Background: CS-56 immunoreactivity reveals a subpopulation of astrocytes in the adult mouse cerebral cortex.
Results: TNR mRNA was detected in CS-56-positive astrocytes, and TNR regulates astrocytic GLAST expression.
Conclusion: TNR-expressing cells are a subpopulation of astrocytes and regulate extracellular glutamate homeostasis.
Significance: Astrocytes in the adult mouse cerebral cortex exhibit a molecular and functional heterogeneity that could become future therapeutic targets in brain injury.
Keywords: Astrocytes, Chondroitin Sulfate, Extracellular Matrix, Glutamate, Tenascin
Abstract
In our previous study, the CS-56 antibody, which recognizes a chondroitin sulfate moiety, labeled a subset of adult brain astrocytes, yielding a patchy extracellular matrix pattern. To explore the molecular nature of CS-56-labeled glycoproteins, we purified glycoproteins of the adult mouse cerebral cortex using a combination of anion-exchange, charge-transfer, and size-exclusion chromatographies. One of the purified proteins was identified as tenascin-R (TNR) by mass spectrometric analysis. When we compared TNR mRNA expression patterns with the distribution patterns of CS-56-positive cells, TNR mRNA was detected in CS-56-positive astrocytes. To examine the functions of TNR in astrocytes, we first confirmed that cultured astrocytes also expressed TNR protein. TNR knockdown by siRNA expression significantly reduced glutamate uptake in cultured astrocytes. Furthermore, expression of mRNA and protein of excitatory amino acid transporter 1 (GLAST), which is a major component of astrocytic glutamate transporters, was reduced by TNR knockdown. Our results suggest that TNR is expressed in a subset of astrocytes and contributes to glutamate homeostasis by regulating astrocytic GLAST expression.
Introduction
Chondroitin sulfate proteoglycans (CSPGs)2 are major components of the extracellular matrix in the brain and play significant roles in cell migration, neurite elongation, pathfinding, and synaptogenesis in the developing brain as well as regeneration and synaptic plasticity in the adult brain (1–4). In the adult brain, CSPGs exhibit a unique distribution pattern, termed perineuronal nets (PNNs). PNNs surround cell bodies and proximal dendrites of GABAergic interneurons and consist of several kinds of CSPGs as well as hyaluronan and cell adhesion molecules (5–9). The appearance of PNNs may terminate the critical period for neuronal plasticity, and the enzymatic digestion of CSPGs by chondroitinase ABC restores neuronal plasticity (10–14).
In previous studies, we examined the distribution of CSPGs in the adult brain using CS-56, an anti-chondroitin sulfate antibody, and found a unique matrix structure distinct from PNNs in the cerebral cortex. Because the patchy morphology of the CS-56-labeled matrix resembled a dandelion clock (the seed head of Taraxacum officinale), we named it the dandelion clock-like structure (DACS) (15). The DACS is conserved in brains of various species, including humans, primates, and domestic pigs but not in mallards (16). CS-56-positive patches surrounded groups of NeuN-positive/GABA-negative neurons (15). At the ultrastructural level, CS-56 immunoreactivity was localized in the cytoplasm and on the plasma membrane of astrocytes, most of which were GFAP-negative.
In this study, we sought to identify core proteins in CS-56-positive CSPGs. We first purified glycoproteins from the adult mouse cerebral cortex by anion-exchange, charge-transfer, and size-exclusion chromatographies. Mass spectrometric analyses revealed several candidates, including lecticans and tenascin-R (TNR). TNR is an extracellular matrix protein with chondroitin sulfate moieties that is expressed primarily in the central nervous system, which prompted us to examine whether it is expressed in the CS-56-positive astrocytes. Double-labeling experiments revealed TNR mRNA expression in those astrocytes. Furthermore, knockdown of TNR gene expression decreased glutamate uptake activity and expression of the excitatory amino acid transporter 1 (GLAST) in cultured astrocytes. Based on the present findings, the functional implications of TNR in neuro-glial interactions are discussed.
EXPERIMENTAL PROCEDURES
Experimental Animals
Adult male C56BL/6 mice (12 weeks old) were purchased from Charles River Laboratory (Yokohama, Japan) and housed in plastic breeding cages under standard laboratory conditions (23 ± 1 °C, 55 ± 5% humidity in a room with a 12-h light-dark cycle) and had access to tap water and food ad libitum. The Animal Care Committee of Nara Medical University approved the protocols for this study in accordance with the policies established in the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Immunohistochemistry
Immunohistochemistry was performed as described previously (17). Briefly, mice were perfused with phosphate-buffered saline (PBS) (pH 7.4) followed by 4% paraformaldehyde in 0.1 m phosphate buffer (pH 7.4). Brains were dissected out and postfixed in the same fixative at 4 °C for 6 h. Coronal sections (70 μm) were cut on a vibratome (Liner slicer Pro. 7, DSK, Kyoto, Japan). The primary antibodies used in this study were as follows: anti-chondroitin sulfate monoclonal antibody (CS-56, 1:300, Millipore, Bedford, MA); anti-S100β protein (S100β, 1:2000, Yanaihara Institute, Shizuoka, Japan); anti-glutamine synthetase (GS, 1:3000, Sigma); anti-GLAST (1:500, Cell Signaling Technology, Beverly, MA); anti-π isoform of glutathione S-transferase (GSTπ, 1:500, Medical and Biological Laboratories, Nagoya, Japan); anti-myelin basic protein (1:500, DAKO, Carpinteria, CA), and anti-NG2 chondroitin sulfate proteoglycan (NG2, 1:200, Millipore). Secondary antibodies were as follows: biotinylated anti-mouse IgM (1:200, Vector Laboratories, Burlingame, CA), Alexa Fluor 546-labeled anti-rabbit IgG and Alexa Fluor 488 streptavidin (1:1000, Invitrogen). For control experiments, normal mouse IgM (Millipore), normal mouse IgG (Millipore), or normal rabbit IgG (Millipore) was used instead of the primary antibodies. Immunofluorescence images were captured using an Olympus FluoView 1000 confocal laser scanning microscope.
Immunoelectron Microscopy
Under deep anesthesia with pentobarbital, mice were perfused with PBS (pH 7.4) followed by 4% paraformaldehyde, 1% glutaraldehyde in 0.1 m phosphate buffer (pH 7.4). After post-fixation in the same fixative at 4 °C for 2 h, coronal sections were prepared using a vibratome. The sections were treated with 25 mm glycine in PBS, blocked with PBS, and immersed in 0.3% Triton X-100 in PBS (PBST) containing 5% normal goat serum at 4 °C overnight. The CS-56 antibody (1:500) was incubated with the sections at 4 °C for 2 days, followed by biotinylated anti-mouse IgM antibody for 2 h at room temperature. For enzyme-labeling procedures, these sections were then incubated with VECTASTAIN Elite ABC reagent (Vector Laboratories) for 1 h at room temperature. After fixation with 1% glutaraldehyde at 4 °C for 10 min, the sections were washed with 50 mm Tris-HCl (pH 7.4) containing 0.15 m NaCl and then incubated with 3,3′-diaminobenzidine tetrachloride and 0.05% H2O2. The sections were fixed with 2% OsO4, and the sections were dehydrated in propylene oxide after going through the alcohol series and embedded in epoxy resin (Oken, Tokyo, Japan). Ultrathin sections were cut using an ultramicrotome (Leica Microsystems, Wetzlar, Germany), mounted on nickel mesh, stained with uranyl acetate, and observed using a JEM 1200EX electron microscope (JEOL, Tokyo, Japan).
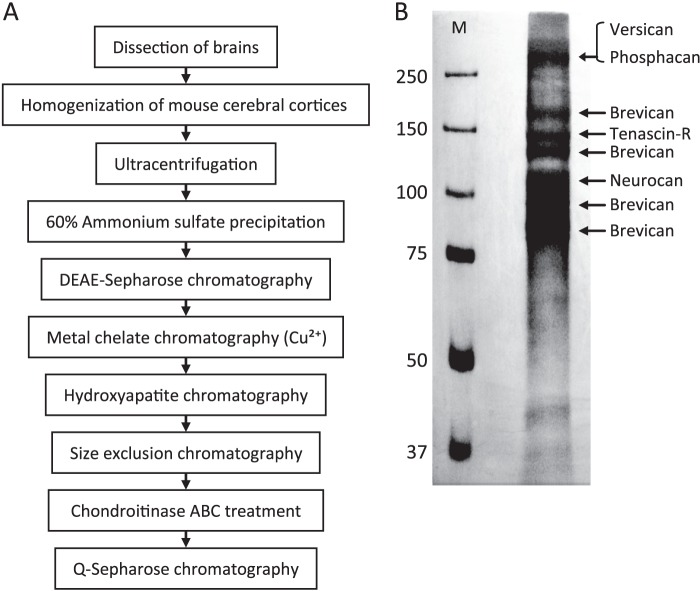
Protein Purification
The purification protocol is shown in Fig. 3A. Cerebral cortices from 50 mice (total weight, 4.4 g) were homogenized in 200 ml of homogenization buffer (50 mm Tris-HCl (pH 7.5), 8 m urea, 0.5% polyethylene glycol p-(1,1,3,3-tetramethylbutyl)-phenyl ether (Triton X-100), 0.1 m NaCl, 1 mm dithiothreitol (DTT), protease inhibitor mixture (Nacalai Tesque, Kyoto, Japan)) using a glass Teflon homogenizer. The homogenates were centrifuged at 100,000 × g for 1 h, and the supernatants were then used as starting material (designated the S100 fraction in Table 1) and precipitated by stepwise treatment with ammonium sulfate. The fraction precipitated at 60% ammonium sulfate saturation was dissolved in 100 ml of homogenization buffer and loaded onto a DEAE-Sepharose FF column (GE Healthcare) and then washed with homogenization buffer containing 0.2 m NaCl. Bound glycoproteins were eluted with the same buffer containing 0.4 m NaCl, and CSPG-rich fractions were then loaded onto a Cu2+-chelating Sepharose FF column (GE Healthcare) and washed with washing buffer (20 mm phosphate buffer (pH 7.5), 0.5 m NaCl, 0.5% Nonidet P-40). The bound proteins were eluted with washing buffer containing 50 mm imidazole, and fractions were subjected to chromatography on hydroxyapatite (Nihon Chemical, Tokyo, Japan). After being washed with washing buffer (10 mm Tris-HCl (pH 7.5), 150 mm NaCl, 0.2% Nonidet P-40), proteins were eluted with washing buffer containing 600 mm K2HPO4. Eluted fractions were loaded at a flow rate of 0.7 ml/min on a Superdex 200 10/300 GL column (GE Healthcare) equilibrated with 10 mm Tris-HCl (pH 8.0), 150 mm NaCl, and 0.02% Nonidet P-40. The CSPG-rich fractions were treated with chondroitinase ABC (Seikagaku Kogyo, Tokyo, Japan) and glycopeptidase F (TaKaRa Bio, Otsu, Japan) and then loaded onto a Q-Sepharose FF column (GE Healthcare) to concentrate fractions and remove chondroitinase ABC and glycopeptidase F. After being subjected to washing buffer (10 mm Tris-HCl (pH 8.0), 150 mm NaCl, 0.05% Nonidet P-40), the bound CSPGs were eluted with washing buffer containing 0.8 m NaCl. To reduce salt concentration, eluted fractions were diluted 3-fold with 10 mm Tris-HCl (pH 8.0), separated on 7.5% polyacrylamide gels, and stained using a two-dimensional silver stain II kit (Cosmo Bio, Tokyo, Japan). Protein concentrations were measured using the two-dimensional Quant kit (GE Healthcare) or CBQCA protein quantitation kit (Invitrogen). CSPG concentrations were measured by dot blot assay using CS-56 antibody. We set the calibration curve by plotting standard CSPG solutions (1–1000 μg/ml). Quantification was done by densitometry of dot blot signals using ImageJ software.
FIGURE 3.
Purification and identification of mouse brain CSPGs. A, purification steps for glycoproteins from the adult mouse cerebral cortex are indicated. B, representative silver-stained polyacrylamide gel of fractionated purified glycoproteins. Lane M, protein size markers (kDa). The protein bands in the right lane were cut out, in-gel-digested, and subjected to MALDI-TOF mass spectrometry. Mass fingerprints for phosphacan, versican, brevican, neurocan, and tenascin-R were obtained, and protein-specific peptides were identified using Mascot software. Band identities are indicated on the right side.
TABLE 1.
Purification of CSPGs from the adult mouse cerebral cortex
| Purification step | Total protein | Yield | CSPG protein | Yield | Ratio (CSPG/total protein) | Purification |
|---|---|---|---|---|---|---|
| mg | % | μg | % | -fold | ||
| S100 fraction | 627.62 | 100.000 | 16300 | 100.00 | 2.60 | 1.00 |
| Ammonium sulfate precipitation | 502.96 | 80.137 | 14899 | 91.40 | 2.96 | 1.14 |
| DEAE-Sepharose | 10.75 | 1.712 | 6250 | 38.34 | 58.15 | 22.39 |
| Metal chelate (Cu2+) | 4.57 | 0.728 | 57a | 0.35a | 1.25 | 0.48 |
| Hydroxyapatite | 0.31 | 0.050 | 157 | 0.96 | 50.60 | 19.48 |
| Size exclusion | 0.05 | 0.009 | 44 | 0.27 | 81.82 | 31.50 |
| Q-Sepharose | 0.02 | 0.004 | NAb | NAb | NAb | NAb |
a We found that these values were much lower than expected for unknown reason(s). A possible cause is that the buffer components such as 0.5% Nonidet P-40 might have prevented protein adsorption to dot blot membranes.
b Chondroitin sulfate chains were removed by chondroitinase ABC and therefore could not be detected by CS-56 antibody. NA means not applicable.
In-gel Digestion and Mass Spectrometry
Gel slices were dehydrated in 300 μl of CH3CN for 10 min and then incubated in 50 μl of reduction buffer (10 mm DTT and 100 mm NH4HCO3) at 56 °C for 30 min. After supernatant removal and dehydration in 300 μl of CH3CN for 10 min, the gel pieces were incubated in 50 μl of 50 mm iodoacetamide in 100 mm NH4HCO3 for 20 min at room temperature. After supernatant removal and dehydration with CH3CN, the dried gel pieces were rehydrated on ice in 50 μl of digestion buffer (50 mm NH4HCO3, 12.5 ng/μl each of lysylendopeptidase (Wako Chemical, Osaka, Japan) and sequencing grade trypsin (Promega, Madison, WI)) for 45 min. The supernatant was replaced with 50 mm NH4HCO3, and the gel pieces were incubated at 37 °C overnight; the supernatant was then collected, and peptides were extracted with 50 μl of extraction buffer (5% (v/v) formic acid, 50% (v/v) CH3CN). The combined supernatants were evaporated in a vacuum centrifuge, and the resulting peptides were dissolved in 0.1% trifluoroacetic acid and adsorbed onto a ZipTip C18 (Millipore). Bound peptides were eluted with 50% CH3CN and 0.1% trifluoroacetic acid. Equal amounts of the resulting peptide solution and a matrix-assisted laser desorption/ionization (MALDI) sample matrix solution (10 mg/ml 2,5-dihydroxybenzoic acid (Wako Chemical) dissolved in 50% CH3CN and 0.1% trifluoroacetic acid) were mixed and placed on the sample target. MALDI time-of-flight (TOF) measurements were carried using a Voyager DE Pro MALDI-TOF mass spectrometer equipped with a nitrogen-pulsed laser (337 nm; Applied Biosystems by Invitrogen). Peptide mass fingerprinting was carried out with a Mascot search for the peptide masses obtained by the mass spectrometer.
In Situ Hybridization
In situ hybridization was performed as described previously (17). Partial cDNAs for mouse TNR, phosphacan, brevican, neurocan, and versican were amplified by reverse transcriptase-PCR using the following primers: partial TNR forward primer, 5′-AAGGGCTACAGTTCAAGACG-3′ (complement of nucleotides (nt) 1256–1275); partial TNR reverse primer, 5′-GCTGTTATCCCATTCCAGGT-3′ (reverse complement of nt 1888–1907); partial phosphacan forward primer, 5′-AATAGCCCAAAGCAGTCTCC-3′ (complement of nt 173–192); partial phosphacan reverse primer, 5′-CTTTCAGAGATGCTAACTGTATCC-3′ (reverse complement of nt 750–773); partial brevican forward primer, 5′-AGGAACTGCAGCTGCCTCAG-3′ (complement of nt 1148–1167); partial brevican reverse primer, 5′-GGTATCGATCGTTGACAAAGTC-3′ (reverse complement of nt 2125–2146); partial neurocan forward primer, 5′-AACTCTACGATGTCTACTGC-3′ (complement of nt 737–756); partial neurocan reverse primer, 5′-TGGAGTAGAACCAAGGGTCT-3′ (reverse complement of nt 1277–1296); partial versican forward primer, 5′-GGAATCACCCACAACTACTCG-3′ (complement of nt 1275–1295); partial versican reverse primer, 5′-CCAAACTCATCTTCAGTCACC-3′ (reverse complement of nt 1893–1913).
In Situ Hybridization Combined with Immunohistochemistry
Sections were processed first for immunohistochemistry and then for in situ hybridization. The CS-56 antibody and biotinylated Wisteria floribunda lectin (WFA, Vector Laboratories; a GABAergic interneuron marker) were detected using a new fuchsin substrate (Nichirei, Tokyo, Japan) based on an alkaline phosphatase-conjugated ABC system (Vector Laboratories). For double fluorescence labeling, the sections were processed first for in situ hybridization and then for immunohistochemistry. Digoxigenin-labeled probes were detected using a 2-hydroxy-3-naphthoic acid-2′-phenylanilide phosphate fluorescent detection set (Roche Diagnostics).
Quantification of TNR mRNA/S100β-double Positive Cells in the Cerebral Cortex
To semi-quantitate the TNR mRNA-positive population in S100β-positive astrocytes in the cerebral cortex, 16-μm sagittal sections (eight sections per mouse) were cut on a cryostat and subjected to sequential TNR in situ hybridization and S100β immunohistochemistry (see above). S100β-positive cells and TNR mRNA/S100β double-positive cells in the cerebral cortex were counted under a fluorescence microscope. The ratios (percentage) of double-positive cells to total S100β-positive astrocytes were calculated in each section and averaged. Standard error was calculated from triplicate counting of three mice.
Cell Cultures
Mouse astrocytes were prepared from a mouse cerebral cortex (C57B6/6J, Charles River Laboratory) by a modification of a previously described method (18). Briefly, the cerebral cortex from a mouse (postnatal day 1) was dissociated using a stainless steel mesh in Dulbecco's modified Eagle's medium (DMEM) (Wako Chemical) with 0.05% deoxyribonuclease I (Roche Diagnostics) at 4 °C. Dissociated cells were sieved through a 70-μm pore-size nylon mesh (BD Biosciences) and seeded in poly-l-lysine (Sigma)-coated culture dishes at a density of 1 × 107 cells/dish in DMEM containing 10% heat-inactivated fetal bovine serum at 37 °C in an atmosphere of 95% air, 5% CO2. After 2 weeks in culture, the cells were passaged with 0.05% trypsin in PBS and plated on poly-l-lysine-coated culture dishes.
Knockdown Experiment Using Small Interference RNA (siRNA)
The following siRNAs against various mouse genes were obtained as MISSION predesigned siRNAs (Sigma): TNR siRNA-1 (SASI_Mm01_00073136) and siRNA-2 (SASI_Mm02_00073137); brevican siRNA-1 (SASI_Mm02_00304314) and siRNA-2 (SASI_Mm02_00311845); neurocan siRNA-1 (SASI_Mm02_00312468) and siRNA-2 (SASI_Mm02_00312469); and versican siRNA-1 (SASI_Mm02_00296627) and siRNA-2 (SASI_Mm02_00296628). MISSION siRNA universal negative control (SIC001; Scrambled siRNA, Sigma) was used as a nontargeting control for the siRNA experiments. Cultured mouse astrocytes were transfected with 150 pmol of each siRNA or scrambled siRNA using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions.
Immunocytochemistry for Cultured Cells
Immunocytochemistry for cultured cells was performed as described previously (17). The primary antibodies used in this study were as follows: anti-TNR (dilution 1:1000, BD Transduction Laboratories), anti-glial fibrillary acidic protein (GFAP, 1:10000, DAKO), anti-S100β (1:2000), and CS-56 (1:300), and the secondary antibodies were biotinylated anti-mouse IgG (1:200), Alexa Fluor 546-labeled anti-rabbit IgG, and Alexa Fluor 488-labeled anti-mouse IgG (1:1000, Invitrogen). TNR was detected using a TSA-Plus fluorescein system (PerkinElmer Life Sciences). Fluorescence images were captured using an Olympus FluoView 1000 confocal laser scanning microscope.
RNA Extraction
Total RNA was prepared from mouse astrocytes (17) using an RNeasy mini kit (Qiagen, Valencia, CA), according to the manufacturer's instructions.
Real Time Reverse Transcriptase-Polymerase Chain Reaction
The total RNA extracts were reverse-transcribed using random primers and a QuantiTect Reverse Transcription kit (Qiagen), according to the manufacturer's instructions. Real time PCR was performed using a LightCycler Quick System 350S (Roche Diagnostics), with SYBR Green real time PCR master mix plus (Toyobo, Osaka, Japan) and the following specific primers: mouse TNR, 5′-CAACTACCAAGACTACCCTCAC-3′ (forward) and 5′-GTCAAGTTCAGTCCTGGCATTC-3′ (reverse); mouse GFAP, 5′-CAAACTGGCTGATGTCTACC-3′ (forward) and 5′-GGTTGGTTTCATCTTGGAGC-3′ (reverse); mouse GS, 5′-TGAGCCCAAGTGTGTGGAAG-3′ (forward) and 5′-TCTCGAAACATGGCAACAGG-3′ (reverse); mouse GLAST, 5′-ATCGTGCAGGTCACTGCAGC-3′ (forward) and 5′-GAAGTGTTTCGTTGGACTGG-3′ (reverse); mouse GLT-1, 5′-ATGACAGCCACCTCAGCTCC-3′ (forward) and 5′-CACTGCTCCCAGGATGACAC-3′ (reverse); mouse brevican, 5′-GTGGGTCCTGATGATCTCTA-3′ (forward) and 5′-CACGTTCCAGACAGTAGTCC-3′ (reverse); mouse neurocan, 5′-GTCTGCAAGAAGGGTACAGT-3′ (forward) and 5′-AGAATCCTTCATCACACTGG-3′ (reverse); and mouse versican, 5′-ACACCAGTCATTCCATTAGC-3′ (forward) and 5′-CTAATCTGCCTGTGATAGCC-3′ (reverse). W. floribunda I fluorescence from the double-stranded PCR products was measured according to the manufacturer's instructions (Roche Diagnostics).
Western Blot Analysis
Western blot analysis was performed as described previously (17). We used anti-GLAST (1:5000, Cell Signaling Technology), anti-GLT-1 (1:5000, Cell Signaling Technology), and anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH, 1:5000, mouse monoclonal, Millipore) antibodies. Immunodetection was performed using the ECL Prime or Select Western blotting detection System (GE Healthcare) with horseradish peroxidase-conjugated antibodies against mouse or rabbit IgG (1:10,000, Cell Signaling Technology) according to the manufacturer's instructions. Semi-quantification was done by densitometry of blot signals using ImageJ software.
Assay for Gliotransmitter Release
Astrocytes were grown on poly-l-lysine-coated 24-well chamber dishes. Cells were washed with Krebs-Ringer solution (KRS, 125 mm NaCl, 2.5 mm KCl, 1.25 mm NaH2PO4, 2 mm CaCl2, 1 mm MgCl2, 25 mm NaHCO3, 25 mm glucose) and incubated for 10 min at 37 °C with one of the following reagents: 3′-O-(4-benzoyl)benzoyl adenosine 5′-triphosphate (BzATP, a P2X agonist, Sigma), (RS)-3,5-dihydroxyphenylglycine (DHPG, a group I metabotropic glutamate receptor agonist, Sigma), trans-1-aminocyclopentane-1,3-diacarboxylic acid (tACPD, an agonist of pan-metabotropic glutamate receptors, Sigma), or baclofen (the γ-aminobutyric acid B receptor agonist, Sigma), all diluted in KRS. The media were then collected for assays of d-serine, ATP, and glutamate (see below).
Glutamate Release Assay
Glutamate was measured based on the glutamate dehydrogenase-catalyzed oxidation of glutamate (19). Astrocyte cultures were treated with one of the agonists (see above) for 10 min, and the media were collected for glutamate measurement. Each culture supernatant (50 μl) was mixed with 50 μl of a substrate mixture (0.2 m Tris-HCl buffer (pH 8.2) containing 20 units/ml glutamate dehydrogenase (Sigma), 2.5 mg/ml nicotinamide adenine dinucleotide (Sigma), 0.1% Triton X-100, 0.25 mg/ml 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (Sigma)) and then incubated for 10 min at 37 °C. The reaction was stopped by adding 100 μl of stop solution (50% dimethylformamide, 20% SDS (pH 4.7)). Absorbance was measured using a microplate reader at a wavelength of 570 nm and a reference of 650 nm. We set the calibration curve by plotting standard glutamate solutions (1–1000 nm).
d-Serine Release Assay
d-Serine was measured using a chemiluminescence assay (20). Astrocyte cultures were treated with one of the agonists (BzATP, DHPG, or tACPD) for 10 min, and the media were collected for d-serine measurements. Ten-microliter samples were mixed with 100 μl of medium containing 0.1 m Tris-HCl (pH 8.8), 20 units/ml peroxidase (Sigma), and 8 μm luminol (Sigma). After 15 min, 10 μl of d-amino acid oxidase (75 units/ml, Sigma) was added. Chemiluminescence kinetics was measured for 1 s at room temperature using a SpectraMax L luminescence microplate reader (Molecular Devices, Sunnyvale, CA). We set the calibration curve by plotting standard d-serine solutions (50–1000 nm).
ATP Release Assay
Extracellular ATP concentrations in culture media were measured with a luciferin-luciferase bioluminescence assay (ATP bioluminescence assay kit CLS 2, Roche Diagnostics). After agonist incubation (BzATP, DHPG, or tACPD) for 10 min, supernatants were collected and mixed with luciferase reagents. We measured chemiluminescence using a SpectraMax L luminescence microplate reader (Molecular Devices). We set the calibration curve by plotting standard ATP solutions (1–1000 nm).
Glutamate Uptake Assay
To measure glutamate uptake, astrocytes were incubated for 10 min in KRS containing 20 nm l-[3H]glutamate (0.5 μCi, PerkinElmer Life Sciences), then immediately washed with ice-cold KRS, and solubilized with 200 μl of 0.2 n NaOH. Protein concentrations in the samples were determined with a Bradford assay (Bio-Rad), and l-[3H]glutamate uptake was normalized to protein content. To block dl-threo-β-benzyloxyaspartate (TBOA, a pan-glutamate transporter inhibitor, Sigma)- or dihydrokainate (DHK, a selective inhibitor of GLT-1, Sigma)-sensitive l-[3H]glutamate uptake, astrocytes were incubated with TBOA or DHK for 15 min before measuring uptake.
Statistical Analysis
Graphical data are presented as the means ± S.E. Student's t test was used to compare mean values for unpaired data. Differences were considered significant when the p value was <0.05.
RESULTS
CS-56 Immunoreactivity Reveals a Subpopulation of Astrocytes in the Adult Mouse Cerebral Cortex
In our previous study, immunohistochemistry using the anti-chondroitin sulfate antibody CS-56 revealed a unique staining pattern for a structure in the adult mouse cerebral cortex, which we termed the DACS (15). To confirm our previous findings, we first performed detailed immunohistochemical analyses with CS-56. As shown in Fig. 1A2, CS-56 immunoreactivity exhibited a patchy pattern in the adult mouse cerebral cortex. To clarify the identity of CS-56-positive cells, we performed double-labeling immunohistochemistry with CS-56 antibody and antibodies against cell markers in the adult mouse cerebral cortex. CS-56 immunoreactive patches often encased nuclei positive for S100β (a mature astrocyte marker that mainly accumulates in astrocytic nuclei in vivo) (Fig. 1B) and partially co-localized with GLAST (an astrocyte marker, Fig. 1C), but other astrocytic markers such as GS (Fig. 1D) and GFAP (data not shown) barely overlapped with CS-56 immunoreactivity. We did not observe CS-56 immunoreactivity in mature oligodendrocytes displaying GST-π (Fig. 1E) or myelin basic protein immunoreactivity (Fig. 1F) or in NG2-positive immature oligodendrocytes (Fig. 1G). We also did not observe co-localization of Iba-1, a microglial marker, with CS-56-positive patches (data not shown). These observations strongly suggested that the CS-56-positive patches represent a subpopulation of astrocytes. Immunoelectron microscopic analyses detected CS-56 immunoreactivity in cells with narrow cytoplasm and irregularly shaped nuclei with rich chromatin, all of which are morphological characteristics of astrocytes (Fig. 2A). In addition, a fine cellular process with CS-56-immunoreactive membrane was juxtaposed with an axon terminal and a postsynaptic spine (Fig. 2B) and CS-56-immunoreactive end feet attached to a capillary (Fig. 2C). These morphological characteristics further indicate that CS-56-positive cells are astrocytes. We also found that 27.9 ± 3.7% of the cultured astrocytes exhibited co-localization of CS-56 and S100β (Fig. 5, B1–B4), confirming that DACS is produced by astrocytes.
FIGURE 1.

CS-56 immunoreactivity localizes to astrocytes but not oligodendrocytes in the adult mouse cerebral cortex. A, representative images of immunostaining with CS-56 antibody in the adult mouse cerebral cortex. Schematic diagram of a coronal section of the mouse brain (A1) is shown, and the fluorescence image of the enclosed region appears in A2. Cx, cerebral cortex; cc, corpus callosum. Scale bar, 300 μm. B–F, representative double immunostaining with CS-56 antibody (B1–G1; green) and cell marker antibodies (B2–G2; red) are shown. Each pair of images was merged and is indicated in the 3rd column (B3–G3). The markers included S100β (B2), GLAST (C2), and GS (D2) mature astrocyte markers; GSTπ (E2) and myelin basic protein (MBP) (F2), mature oligodendrocyte markers; and NG2 (G2), an immature oligodendrocyte marker. For control staining, normal mouse IgM (H), normal mouse IgG (I), or normal rabbit IgG (J) was used instead of the primary antibodies. The inset in C3 is an enlargement of the enclosed region. Scale bar, 100 μm. Note that the CS-56-immunoreactive patches closely encased S100β-positive astrocytic nuclei (arrows in B1–B3) and partially co-localized with GLAST-positive astrocytes (enlarged region in C3), demonstrating their astrocytic origin. However, two other astrocytic markers, GS (D) and GFAP (data not shown), barely overlapped with CS-56 immunoreactivity. As shown in E1–E3 and F1–F3, mature oligodendrocyte markers did not co-localize with CS-56 immunoreactivity; likewise, the immature oligodendrocyte marker NG2 did not (G1–G3) overlap with CS-56-positive patches.
FIGURE 2.
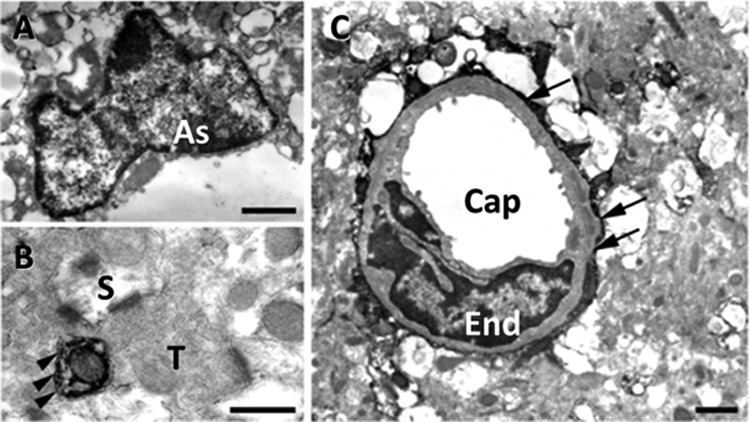
Representative electron micrographs of CS-56 immunoreactivity in the adult mouse cerebral cortex. A, CS-56 immunoreactivity was detected in a cell with narrow cytoplasm and an irregularly shaped nucleus with rich chromatin, all of which are morphological characteristics of the astrocyte (As). B, fine cellular process with CS-56-immunoreactive membrane (arrowheads) is juxtaposed with an axonal terminal (T) and a postsynaptic spine (S). C, CS-56-immunoreactive processes are attached to a capillary (Cap) with an endothelial nucleus (End) in the form of end feet (arrows). Scale bars, A and C are1 μm; B is 500 nm.
FIGURE 5.
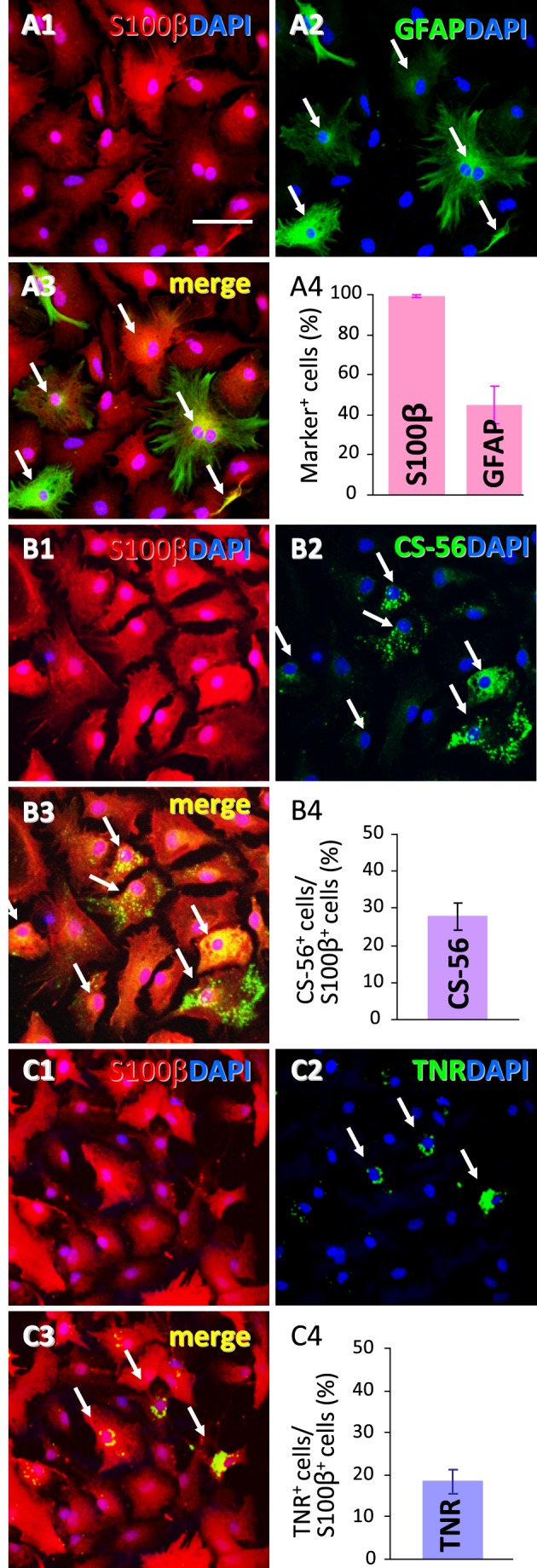
TNR immunoreactivity in cultured astrocytes. A, representative confocal images of anti-S100β (A1, red) and anti-GFAP (A2, green) double-immunostaining in cultured astrocytes (A3, merged image of A1 and A2). We determined the number of S100β and GFAP immunoreactive cells and then calculated their ratios to total cells (DAPI-positive cells) (A4). Values are shown as the mean ± S.E. of three different experiments. B, representative confocal images of double-immunostaining with anti-S100β (B1, red) and CS-56 (B2, green) in cultured astrocytes. We measured the ratio of CS-56-positive cells to S100β-positive astrocytes (B4). Values are shown as the mean ± S.E. of three different experiments. C, representative confocal images of double labeling with anti-S100β (C1, red) and anti-TNR (C2, green) antibodies in cultured astrocytes. We determined the ratio of TNR-immunoreactive cells to S100β-positive cells (C4). Values are shown as the mean ± S.E. of three different experiments. Scale bar, 50 μm. Note that TNR-positive cells composed a subpopulation of cultured astrocytes expressing S100β (arrows in C).
Purification of Glycoproteins from the Adult Mouse Cerebral Cortex
CSPGs consist of a core protein and sulfated glycosaminoglycan chains. However, because the CS-56 antibody recognizes sulfated glycosaminoglycans, the core protein(s) of CS-56-positive CSPGs remain elusive. We therefore attempted to identify the core proteins of DACS-specific CSPGs. In preliminary experiments, CS-56 immunoaffinity chromatography was employed to purify CSPGs from the adult mouse cerebral cortex. However, this method yielded insufficient proteoglycans (data not shown), perhaps because CS-56 antibody is an IgM and its affinity may be weak. We therefore devised a novel purification protocol, as depicted in Fig. 3A, and whose purification efficacy is indicated in Table 1. The combination of ion-exchange chromatography and Cu2+-chelating chromatography efficiently enriched CSPGs, as judged by dot blot assay using the CS-56 antibody (data not shown). We further purified high molecular weight glycoproteins using size-exclusion chromatography and then treated them with chondroitinase ABC. These partially purified and chondroitinase ABC-treated fractions were then electrophoresed on a preparative gradient polyacrylamide gel, from which silver-stained bands were excised and subjected to MALDI-TOF mass spectrometry (Fig. 3B). In addition to conventional lecticans such as brevican, phosphacan, versican, and neurocan, TNR was identified by mass spectrometry (Fig. 3B). Because TNR is known to be a CSPG (21, 22), and chondroitinase ABC digestion of protein immunoprecipitated with anti-TNR antibody yielded detectable ΔDi-4S and ΔDi-6S glycosaminoglycan disaccharides (data not shown), TNR was included in the candidate list of CSPGs that have covalently linked sulfated glycosaminoglycans. TNR was of particular interest because the distribution patterns of other conventional lecticans overlapped little with that of the DACS (see below).
TNR mRNA Co-localizes with CS-56-positive Astrocytes
Next, we compared the distribution patterns of mRNAs encoding core proteins of candidate CSPGs with those of CS-56-positive patches. Although few of the mRNAs of the identified lecticans (versican, neurocan, phosphacan, and brevican) were specifically detected in CS-56-positive cells (data not shown), TNR mRNA was localized in the cytoplasm of CS-56-positive cells in the adult mouse cerebral cortex (Fig. 4A). A previous study localized TNR protein to WFA-positive GABAergic interneurons and oligodendrocytes (23). Because CS-56-positive patches are likely to be a subpopulation of astrocytes (see above), the TNR mRNA expression pattern is discrepant with the previous study. One possible explanation is that TNR-synthesizing cells and cells associated with secreted TNR may not be identical. To elucidate the identity of TNR mRNA-positive cells, in situ hybridization for TNR mRNA and immunostaining for the astrocyte marker S100β, the mature oligodendrocyte marker GSTπ, and the GABAergic interneuron marker WFA were simultaneously performed in the adult mouse cerebral cortex. TNR mRNA localized in S100β-positive astrocytes (Fig. 4C) but not in GSTπ-positive oligodendrocytes (Fig. 4D). Furthermore, WFA-positive GABAergic interneurons were distinct from TNR mRNA-positive cells (Fig. 4B). We counted the numbers of TNR mRNA/S100β double-positive cells and of S100β-positive astrocytes (24 brain sections/three mice, eight sections/mouse). TNR mRNA was found in 8.73 ± 0.74% (mean ± S.E., triplicate experiments using three mice) of the S100β-positive astrocytes. The percentage seemed consistent with the sparse distribution pattern of the DACS (Fig. 1A2). Taken together, the present findings suggested that the TNR mRNA-positive cells were a subpopulation of astrocytes and synthesized the unique matrix structure termed the DACS.
FIGURE 4.

TNR mRNA is localized in CS-56-positive astrocytes in the adult mouse cerebral cortex. In situ hybridization for TNR mRNA (arrows in A and B) and immunostaining with CS-56 antibody (A, red) or WFA (B, red) was performed in single sections of the adult mouse cerebral cortex. Note that halo-like CS-56 immunoreactivity surrounded the TNR mRNA-containing cell bodies, although WFA-positive perineuronal nets did not. To identify TNR mRNA-positive cells, in situ hybridization for TNR mRNA (arrows in C1 and D1, red), immunostaining with the astrocyte marker S100β (C2, green), and the mature oligodendrocyte marker GSTπ (D2, green) were simultaneously performed in the adult mouse cerebral cortex. The TNR mRNA-positive cells were a subpopulation of astrocytes with nuclear S100β immunoreactivity (arrows in C). GSTπ-positive oligodendrocytes were distinct from TNR mRNA-positive cells (arrow in D). Scale bars in A and B are 100 μm; C and D are 50 μm.
TNR Is Expressed in Cultured Astrocytes and Involved in Glutamate Uptake
We next examined the expression of TNR in cultured astrocytes, which are amenable to gene manipulation and pharmacological experiments. In the present culture conditions, nearly all cultured astrocytes (99.3 ± 0.2%) were positive for S100β immunoreactivity (Fig. 5A), whereas more than half (45.2 ± 9.0%) were negative for GFAP immunoreactivity (Fig. 5A). A subpopulation of S100β-positive astrocytes (18.1 ± 3.3%) expressed TNR immunoreactivity (Fig. 5C), which was comparable with our in vivo findings (Fig. 4). The morphologies of marker-positive and -negative astrocytes were indistinguishable by phase contrast observation (data not shown).
Astrocytes can modulate synaptic plasticity by releasing gliotransmitters (24) and by taking up excess amounts of glutamate from synaptic clefts (25). We speculated that TNR might also be involved in such astrocytic functions, and we examined gliotransmitter functions, including glutamate, d-serine, and ATP release as well as glutamate uptake activity in cultured astrocytes, with or without siRNA-mediated knockdown of TNR gene expression (Fig. 6). We used two distinct siRNA sequences targeting TNR mRNA to ensure that the effects were not off-target. We examined whether TNR expression was effectively knocked down using RT-PCR (Fig. 7A). The endogenous expression of TNR mRNA was clearly down-regulated to a greater extent in the TNR siRNA-1- and siRNA-2-transfected astrocytes than in scrambled siRNA-treated astrocytes (TNR siRNA-1, 7.0 ± 0.1%; siRNA-2, 5.0 ± 0.2%) (Fig. 7A).
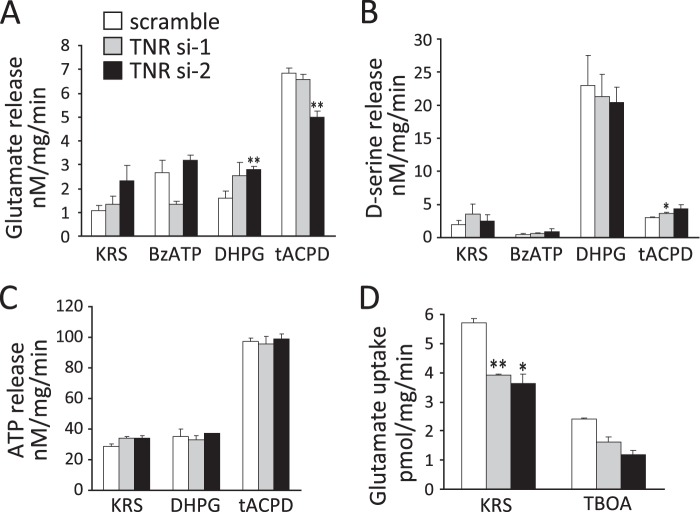
FIGURE 6.
Knockdown of TNR expression decreases glutamate uptake in cultured astrocytes. Glutamate (A), d-serine (B), and ATP (C) release assays were performed with cultured astrocytes transfected with scrambled or two different TNR siRNAs (si-1 and si-2) for 72 h. Gliotransmitters were measured after treating cells for 10 min with BzATP (500 μm), DHPG (50 μm), or tACPD (500 μm) in KRS. D, glutamate uptake was determined after incubating with 20 nm l-[3H]glutamate in KRS for 10 min. In a separate set of glutamate uptake experiments, cells were preincubated with the pan-glutamate transporter antagonist TBOA (300 μm) for 15 min, and glutamate uptake was then examined as noted above. Values are shown as the mean ± S.E. of nine cell samples from three different experiments. *, p < 0.05; **, p < 0.01, significantly different from value obtained for cells transfected with scrambled siRNA. Note that glutamate uptake activity was decreased by TNR knockdown.
FIGURE 7.
Knockdown of TNR expression decreases GLAST expression in cultured astrocytes. A, real time PCR analysis of mRNA expression of astrocyte-specific genes in cultured astrocytes transfected with scrambled or TNR-specific siRNAs (si-1 and si-2) for 72 h. B, representative Western blot analysis of GLAST, GLT-1, and GAPDH proteins. GAPDH was used as a loading control. Lane M, protein size markers (in kDa). The positions of monomer (arrow) and dimer (arrowhead) bands of GLAST are indicated. Numbers indicate independent cell samples. C and D, semi-quantitative densitometric data for GLAST (C) and GLT-1 (D) expression are shown. Values are shown as the mean ± S.E. of nine cell samples from three different experiments. *, p < 0.05; **, p < 0.01, significantly different from value obtained for cells transfected with scrambled (scra) siRNA. Note that TNR siRNA expression resulted in a marked decrease in GLAST expression but not GLT-1 expression. E, representative double immunostaining of cultured astrocytes with TNR and GLAST antibodies. Scale bar, 50 μm. TNR and GLAST were co-localized in cultured astrocytes (arrow). F, real time PCR analysis of mRNA expression of proteoglycan and GLAST genes in cultured astrocytes transfected with scrambled siRNA or proteoglycan-specific siRNAs (two, si-1 and si-2, for each proteoglycan) for 72 h. Note that conventional lecticans were not involved in the regulation of GLAST expression.
We first examined whether pharmacological agonists of receptors on astrocytes can trigger the release of gliotransmitters in the present culture system. Astrocytes express various receptors such as metabotropic glutamate receptor, GABAB receptors, and purinergic receptors and respond to glutamate, GABA, and ATP, respectively (24). Such astrocytic receptor activation induces intracellular Ca2+ elevation and then evokes release of gliotransmitters (26–28). As shown in Fig. 6, 500 μm tACPD (an agonist of pan-metabotropic glutamate receptors), 50 μm DHPG (a group I metabotropic glutamate receptor agonist), and 500 μm BzATP (a selective P2X purinergic agonist) evoked release of gliotransmitters. However, baclofen, an agonist for the GABAB receptors, did not induce release of gliotransmitters at a concentration range of 1–500 μm (data not shown). We then examined whether gliotransmitter release from cultured astrocytes was influenced by TNR expression (Fig. 6, A–C). Treatment with 500 μm tACPD facilitated the release of glutamate (KRS, 1.10 ± 0.21 nm/mg/min; tACPD, 6.82 ± 0.24 nm/mg/min; p = 0.001) (Fig. 6A) and ATP (KRS, 28.7 ± 1.7 nm/mg/min; tACPD, 97.2 ± 2.5 nm/mg/min, p = 0.001) (Fig. 6C) from astrocytes. d-Serine release was stimulated by 50 μm DHPG (KRS, 2.0 ± 0.6 nm/mg/min; DHPG, 23.0 ± 4.5 nm/mg/min; p = 0.037) (Fig. 6B). However, knockdown of TNR expression did not influence release of gliotransmitters from cultured astrocytes (Fig. 6, A–C). Glutamate and d-serine release were affected only in TNR siRNA-2- but not in siRNA-1-transfected astrocytes as compared with scrambled siRNA-transfected astrocytes (Fig. 6, A and B). These changes may be induced by off-target effects and were not specifically modulated by TNR.
Next, we examined whether glutamate uptake in cultured astrocytes was influenced by TNR knockdown. Two major glutamate transporters, GLAST and excitatory amino acid transporter 2 (GLT-1), regulate glutamate uptake in astrocytes and thereby maintain physiological extracellular glutamate concentrations (25, 29). We first tested whether glutamate transporters were functional in the present culture system. l-[3H]Glutamate accumulation in astrocytes increased in a dose- and time-dependent manner (Km = 238.61 nm and Vmax = 23.15 pmol/mg/min, data not shown). We examined the effects of pharmacological transporter inhibitors on glutamate uptake in cultured astrocytes. Pretreatment with DHK, a selective inhibitor of GLT-1, scarcely influenced l-[3H]glutamate accumulation in astrocytes (data not shown). However, pretreatment with TBOA, a pan-glutamate transporter antagonist, inhibited l-[3H]glutamate accumulation in a dose-dependent manner (IC50 = 128.08 μm, data not shown). These results suggest that GLAST but not GLT-1 primarily regulates glutamate uptake activity in the present culture system. We then examined glutamate uptake activity in TNR-knockdown astrocytes. As shown in Fig. 6D, TNR knockdown resulted in a significant decrease of glutamate uptake (scrambled siRNA, 5.70 ± 0.15 pmol/mg/min; TNR siRNA-1, 3.92 ± 0.03 pmol/mg/min; p = 0.001; siRNA-2, 3.63 ± 0.33 pmol/mg/min; p = 0.001). Glutamate uptake activity was inhibited by pretreatment with 300 μm TBOA (KRS, 5.70 ± 0.15 pmol/mg/min; TBOA, 2.42 ± 0.03 pmol/mg/min; p = 0.001, Fig. 6D).
TNR Regulates GLAST Expression in Cultured Astrocytes
The decrease in glutamate uptake in TNR-knockdown astrocytes suggested that glutamate transporter expression might be regulated by TNR. To test this hypothesis, we examined the expression of GLAST mRNA in TNR siRNA-treated astrocytes and compared it with that in control (scrambled siRNA-treated) astrocytes. The level of GLAST mRNA was significantly lower in the TNR-knockdown astrocytes than in the control astrocytes (TNR siRNA-1, 73 ± 4%, p = 0.001; siRNA-2, 60 ± 4%, p = 0.01) (Fig. 7A). To examine the possibility that the conventional lecticans identified in Fig. 3 might also regulate GLAST expression, we performed knockdown experiments with specific siRNAs for versican, neurocan, and brevican mRNAs (Fig. 7F). We did not test phosphacan siRNAs, because the expression level of phosphacan was very low in the adult mouse cerebral cortex (data not shown). Knockdown using two different siRNAs for each lectican effectively and specifically lowered the expression of the target lectican (Fig. 7F), whereas GLAST expression was little affected by these siRNAs, with the exception of neurocan siRNA-2 (Fig. 7F). Because neurocan siRNA-1 did not influence the GLAST expression level, we considered that the effect of neurocan siRNA-2 was off-target, and we concluded that conventional lecticans are not involved in the regulation of GLAST expression. To further confirm the role of TNR in GLAST expression, we examined GLAST protein levels using Western blot analysis in the TNR-knockdown and control astrocytes. Consistent with the mRNA expression patterns, the GLAST protein level in the TNR-knockdown astrocytes was significantly lower than that in scrambled siRNA-treated astrocytes (TNR siRNA-1, 61 ± 4%, p = 0.032; siRNA-2, 39 ± 7%, p = 0.010) (Fig. 7, B and C). In contrast, GLT-1 mRNA was increased only by one of the two siRNAs (TNR siRNA-1, 92 ± 7% p = 0.271; siRNA-2, 119 ± 3%, p = 0.010) (Fig. 7A), and GLT-1 protein was influenced by neither of the siRNAs as compared with scrambled siRNA-treated astrocytes (TNR siRNA-1, 74 ± 16%, p = 0.059; siRNA-2, 52 ± 35%, p = 0.081) (Fig. 7, B and D). In cultured astrocytes, the expression level of GLT-1 is known to be very low, and DHK, a GLT-1- specific inhibitor, does not influence glutamate uptake activity (30, 31). We also found that DHK did not change the glutamate uptake activity in our culture system (data not shown). Taken together, the present results do not negate the involvement of TNR in GLT-1 expression but suggested that the influence in GLT-1 expression was minimal. Finally, we confirmed that TNR and GLAST indeed co-localized in cultured astrocytes (Fig. 7E).
DISCUSSION
In this study, we purified glycoproteins from the adult cerebral cortex and sought to identify constituents of the CS-56-labeled extracellular matrix DACS, which decorates a subset of astrocytes (15). Among the sequenced candidate glycoproteins examined in this study, TNR mRNA co-localized well with the CS-56-immunoreactive astrocytes, indicating that TNR is a component of the DACS. TNR, a member of the tenascin family, is predominantly expressed in the central and peripheral nervous systems (32, 33). Previous studies have shown that brain TNR is localized in PNNs (8, 34, 35) and specifically decorates inhibitory interneurons (14). The present results are not incompatible with those findings, because PNNs are composed of a variety of CSPGs, such as aggrecan, brevican, phosphacan, and neurocan, in addition to TNR and hyaluronan (36). The DACS likely contains lecticans and hyaluronan in addition to TNR. Thus, PNNs and the DACS are not completely distinct structures but can share components such as TNR.
Knock-out mice revealed the roles of TNR in the central nervous system; abnormal aggregation of CSPGs mixed with hyaluronan was observed in the mutant mice, suggesting that TNR provides molecular scaffolding for CSPGs (37–39). The molecular mechanisms underlying the scaffolding activity include protein-protein interactions between TNR and lecticans (40–43). In addition to these protein-protein interactions, TNR is involved in regulating the action potential conduction of neurons (39) and in neuronal excitability (44, 45). Those studies primarily focused on neuronal functions using TNR-knock-out mice, although the present findings further implicate TNR in glial functions, as it was found in a subset of astrocytes and regulated expression levels of the glutamate transporter GLAST, which takes up glutamate from synaptic clefts and thereby modulates glutamatergic neurotransmission (46–48). Together with GLT-1, astrocytic transporters play homeostatic roles in neuron-glia interactions. These homeostatic mechanisms are also important in the case of neuronal injury and ischemia, because excess glutamate harms neurons in such pathological conditions (49). In this regard, part of the enhanced excitability of neurons in TNR-knock-out mice (44, 45, 50) may be attributed to a reduction of GLAST expression levels. Consistent with this possibility, TNR is up-regulated in astrogliosis (51, 52).
The DACS becomes visible as postnatal cortical development proceeds (16). Expression levels of astrocytic glutamate transporters (GLAST and GLT-1) increase during postnatal development (53, 54), whereas null mutations of these transporters impair metabolic cross-talk between astrocytes and neurons, and thereby affect synaptic activities (55). The developmental pattern of DACS expression, including that of TNR, may be linked to the functional maturation of cortical astrocytes through regulation of astrocytic GLAST expression. In this context, the regulatory mechanisms of GLAST by TNR are of particular interest. Currently, signaling pathways involved in this regulation are undefined. Because knockdown of TNR resulted in the rapid reductions in GLAST mRNA and protein in this study, the mechanisms underlying GLAST regulation are unlikely to be complex. Astrocytes may have a receptor for TNR that regulates GLAST expression levels, possibly by changing calcium ion levels, because GLAST expression is responsive to cellular Ca2+ concentration (56). Alternatively, TNR may raise the concentration of fibroblast growth factor-2 in the extracellular microenvironment, similar to tenascin-C (57). The increased fibroblast growth factor-2 may then induce GLAST expression in astrocytes (58).
Neuron-glia interactions mediated by the TNR-containing DACS are likely important in pathological conditions such as cerebral ischemia or mechanical injuries, as well as for normal development and maintenance of brain functions, because glutamate toxicity is critical in such pathological conditions. The close relationship between TNR and GLAST expression in astrocytes suggests TNR as a potential therapeutic tool for disorders affecting the central nervous system.
This work was supported by Japan Society for the Promotion of Science KAKENHI Grant 24592141.
- CSPG
- chondroitin sulfate proteoglycan
- TNR
- tenascin-R
- PNN
- perineuronal net
- DACS
- DAndelion Clock-like Structure
- CS-56
- anti-chondroitin sulfate monoclonal antibody
- GLAST
- excitatory amino acid transporter 1
- S100β
- S100β protein
- GS
- glutamine synthetase
- GSTπ
- π isoform of glutathione S-transferase
- NG2
- NG2 chondroitin sulfate proteoglycan
- WFA
- Wisteria floribunda lectin
- GFAP
- glial fibrillary acidic protein
- KRS
- Krebs-Ringer solution
- BzATP
- 3′-O-(4-benzoyl)benzoyl adenosine 5′-triphosphate
- GLT-1
- excitatory amino acid transporter 2
- DHPG
- (RS)-3,5-dihydroxyphenylglycine
- tACPD
- trans-1-aminocyclopentane-1,3-diacarboxylic acid
- TBOA
- dl-threo-β-benzyloxyaspartate
- DHK
- dihydrokainate
- nt
- nucleotide.
REFERENCES
- 1. Galtrey C. M., Fawcett J. W. (2007) The role of chondroitin sulfate proteoglycans in regeneration and plasticity in the central nervous system. Brain Res. Rev. 54, 1–18 [DOI] [PubMed] [Google Scholar]
- 2. Rhodes K. E., Fawcett J. W. (2004) Chondroitin sulphate proteoglycans: preventing plasticity or protecting the CNS? J. Anat. 204, 33–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carulli D., Laabs T., Geller H. M., Fawcett J. W. (2005) Chondroitin sulfate proteoglycans in neural development and regeneration. Curr. Opin. Neurobiol. 15, 116–120 [DOI] [PubMed] [Google Scholar]
- 4. Wilson M. T., Snow D. M. (2000) Chondroitin sulfate proteoglycan expression pattern in hippocampal development: potential regulation of axon tract formation. J. Comp. Neurol. 424, 532–546 [DOI] [PubMed] [Google Scholar]
- 5. Brückner G., Szeöke S., Pavlica S., Grosche J., Kacza J. (2006) Axon initial segment ensheathed by extracellular matrix in perineuronal nets. Neuroscience 138, 365–375 [DOI] [PubMed] [Google Scholar]
- 6. Brückner G., Grosche J., Hartlage-Rübsamen M., Schmidt S., Schachner M. (2003) Region and lamina-specific distribution of extracellular matrix proteoglycans, hyaluronan and tenascin-R in the mouse hippocampal formation. J. Chem. Neuroanat. 26, 37–50 [DOI] [PubMed] [Google Scholar]
- 7. Carulli D., Rhodes K. E., Fawcett J. W. (2007) Upregulation of aggrecan, link protein 1, and hyaluronan synthases during formation of perineuronal nets in the rat cerebellum. J. Comp. Neurol. 501, 83–94 [DOI] [PubMed] [Google Scholar]
- 8. Viggiano D. (2000) The two faces of perineuronal nets. Neuroreport 11, 2087–2090 [DOI] [PubMed] [Google Scholar]
- 9. Wintergerst E. S., Vogt Weisenhorn D. M., Rathjen F. G., Riederer B. M., Lambert S., Celio M. R. (1996) Temporal and spatial appearance of the membrane cytoskeleton and perineuronal nets in the rat neocortex. Neurosci. Lett. 209, 173–176 [DOI] [PubMed] [Google Scholar]
- 10. Balmer T. S., Carels V. M., Frisch J. L., Nick T. A. (2009) Modulation of perineuronal nets and parvalbumin with developmental song learning. J. Neurosci. 29, 12878–12885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dityatev A., Brückner G., Dityateva G., Grosche J., Kleene R., Schachner M. (2007) Activity-dependent formation and functions of chondroitin sulfate-rich extracellular matrix of perineuronal nets. Dev. Neurobiol. 67, 570–588 [DOI] [PubMed] [Google Scholar]
- 12. Lander C., Kind P., Maleski M., Hockfield S. (1997) A family of activity-dependent neuronal cell-surface chondroitin sulfate proteoglycans in cat visual cortex. J. Neurosci. 17, 1928–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nowicka D., Soulsby S., Skangiel-Kramska J., Glazewski S. (2009) Parvalbumin-containing neurons, perineuronal nets and experience-dependent plasticity in murine barrel cortex. Eur. J. Neurosci. 30, 2053–2063 [DOI] [PubMed] [Google Scholar]
- 14. Pizzorusso T., Medini P., Berardi N., Chierzi S., Fawcett J. W., Maffei L. (2002) Reactivation of ocular dominance plasticity in the adult visual cortex. Science 298, 1248–1251 [DOI] [PubMed] [Google Scholar]
- 15. Hayashi N., Tatsumi K., Okuda H., Yoshikawa M., Ishizaka S., Miyata S., Manabe T., Wanaka A. (2007) DACS, novel matrix structure composed of chondroitin sulfate proteoglycan in the brain. Biochem. Biophys. Res. Commun. 364, 410–415 [DOI] [PubMed] [Google Scholar]
- 16. Horii-Hayashi N., Tatsumi K., Matsusue Y., Okuda H., Okuda A., Hayashi M., Yano H., Tsuboi A., Nishi M., Yoshikawa M., Wanaka A. (2010) Chondroitin sulfate demarcates astrocytic territories in the mammalian cerebral cortex. Neurosci. Lett. 483, 67–72 [DOI] [PubMed] [Google Scholar]
- 17. Okuda H., Miyata S., Mori Y., Tohyama M. (2007) Mouse Prickle1 and Prickle2 are expressed in postmitotic neurons and promote neurite outgrowth. FEBS Lett. 581, 4754–4760 [DOI] [PubMed] [Google Scholar]
- 18. Yamamoto A., Nakamura Y., Kobayashi N., Iwamoto T., Yoshioka A., Kuniyasu H., Kishimoto T., Mori T. (2007) Neurons and astrocytes exhibit lower activities of global genome nucleotide excision repair than do fibroblasts. DNA Repair 6, 649–657 [DOI] [PubMed] [Google Scholar]
- 19. Abe K., Abe Y., Saito H. (2000) Evaluation of l-glutamate clearance capacity of cultured rat cortical astrocytes. Biol. Pharm. Bull. 23, 204–207 [DOI] [PubMed] [Google Scholar]
- 20. Zhuang Z., Yang B., Theus M. H., Sick J. T., Bethea J. R., Sick T. J., Liebl D. J. (2010) EphrinBs regulate d-serine synthesis and release in astrocytes. J. Neurosci. 30, 16015–16024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Probstmeier R., Braunewell K., Pesheva P. (2000) Involvement of chondroitin sulfates on brain-derived tenascin-R in carbohydrate-dependent interactions with fibronectin and tenascin-C. Brain Res. 863, 42–51 [DOI] [PubMed] [Google Scholar]
- 22. Woodworth A., Pesheva P., Fiete D., Baenziger J. U. (2004) Neuronal-specific synthesis and glycosylation of tenascin-R. J. Biol. Chem. 279, 10413–10421 [DOI] [PubMed] [Google Scholar]
- 23. Wintergerst E. S., Rathjen F. G., Schwaller B., Eggli P., Celio M. R. (2001) Tenascin-R associates extracellularly with parvalbumin immunoreactive neurones but is synthesised by another neuronal population in the adult rat cerebral cortex. J. Neurocytol. 30, 293–301 [DOI] [PubMed] [Google Scholar]
- 24. Hamilton N. B., Attwell D. (2010) Do astrocytes really exocytose neurotransmitters? Nat. Rev. Neurosci. 11, 227–238 [DOI] [PubMed] [Google Scholar]
- 25. Coulter D. A., Eid T. (2012) Astrocytic regulation of glutamate homeostasis in epilepsy. Glia 60, 1215–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kang J., Jiang L., Goldman S. A., Nedergaard M. (1998) Astrocyte-mediated potentiation of inhibitory synaptic transmission. Nat. Neurosci. 1, 683–692 [DOI] [PubMed] [Google Scholar]
- 27. Anderson C. M., Bergher J. P., Swanson R. A. (2004) ATP-induced ATP release from astrocytes. J. Neurochem. 88, 246–256 [DOI] [PubMed] [Google Scholar]
- 28. Henneberger C., Papouin T., Oliet S. H., Rusakov D. A. (2010) Long-term potentiation depends on release of d-serine from astrocytes. Nature 463, 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Verkhratsky A., Kirchhoff F. (2007) Glutamate-mediated neuronal-glial transmission. J. Anat. 210, 651–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Swanson R. A., Liu J., Miller J. W., Rothstein J. D., Farrell K., Stein B. A., Longuemare M. C. (1997) Neuronal regulation of glutamate transporter subtype expression in astrocytes. J. Neurosci. 17, 932–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schlag B. D., Vondrasek J. R., Munir M., Kalandadze A., Zelenaia O. A., Rothstein J. D., Robinson M. B. (1998) Regulation of the glial Na+-dependent glutamate transporters by cyclic AMP analogs and neurons. Mol. Pharmacol. 53, 355–369 [DOI] [PubMed] [Google Scholar]
- 32. Chiquet-Ehrismann R. (1995) Tenascins, a growing family of extracellular matrix proteins. Experientia 51, 853–862 [DOI] [PubMed] [Google Scholar]
- 33. Chiquet-Ehrismann R., Hagios C., Matsumoto K. (1994) The tenascin gene family. Perspect. Dev. Neurobiol. 2, 3–7 [PubMed] [Google Scholar]
- 34. Hagihara K., Miura R., Kosaki R., Berglund E., Ranscht B., Yamaguchi Y. (1999) Immunohistochemical evidence for the brevican-tenascin-R interaction: colocalization in perineuronal nets suggests a physiological role for the interaction in the adult rat brain. J. Comp. Neurol. 410, 256–264 [PubMed] [Google Scholar]
- 35. Bukalo O., Schachner M., Dityatev A. (2001) Modification of extracellular matrix by enzymatic removal of chondroitin sulfate and by lack of tenascin-R differentially affects several forms of synaptic plasticity in the hippocampus. Neuroscience 104, 359–369 [DOI] [PubMed] [Google Scholar]
- 36. Carulli D., Rhodes K. E., Brown D. J., Bonnert T. P., Pollack S. J., Oliver K., Strata P., Fawcett J. W. (2006) Composition of perineuronal nets in the adult rat cerebellum and the cellular origin of their components. J. Comp. Neurol. 494, 559–577 [DOI] [PubMed] [Google Scholar]
- 37. Brückner G., Grosche J., Schmidt S., Härtig W., Margolis R. U., Delpech B., Seidenbecher C. I., Czaniera R., Schachner M. (2000) Postnatal development of perineuronal nets in wild-type mice and in a mutant deficient in tenascin-R. J. Comp. Neurol. 428, 616–629 [DOI] [PubMed] [Google Scholar]
- 38. Haunsø A., Ibrahim M., Bartsch U., Letiembre M., Celio M. R., Menoud P. (2000) Morphology of perineuronal nets in tenascin-R and parvalbumin single and double knockout mice. Brain Res. 864, 142–145 [DOI] [PubMed] [Google Scholar]
- 39. Weber P., Bartsch U., Rasband M. N., Czaniera R., Lang Y., Bluethmann H., Margolis R. U., Levinson S. R., Shrager P., Montag D., Schachner M. (1999) Mice deficient for tenascin-R display alterations of the extracellular matrix and decreased axonal conduction velocities in the CNS. J. Neurosci. 19, 4245–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Aspberg A., Binkert C., Ruoslahti E. (1995) The versican C-type lectin domain recognizes the adhesion protein tenascin-R. Proc. Natl. Acad. Sci. U.S.A. 92, 10590–10594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Aspberg A., Miura R., Bourdoulous S., Shimonaka M., Heinegârd D., Schachner M., Ruoslahti E., Yamaguchi Y. (1997) The C-type lectin domains of lecticans, a family of aggregating chondroitin sulfate proteoglycans, bind tenascin-R by protein-protein interactions independent of carbohydrate moiety. Proc. Natl. Acad. Sci. U.S.A. 94, 10116–10121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu Y. J., La Pierre D. P., Wu J., Yee A. J., Yang B. B. (2005) The interaction of versican with its binding partners. Cell Res. 15, 483–494 [DOI] [PubMed] [Google Scholar]
- 43. Xiao Z. C., Bartsch U., Margolis R. K., Rougon G., Montag D., Schachner M. (1997) Isolation of a tenascin-R binding protein from mouse brain membranes. A phosphacan-related chondroitin sulfate proteoglycan. J. Biol. Chem. 272, 32092–32101 [DOI] [PubMed] [Google Scholar]
- 44. Brenneke F., Bukalo O., Dityatev A., Lie A. (2004) Mice deficient for the extracellular matrix glycoprotein tenascin-R show physiological and structural hallmarks of increased hippocampal excitability, but no increased susceptibility to seizures in the pilocarpine model of epilepsy. Neuroscience 124, 841–855 [DOI] [PubMed] [Google Scholar]
- 45. Gurevicius K., Gureviciene I., Valjakka A., Schachner M., Tanila H. (2004) Enhanced cortical and hippocampal neuronal excitability in mice deficient in the extracellular matrix glycoprotein tenascin-R. Mol. Cell. Neurosci. 25, 515–523 [DOI] [PubMed] [Google Scholar]
- 46. Schousboe A. (2003) Role of astrocytes in the maintenance and modulation of glutamatergic and GABAergic neurotransmission. Neurochem. Res. 28, 347–352 [DOI] [PubMed] [Google Scholar]
- 47. Bezzi P., Vesce S., Panzarasa P., Volterra A. (1999) Astrocytes as active participants of glutamatergic function and regulators of its homeostasis. Adv. Exp. Med. Biol. 468, 69–80 [DOI] [PubMed] [Google Scholar]
- 48. Anderson C. M., Swanson R. A. (2000) Astrocyte glutamate transport: review of properties, regulation, and physiological functions. Glia 32, 1–14 [PubMed] [Google Scholar]
- 49. Yi J. H., Hazell A. S. (2006) Excitotoxic mechanisms and the role of astrocytic glutamate transporters in traumatic brain injury. Neurochem. Int. 48, 394–403 [DOI] [PubMed] [Google Scholar]
- 50. Morellini F., Sivukhina E., Stoenica L., Oulianova E., Bukalo O., Jakovcevski I., Dityatev A., Irintchev A., Schachner M. (2010) Improved reversal learning and working memory and enhanced reactivity to novelty in mice with enhanced GABAergic innervation in the dentate gyrus. Cereb. Cortex 20, 2712–2727 [DOI] [PubMed] [Google Scholar]
- 51. Brenneke F., Schachner M., Elger C. E., Lie A. A. (2004) Up-regulation of the extracellular matrix glycoprotein tenascin-R during axonal reorganization and astrogliosis in the adult rat hippocampus. Epilepsy Res. 58, 133–143 [DOI] [PubMed] [Google Scholar]
- 52. Gutowski N. J., Newcombe J., Cuzner M. L. (1999) Tenascin-R and -C in multiple sclerosis lesions: relevance to extracellular matrix remodelling. Neuropathol. Appl. Neurobiol. 25, 207–214 [DOI] [PubMed] [Google Scholar]
- 53. Furuta A., Rothstein J. D., Martin L. J. (1997) Glutamate transporter protein subtypes are expressed differentially during rat CNS development. J. Neurosci. 17, 8363–8375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kugler P., Schleyer V. (2004) Developmental expression of glutamate transporters and glutamate dehydrogenase in astrocytes of the postnatal rat hippocampus. Hippocampus 14, 975–985 [DOI] [PubMed] [Google Scholar]
- 55. Voutsinos-Porche B., Bonvento G., Tanaka K., Steiner P., Welker E., Chatton J. Y., Magistretti P. J., Pellerin L. (2003) Glial glutamate transporters mediate a functional metabolic crosstalk between neurons and astrocytes in the mouse developing cortex. Neuron 37, 275–286 [DOI] [PubMed] [Google Scholar]
- 56. Liu Y. P., Yang C. S., Tzeng S. F. (2008) Inhibitory regulation of glutamate aspartate transporter (GLAST) expression in astrocytes by cadmium-induced calcium influx. J. Neurochem. 105, 137–150 [DOI] [PubMed] [Google Scholar]
- 57. Karus M., Denecke B., ffrench-Constant C., Wiese S., Faissner A. (2011) The extracellular matrix molecule tenascin C modulates expression levels and territories of key patterning genes during spinal cord astrocyte specification. Development 138, 5321–5331 [DOI] [PubMed] [Google Scholar]
- 58. Figiel M., Maucher T., Rozyczka J., Bayatti N., Engele J. (2003) Regulation of glial glutamate transporter expression by growth factors. Exp. Neurol. 183, 124–135 [DOI] [PubMed] [Google Scholar]