1 and gh5-1 were normalized to 1 when induced with 1% sucrose. Expression levels of actin (NCU04173) were used as an endogenous control in all samples. Each strain was grown in triplicate, and error bars indicate 1 S.D. B, growth phenotype of the Δ3βG, Δ3βGΔcdt-1, Δ3βGΔcdt-2, and Δ3βGΔcdt-1Δcdt-2 strains when grown on cellobiose or Avicel. Strains were grown for 72 h on 2% cellobiose or 96 h on 2% Avicel.
Point Mutants in CDT-1 and CDT-2 Significantly Decrease Cellobiose Uptake, but Still Allow Cellulase Gene Induction
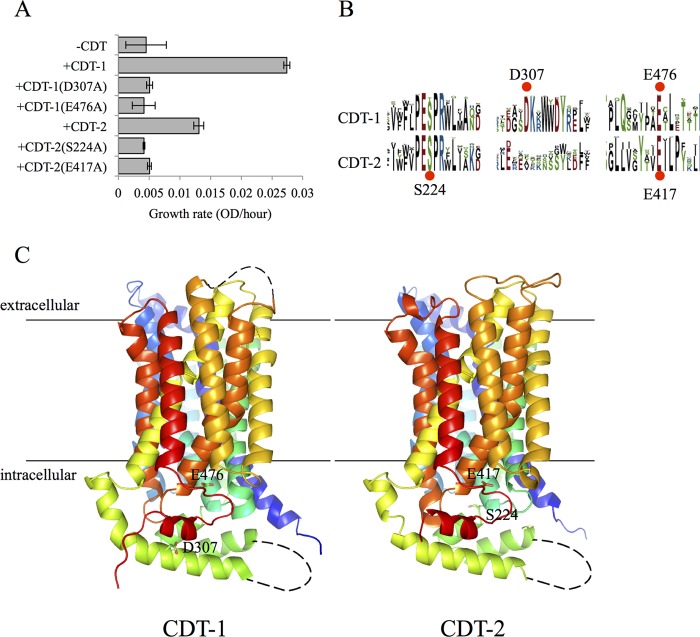
When heterologously expressed in S. cerevisiae, CDT-1 and CDT-2 have been shown to be high-affinity transporters, with CDT-1 acting as a proton symporter and CDT-2 acting as a facilitator (33). To identify functionally important residues within the cellodextrin transporters, CDT-1 and CDT-2, several conserved amino acids were individually mutated to alanine. The functional consequence of each mutation was determined by monitoring the growth of S. cerevisiae strains expressing the intracellular β-glucosidase, GH1-1, and mutant CDT-1 or CDT-2 transporters, using cellobiose as a sole carbon source, as described previously (22) (Fig. 4A). From these libraries of mutant CDT-1 and CDT-2 (34), several completely abolished growth of S. cerevisiae on cellodextrins, implying that these residues were required for cellobiose transport. A comparison of these residues indicates that the sites we chose to mutate are highly conserved (Fig. 4B).
FIGURE 4.
Identification of non-transporting CDT-1 and CDT-2 point mutants. A, cellobiose mediated growth rates of S. cerevisiae transformed with CDT-1 or CDT-2 bearing alanine substitutions and GH1–1. Conserved amino acids in CDT-1 and CDT-2 were separately mutated to alanine. A construct containing a mutation at one position was transformed into S. cerevisiae alone with the β-glucosidase, GH1–1. These strains were grown with cellobiose as the sole carbon source. The growth rates (increase in the OD at 600 nm per hours) were measured and compared with strains with a wild type CDT and GH1–1 or no transporter and GH1–1. Each sample was performed in duplicate and error bars represent the standard deviation. B, sequence logos for CDT-1 and CDT-2 were retrieved from the NCBI BLAST server with an E value cutoff of 1E−130 and aligned using the MUSCLE algorithm (47). The alignment logos were created with WebLogo (48). The size of the residue letter is proportional to its frequency of appearance at the given position. C, homology models of CDT-1 and CDT-2. Homology models of CDT-1 and CDT-2 were built by Modeler v9.12 (49) using a xylose transporter XylE structure; PDB code 4GBZ (50) as the template. The black lines indicate the transmembrane region assigned by comparing with XylE structure. Structures were colored in rainbow with N terminus in blue and C terminus in red. The mutated residues are shown as sticks. The figure was prepared with the molecular visualization software PyMol (52).
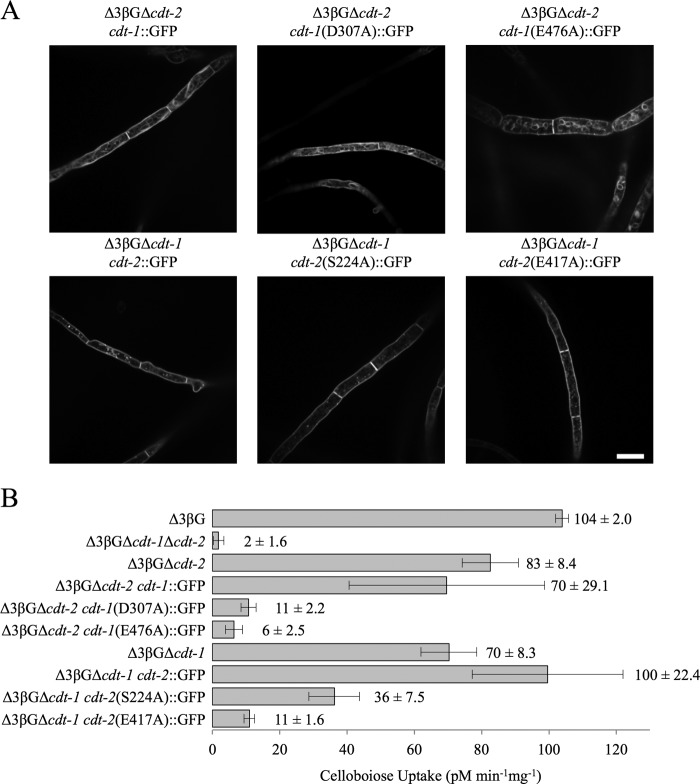
The fact that deletion of cdt-1 and cdt-2 eliminated nearly all growth of N. crassa on cellobiose or Avicel (Fig. 3B) allowed us to probe the transport properties of these transporters in their native host. The wild type cellodextrin transporters and point mutants were GFP-tagged and individually transformed into N. crassa, replacing the wild type copy of cdt-1 or cdt-2 at its resident locus with its GFP-tagged form (“Experimental Procedures”). Only those strains in which mutant CDT-1 or CDT-2 localized properly to the membrane (Fig. 5A) were further tested for cellobiose uptake (Fig. 5B) and cellulase induction (Fig. 6, A and B). Comparing cellobiose uptake in the Δ3βG background strain to that in the strain lacking both cdt-1 and cdt-2 confirmed that CDT-1 and CDT-2 are the major transporters responsible for cellobiose uptake in N. crassa (Fig. 5B). Strains carrying a deletion of each transporter individually showed a slight decrease in cellobiose uptake, but as shown earlier (Fig. 3) did not significantly affect the cellulase gene induction by cellobiose. GFP-tagged CDT-1 or CDT-2 transporters also did not significantly alter cellobiose uptake or the ability to induce cellulases in response to cellobiose or Avicel as compared with wild type (Fig. 5, A and B). Thus, the presence of either cdt-1 or cdt-2, with or without a GFP tag, was sufficient to allow induction of cellulose genes in response to either crystalline cellulose or physiologically relevant concentrations of cellobiose.
FIGURE 5.
Uptake of cellobiose by N. crassa strains expressing untagged and/or GFP-tagged cellodextrin transporters. A, fluorescence microscopy of WT and GFP-tagged point mutants after 24 h induction with 2% cellobiose. Scale bar: 10 μm. B, cellobiose uptake in the Δ3βG, Δ3βGΔcdt-1Δcdt-2, Δ3βGΔcdt-2, Δ3βGΔcdt-2 cdt-1::GFP, Δ3βGΔcdt-2 cdt-1(D307A)::GFP, Δ3βGΔcdt-2 cdt-1(E476A)::GFP, Δ3βGΔcdt-1, Δ3βGΔcdt-1 cdt-2::GFP, Δ3βGΔcdt-1 cdt-2(S224A)::GFP, and Δ3βGΔcdt-1 cdt-2(E417A)::GFP strains after 24 h induction with 2% cellobiose.
FIGURE 6.
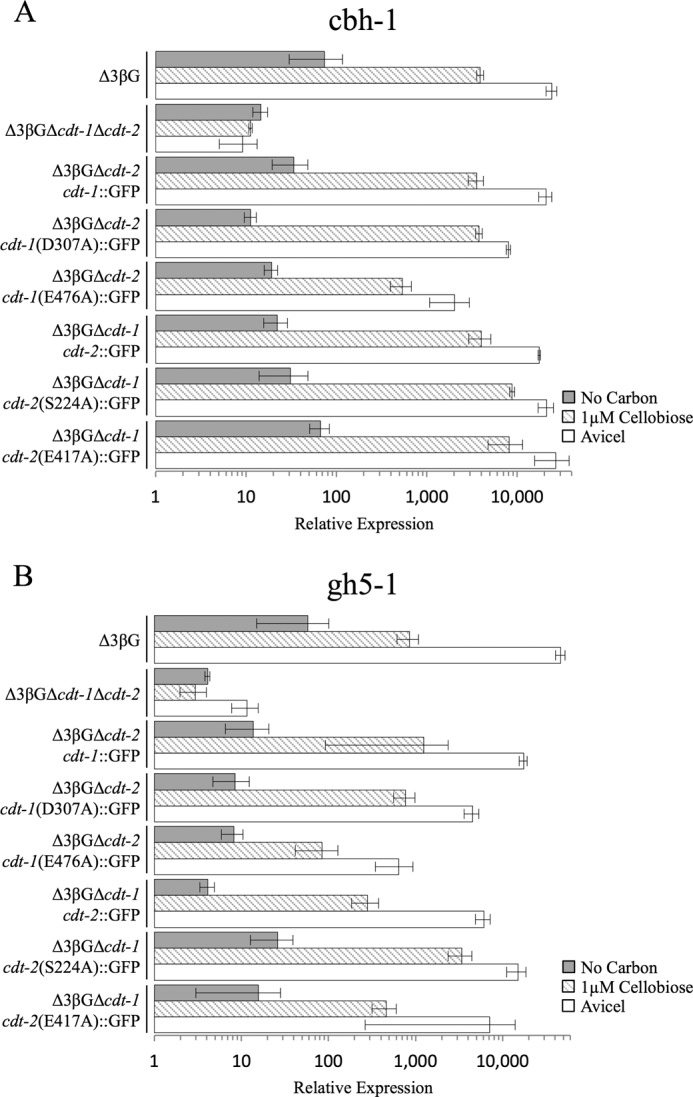
Cellulase gene expression levels in Δ3βG and Δ3βG-cellodextrin transporter mutant strains after induction with cellobiose or Avicel. Gene expression of the cellulases (A) cbh-1 or (B) gh5-1 after a 4-h induction with 1 μm cellobiose, 1% Avicel or no carbon (starvation) in the Δ3βG, Δ3βGΔcdt-1Δcdt-2, Δ3βGΔcdt-2, Δ3βGΔcdt-2 cdt-1::GFP, Δ3βGΔcdt-2 cdt-1(D307A)::GFP, Δ3βGΔcdt-2 cdt-1(E476A)::GFP, Δ3βGΔcdt-1, Δ3βGΔcdt-1 cdt-2::GFP, Δ3βGΔcdt-1 cdt-2(S224A)::GFP, and Δ3βGΔcdt-1 cdt-2(E417A)::GFP strains. Gene expression levels of cbh-1 and gh5-1 were normalized to 1 when induced with 1% sucrose. Expression levels of actin (NCU04173) were used as an endogenous control in all samples. Each strain was grown in triplicate, and error bars indicate 1 S.D.
As predicted by the S. cerevisiae growth assays (Fig. 4A), expression of three of the four defective transporters in N. crassa resulted in strains in which cellobiose uptake was almost completely abolished (Fig. 5B). We hypothesized that if N. crassa senses cellobiose intracellularly, abolishing transport should also abolish cellulase gene induction in a similar fashion to what was observed in the strain lacking both transporters (Δ3βGΔcdt-1Δcdt-2) (Fig. 3A). We therefore examined the ability of cellobiose or Avicel to induce cellulases in N. crassa strains expressing the defective transporters. To our surprise, three of the four point mutants still promoted induction of cbh-1 and gh5-1 in response to either cellobiose or Avicel at levels indistinguishable from wild type (Fig. 6). Particularly noteworthy, the Δ3βGΔcdt-1 cdt-2 (E417A)::GFP strain had a 9-fold reduced rate of cellobiose import (Fig. 5B), yet had an indistinguishable cellulase induction profile when compared with WT cdt-2 tagged with GFP (Fig. 6, strain Δ3βGΔcdt-1 cdt-2::GFP). The fourth point mutant, Δ3βGΔcdt-1 cdt-2 (E476A)::GFP showed a decrease in induction of cbh-1 and gh5-1 in response to either cellobiose or Avicel as compared with wild-type with GFP-tagged cdt-2, although the induction of cbh-1 and gh5-1was still far above that observed for starvation/derepression, as shown by expression levels in the Δ3βGΔcdt-1 Δcdt-2 mutant (Fig. 6).
DISCUSSION
In this study, we identified two cellodextrin transporters in the model cellulolytic fungus N. crassa (cdt-1 and cdt-2) that are essential for the induction of cellulase gene expression in response to cellulose. Furthermore, in a strain lacking three β-glucosidase genes, one or both transporters are required for sensing the cellulose-derived product cellobiose. Deletion of either cdt-1 or cdt-2 does not affect the ability of cellulose or cellobiose to induce cellulase gene expression or activity, indicating that both transporters contribute to cellobiose sensing. However, based on the growth defect for a strain lacking cdt-2 (22) (Fig. 1B), we conclude that cdt-2 is crucial for sustained growth on cellulose.
Recently, two studies in T. reesei identified transporters involved in lactose uptake and strains lacking these lactose transporters no longer induce cellulases in response to lactose. However, because these strains are still able to induce cellulases in response to sophorose (10) or crystalline cellulose (11), these results imply that cellulase signaling via lactose either occurs through an alternative transporter or an alternative mechanism unrelated to the one used during growth on plant cell wall substrates. In a recent study using sophorose, cellobiose or crystalline cellulose as inducers, T. reesei was shown to use a “transceptor” Stp1, a homologue of the receptors Rgt2 and Snf3 in S. cerevisiae, to regulate the carbon catabolite repression (12). Nutrient transceptors, transporter-like proteins with a receptor function, have been identified and characterized in several systems including E. coli (glucose-6-phosphate sensing (35)), S. cerevisiae (nitrogen (36), phosphate (37), and glucose sensing (38)), Drosophila (amino acid sensing (39)), Arabidopsis (nitrate sensing (40)), as well as in the fungus Ustilago maydis (41) and Candida albicans (42) for the control of filamentation. Notably, in T. reesei, the transceptor Stp1 seems to act independently of the sophorose induction mechanism. By contrast, the T. reesei protein Crt1 (Trire2:3405) may act directly as a receptor for cellobiose and sophorose, as its deletion does not impact cellobiose or sophorose import, but abolishes cellulase induction (12). While the studies performed in T. reesei utilize single transporter deletion strains to examine transporters involved in cellulase signaling, the data presented in this study utilizes strains containing several deletions as well as site directed mutants to determine how transport of cellobiose versus signaling though transceptors mediates the induction of cellulases.
In contrast to T. reesei, N. crassa uses an alternative mechanism for sensing cellulose. On both crystalline cellulose and cellobiose (the major soluble end product of cellulases) the two transporters CDT-1 and CDT-2 are solely responsible for the ability to of N. crassa to sense and grow on cellulose in plant cell walls. Both CDT-1 and CDT-2 seem to act as “transceptors” with the ability for robust cellodextrin transport (21) (Fig. 5B) and cellulase gene induction by cellobiose (Fig. 6). While further characterization must be completed to map the structural determinants in CDT-1 and CDT-2 to fully separate nutrient sensing from transport, our studies looking at two point mutations in each transporter have helped to begin to separate transport from signaling and lead to the model presented in Fig. 7. Based on our homology models to the xylose transporter XylE (Fig. 4C) homologous point mutants in CDT-1 (E476A) and CDT-2 (E417A) fall within an intracellular unstructured loop region and may block the import of cellobiose by preventing a required conformational change in the protein. The other point mutants in CDT-1 (D307A) and CDT-2 (S224A) fall within an intracellular α-helix and an intracellular transition between an unstructured loop and α-helix, respectively. Their role in blocking the import of cellobiose is less easy to predict due to their further distance from the transmembrane region.
FIGURE 7.
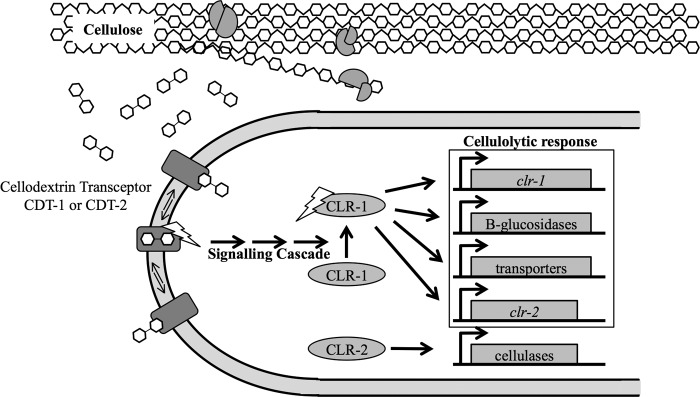
Transporting transceptor model for cellulase induction in N. crassa. Cellobiose is released by cellulases and is recognized by N. crassa through either CDT-1 or CDT-2.
The data presented in this study significantly adds to the model for the induction of cellulases in filamentous fungi, however several questions remain. Although the mutations identified in this study begin to define regions of CDT-1 and CDT-2 required for transport versus nutrient signaling, support for a role for CDT-1 and CDT-2 as transceptors will additionally require the identification of the mechanism of intracellular signaling that ultimately results in the activation of the transcription factor CLR-1, the major transcriptional regulator required for induction of cellulase genes, and CLR-2, which also regulates cellulase gene expression in N. crassa (Fig. 7) (43, 44). Given the widespread presence of cellodextrin transporters in diverse fungi (22), such signaling networks may also function in a wide range of filamentous fungi of ecological or industrial interest for biofuels applications, such as the related thermotolerant fungus Myceliophthora thermophila (45, 46).
This work was supported in part by National Institutes of Health Grant P01 GM068087. This work was also supported by a grant from the Energy Biosciences Institute (to N. L. G. and J. H. D. C.).
- MFS
- major facilitator superfamily
- CDT
- cellodextrin transporter
- hph
- hygromycin phosphotransferase.
REFERENCES
- 1. Wyman C. E. (1999) BIOMASS ETHANOL: Technical Progress, Opportunities, and Commercial Challenges. Annu. Rev. Energy Environ. 24, 189–226 [Google Scholar]
- 2. Rubin E. M. (2008) Genomics of cellulosic biofuels. Nature 454, 841–845 [DOI] [PubMed] [Google Scholar]
- 3. Carroll A., Somerville C. (2009) Cellulosic biofuels. Annu. Rev. Plant Biol. 60, 165–182 [DOI] [PubMed] [Google Scholar]
- 4. Somerville C. (2007) Biofuels. Curr. Biol. 17, R115–R119 [DOI] [PubMed] [Google Scholar]
- 5. Himmel M. E., Bayer E. A. (2009) Lignocellulose conversion to biofuels: current challenges, global perspectives. Curr. Opin. Biotechnol. 20, 316–317 [DOI] [PubMed] [Google Scholar]
- 6. Himmel M. E., Ding S. Y., Johnson D. K., Adney W. S., Nimlos M. R., Brady J. W., Foust T. D. (2007) Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315, 804–807 [DOI] [PubMed] [Google Scholar]
- 7. Cherry J. R., Fidantsef A. L. (2003) Directed evolution of industrial enzymes: an update. Curr. Opin. Biotechnol. 14, 438–443 [DOI] [PubMed] [Google Scholar]
- 8. Glass N. L., Schmoll M., Cate J. H., Coradetti S. (2013) Plant cell wall deconstruction by ascomycete fungi. Annu. Rev. Microbiol. 67, 477–498 [DOI] [PubMed] [Google Scholar]
- 9. Kubicek C. P., Messner R., Gruber F., Mandels M., Kubicek-Pranz E. M. (1993) Triggering of cellulase biosynthesis by cellulose in Trichoderma reesei. Involvement of a constitutive, sophorose-inducible, glucose- inhibited β-diglucoside permease. J. Biol. Chem. 268, 19364–19368 [PubMed] [Google Scholar]
- 10. Ivanova C., Bååth J. A., Seiboth B., Kubicek C. P. (2013) Systems Analysis of Lactose Metabolism in Trichoderma reesei Identifies a Lactose Permease That Is Essential for Cellulase Induction. PLoS One 8, e62631 10.1371/journal.pone.0062631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Porciuncula Jde O., Furukawa T., Shida Y., Mori K., Kuhara S., Morikawa Y., Ogasawara W. (2013) Identification of Major Facilitator Transporters Involved in Cellulase Production during Lactose Culture of Trichoderma reesei PC-3–7. Biosci. Biotechnol. Biochem. 77, 1014–1022 [DOI] [PubMed] [Google Scholar]
- 12. Zhang W., Kou Y., Xu J., Cao Y., Zhao G., Shao J., Wang H., Wang Z., Bao X., Chen G., Liu W. (2013) Two major facilitator superfamily sugar transporters from Trichoderma reesei and their roles in induction of cellulase biosynthesis. J. Biol. Chem. 10.1074/jbc.M113.505826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thevelein J. M., Voordeckers K. (2009) Functioning and evolutionary significance of nutrient transceptors. Mol. Biol. Evol. 26, 2407–2414 [DOI] [PubMed] [Google Scholar]
- 14. Kriel J., Haesendonckx S., Rubio-Texeira M., Van Zeebroeck G., Thevelein J. M. (2011) From transporter to transceptor: signaling from transporters provokes re-evaluation of complex trafficking and regulatory controls: endocytic internalization and intracellular trafficking of nutrient transceptors may, at least in part, be governed by their signaling function. Bioessays 33, 870–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Davis R. H., Perkins D. D. (2002) Neurospora: a model of model microbes. Nat. Rev. Genet. 3, 397–403 [DOI] [PubMed] [Google Scholar]
- 16. Jacobson D. J., Powell A. J., Dettman J. R., Saenz G. S., Barton M. M., Hiltz M. D., Dvorachek W. H., Jr., Glass N. L., Taylor J. W., Natvig D. O. (2004) Neurospora in temperate forests of western North America. Mycologia 96, 66–74 [PubMed] [Google Scholar]
- 17. Jacobson D. J., Dettman J. R., Adams R. I., Boesl C., Sultana S., Roenneberg T., Merrow M., Duarte M., Marques I., Ushakova A., Carneiro P., Videira A., Navarro-Sampedro L., Olmedo M., Corrochano L. M., Taylor J. W. (2006) New findings of Neurospora in Europe and comparisons of diversity in temperate climates on continental scales. Mycologia 98, 550–559 [DOI] [PubMed] [Google Scholar]
- 18. Perkins D. D., Turner B. C., Barry E. G. (1976) Strains of Neurospora collected from nature. Evolution 30, 281–313 [DOI] [PubMed] [Google Scholar]
- 19. Turner B. C., Perkins D. D., Fairfield A. (2001) Neurospora from natural populations: A global study. Fungal Genet. Biol. 32, 67–92 [DOI] [PubMed] [Google Scholar]
- 20. Znameroski E. A., Glass N. L. (2013) Using a model filamentous fungus to unravel mechanisms of lignocellulose deconstruction. Biotechnol. Biofuels 6, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tian C., Beeson W. T., Iavarone A. T., Sun J., Marletta M. A., Cate J. H., Glass N. L. (2009) Systems analysis of plant cell wall degradation by the model filamentous fungus Neurospora crassa. Proc. Natl. Acad. Sci. U.S.A. 106, 22157–22162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Galazka J. M., Tian C., Beeson W. T., Martinez B., Glass N. L., Cate J. H. (2010) Cellodextrin transport in yeast for improved biofuel production. Science 330, 84–86 [DOI] [PubMed] [Google Scholar]
- 23. Znameroski E. A., Coradetti S. T., Roche C. M., Tsai J. C., Iavarone A. T., Cate J. H., Glass N. L. (2012) Induction of lignocellulose-degrading enzymes in Neurospora crassa by cellodextrins. Proc. Natl. Acad. Sci. U.S.A. 109, 6012–6017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McCluskey K. (2003) The Fungal Genetics Stock Center: from molds to molecules. Adv. Appl. Microbiol. 52, 245–262 [DOI] [PubMed] [Google Scholar]
- 25. Westergaard M., Mitchell H. K. (1947) Neurospora V. A synthetic medium favoring sexual reproduction. Am. J. Bot. 34, 573–577 [Google Scholar]
- 26. Vogel H. J. (1956) A convenient growth medium for Neurospora (medium N). Microb. Genet. Bullet. 13, 42–43 [Google Scholar]
- 27. Dementhon K., Iyer G., Glass N. L. (2006) VIB-1 is required for expression of genes necessary for programmed cell death in Neurospora crassa. Eukaryot Cell 5, 2161–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Honda S., Selker E. U. (2009) Tools for fungal proteomics: multifunctional neurospora vectors for gene replacement, protein expression and protein purification. Genetics 182, 11–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ninomiya Y., Suzuki K., Ishii C., Inoue H. (2004) Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. U.S.A. 101, 12248–12253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Colot H. V., Park G., Turner G. E., Ringelberg C., Crew C. M., Litvinkova L., Weiss R. L., Borkovich K. A., Dunlap J. C. (2006) A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. U.S.A. 103, 10352–10357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Margolin B. S., Freitag M., Selker E. U. (1997) Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet. Newslett. 44, 34–36 [Google Scholar]
- 32. Wu W., Kasuga T., Nguyen L., Fan Z. (2013) The effects of each β-glucosidase gene deletion on cellulase gene regulation in Neurospora crassa. A Van Leeuw J. Microb. published online, 10.1007/s10482-013-9972-7 [DOI] [Google Scholar]
- 33. Kim H., Lee W. H., Galazka J. M., Cate J. H., Jin Y. S. (2013) Analysis of cellodextrin transporters from Neurospora crassa in Saccharomyces cerevisiae for cellobiose fermentation. Appl. Microbiol. Biotechnol. 10.1007/s00253-013-5339-2 [DOI] [PubMed] [Google Scholar]
- 34. Galazka J. M. (2011) Cellodextrin Transporters of Neurospora crassa and their Utility in Saccharomyces cerevisiae During a Biofuel Production Process. Ph.D., University of California [Google Scholar]
- 35. Schwöppe C., Winkler H. H., Neuhaus H. E. (2003) Connection of transport and sensing by UhpC, the sensor for external glucose-6-phosphate in Escherichia coli. Eur. J. Biochem. 270, 1450–1457 [DOI] [PubMed] [Google Scholar]
- 36. Van Nuland A., Vandormael P., Donaton M., Alenquer M., Lourenço A., Quintino E., Versele M., Thevelein J. M. (2006) Ammonium permease-based sensing mechanism for rapid ammonium activation of the protein kinase A pathway in yeast. Mol. Microbiol. 59, 1485–1505 [DOI] [PubMed] [Google Scholar]
- 37. Giots F., Donaton M. C., Thevelein J. M. (2003) Inorganic phosphate is sensed by specific phosphate carriers and acts in concert with glucose as a nutrient signal for activation of the protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 47, 1163–1181 [DOI] [PubMed] [Google Scholar]
- 38. Kraakman L., Lemaire K., Ma P., Teunissen A. W., Donaton M. C., Van Dijck P., Winderickx J., de Winde J. H., Thevelein J. M. (1999) A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol. Microbiol. 32, 1002–1012 [DOI] [PubMed] [Google Scholar]
- 39. Goberdhan D. C., Meredith D., Boyd C. A., Wilson C. (2005) PAT-related amino acid transporters regulate growth via a novel mechanism that does not require bulk transport of amino acids. Development 132, 2365–2375 [DOI] [PubMed] [Google Scholar]
- 40. Walch-Liu P., Forde B. G. (2008) Nitrate signalling mediated by the NRT1.1 nitrate transporter antagonises L-glutamate-induced changes in root architecture. Plant J. 54, 820–828 [DOI] [PubMed] [Google Scholar]
- 41. Smith D. G., Garcia-Pedrajas M. D., Gold S. E., Perlin M. H. (2003) Isolation and characterization from pathogenic fungi of genes encoding ammonium permeases and their roles in dimorphism. Mol. Microbiol. 50, 259–275 [DOI] [PubMed] [Google Scholar]
- 42. Biswas K., Morschhäuser J. (2005) The Mep2p ammonium permease controls nitrogen starvation-induced filamentous growth in Candida albicans. Mol. Microbiol. 56, 649–669 [DOI] [PubMed] [Google Scholar]
- 43. Coradetti S. T., Craig J. P., Xiong Y., Shock T., Tian C., Glass N. L. (2012) Conserved and essential transcription factors for cellulase gene expression in ascomycete fungi. Proc. Natl. Acad. Sci. U.S.A. 109, 7397–7402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Coradetti S. T., Xiong Y., Glass N. L. (2013) Analysis of a conserved cellulase transcriptional regulator reveals inducer-independent production of cellulolytic enzymes in Neurospora crassa. Microbiologyopen 2, 595–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rosgaard L., Pedersen S., Cherry J. R., Harris P., Meyer A. S. (2006) Efficiency of new fungal cellulase systems in boosting enzymatic degradation of barley straw lignocellulose. Biotechnol. Prog. 22, 493–498 [DOI] [PubMed] [Google Scholar]
- 46. Berka R. M., Grigoriev I. V., Otillar R., Salamov A., Grimwood J., Reid I., Ishmael N., John T., Darmond C., Moisan M. C., Henrissat B., Coutinho P. M., Lombard V., Natvig D. O., Lindquist E., Schmutz J., Lucas S., Harris P., Powlowski J., Bellemare A., Taylor D., Butler G., de Vries R. P., Allijn I. E., van den Brink J., Ushinsky S., Storms R., Powell A. J., Paulsen I. T., Elbourne L. D. H., Baker S. E., Magnuson J., Laboissiere S., Clutterbuck A. J., Martinez D., Wogulis M., de Leon A. L., Rey M. W., Tsang A. (2011) Comparative genomic analysis of the thermophilic biomass-degrading fungi Myceliophthora thermophila and Thielavia terrestris. Nat. Biotechnol. 29, 922-U222 [DOI] [PubMed] [Google Scholar]
- 47. Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schneider T. D., Stephens R. M. (1990) Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Eswar N., Webb B., Marti-Renom M. A., Madhusudhan M. S., Eramian D., Shen M. Y., Pieper U., Sali A. (2007) Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. Chapter 2, Unit 2 9 [DOI] [PubMed] [Google Scholar]
- 50. Sun L., Zeng X., Yan C., Sun X., Gong X., Rao Y., Yan N. (2012) Crystal structure of a bacterial homologue of glucose transporters GLUT1–4. Nature 490, 361–366 [DOI] [PubMed] [Google Scholar]
- 51. Lee S. B., Milgroom M. G., Taylor J. W. (1988) A rapid, high yield mini-prep method for isolation of total genomic DNA from fungi. Fungal Genetic Newsletter 35, 23 [Google Scholar]
- 52. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]