Background: Thermodynamically unstable magnesium calcite is deposited in the shell of pearl oysters at ambient pressure.
Results: The novel acidic matrix protein PfN44 interacts with magnesium to inhibit the deposition of aragonite.
Conclusion: PfN44 participates in shell formation by inhibiting aragonite formation.
Significance: Results of this study suggest a connection between the matrix protein and magnesium.
Keywords: Biomaterials, Biomineralization, Cloning, Extracellular Matrix Proteins, Protein Chemistry
Abstract
Magnesium is widely used to control calcium carbonate deposition in the shell of pearl oysters. Matrix proteins in the shell are responsible for nucleation and growth of calcium carbonate crystals. However, there is no direct evidence supporting a connection between matrix proteins and magnesium. Here, we identified a novel acidic matrix protein named PfN44 that affected aragonite formation in the shell of the pearl oyster Pinctada fucata. Using immunogold labeling assays, we found PfN44 in both the nacreous and prismatic layers. In shell repair, PfN44 was repressed, whereas other matrix proteins were up-regulated. Disturbing the function of PfN44 by RNAi led to the deposition of porous nacreous tablets with overgrowth of crystals in the nacreous layer. By in vitro circular dichroism spectra and fluorescence quenching, we found that PfN44 bound to both calcium and magnesium with a stronger affinity for magnesium. During in vitro calcium carbonate crystallization and calcification of amorphous calcium carbonate, PfN44 regulated the magnesium content of crystalline carbonate polymorphs and stabilized magnesium calcite to inhibit aragonite deposition. Taken together, our results suggested that by stabilizing magnesium calcite to inhibit aragonite deposition, PfN44 participated in P. fucata shell formation. These observations extend our understanding of the connections between matrix proteins and magnesium.
Introduction
Many organisms must control mineral formation through biomineralization to live. A main issue in the study of biomineralization is an understanding of the structure of minerals and the functions of macromolecules in mineral formation (1–3). Calcium carbonate is widely used by metazoan taxa, including sea urchins, sponges, crustacean, mollusks, and ascidians, to deposit biominerals for protection (1, 2, 4, 5). At ambient pressure, calcium carbonate forms five crystalline polymorphs: calcite, aragonite, and vaterite in the anhydrous phase and monohydrocalcite and ikaite in the hydrate phase. Calcium carbonate also forms amorphous forms including amorphous calcium carbonate (ACC),3 which is widely accepted as the precursor in biomineralization (6–11). Biomineralization in the pearl oyster Pinctada fucata is a focus of calcium carbonate crystallization studies (12, 13). A substantial amount of work has explored the relationship between macromolecules and calcium carbonate crystallization, but information on the molecular mechanisms of crystallization is still limited (14–16).
The P. fucata shell is composed of two mineralized layers, i.e. the outer prismatic layer and the inner nacreous layer. The main components of these two layers are calcium carbonate crystals that are deposited as calcite in the prismatic layer and aragonite in the nacreous layer (17). The proteins in the shell, which are also referred to as matrix proteins, are believed to play important roles in the control of calcium carbonate nucleation and growth (18–21). These matrix proteins are currently being isolated from <5% (w/w) of the shell (6, 22–29). The acidic matrix proteins, which have cation binding capacity, are believed to be the main proteins involved in control of calcium carbonate crystallization and shell formation (10, 11, 30, 31). Previous studies reported that these acidic proteins control the polymorphism and morphology of calcium carbonate in vitro (26, 28, 32).
In contrast to classical stepwise growth via the terrace-ledge-kink model of crystal growth, the deposition of biominerals begins with ACC as an intermediate that is transformed to crystalline carbonate polymorphs such as calcite or aragonite (33). Most naturally occurring ACC contains magnesium, and ACC crystallization is under control of biomolecules (34). Magnesium has a strong influence on calcium carbonate precipitation. Magnesium is incorporated into calcite when the magnesium content is low or induces the formation of aragonite when magnesium content is high (35). Calcite with high magnesium content is not stable and is generally not formed under ambient conditions. However, in many marine organisms, deposited calcite has a high incorporation of magnesium (36). The topic of the mechanism by which the biologically controlled deposition of the thermodynamically unstable magnesium calcite in marine organisms is very interesting.
However, because of the lack of efficient technology for analyzing shell proteins, only a few matrix proteins have been purified and analyzed (13, 37, 38). During shell growth, calcite in the prismatic layer and aragonite in the nacreous layer are under tight control of the matrix proteins (6, 17, 39–41). Different matrix proteins are involved in the formation of different layers (42–44). To isolate matrix proteins involved in shell formation, three suppression substrate hybridization libraries were constructed (45). In this study we identified an acidic protein, PfN44, in the shell using the suppression substrate hybridization libraries and the mantle tissue transcriptome. The nature and functions of PfN44 were analyzed.
Ethics Statement
All rabbits were raised under standardized pathogen-free conditions in the Animal Care Facility at Beijing Hospital. The study protocol for the experimental use of the animals was approved by the Ethics Committee of National Center for Clinical Laboratories.
MATERIALS AND METHODS
Animals
P. fucata were purchased from the Marine Biology Research Station at Daya Bay, CAS, Shenzhen, China. They were maintained in glass aquaria filled with aerated artificial seawater (Sude Instant Sea Salt, 3% at 20 °C) for 3 days before the experiments.
Primers
Refer to Table 1 for primer details.
TABLE 1.
Primers used in this study
F, forward; R, reverse; q, quantitative.
| Primers used to clone PfN44 to the expression vector | |
| PfN44-e-5 | GATCCATGGGCTTTTGTTACAAAACATTTGG |
| PfN44-e-3 | CATCTCGAGAAATCCCGGGAAATCAGCAAATTTC |
| Primers used for RACE | |
| RACE3 | TTCCCTCTACTATGAAGACAAG |
| RACE5 | GCCGTTGATTGGCTGTCC |
| LONG | CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT |
| SHORT | CTAATACGACTCACTATAGGGC |
| NUP | AAGCAGTGGTATCAACGCAGAGT |
| Primers used for full-length clone | |
| FULL1 | AATCTCATACGGAGAAATTGGTGT |
| FULL2 | GAATGACACACATTTTTTATTGTA |
| Primers used for RNAi | |
| RNAi-P1 | CTAATACGACTCACTATAGGGAGAAGGAATTCTATTTTTGATGC |
| RNAi-P2 | CTAATACGACTCACTATAGGGAGAAGGGTTTATTGTAATGTTCG |
| Primers used for RT-PCR | |
| RT-PfN44-F | GAGGTCGGAGAAGTAAG |
| RT-PfN44-R | GAACACGATGGTAAAAA |
| RT-GAPDH-F | GCCGAGTATGTGGTAGAATC |
| RT-GAPDH-R | CACTGTTTTCTGGGTAGCTG |
| Primers for real time | |
| qβ-actin-f | CTCCTCACTGAAGCCCCCCTC |
| qβ-actin-r | ATGGCTGGAATAGGGATTCTGG |
| qKRMP-F | AAGAAATGTCACCCTTGGGATTGG |
| qKRMP-R | AATCATCGCCACCATATCCATCG |
| qNacrein-F | GGCTTTGGCGACGAACCGGA |
| qNacrein-R | ACACGGGGGAGTGGTCAGGG |
| qPfN44-F | TTGCTGATGGATGTGAAG |
| qPfN44-R | CTGTATATTGCCGTTGATTG |
Cloning and Bioinformatics Analysis of the Complete PfN44 cDNA Sequence
Total RNA was extracted from the mantle tissue of P. fucata using TRIzol (Invitrogen) according to the manufacturer's instructions. Rapid amplification of cDNA ends (RACE) was performed using a SMARTerTM RACE cDNA Amplification kit (Clontech). RACE3 and RACE5 primers were used with primers supplied with the 3′- and 5′-RACE kits. The full-length sequence was confirmed using primers FULL1 and FULL2. The deduced amino acid sequence of PfN44 was determined bioinformatically using the ORF Finder online (www.ncbi.nlm.nih.gov). The signal peptide was predicted using SignalP 3.0.
Detection of PfN44 in Shell Matrix
Primers PfN44-e-5/PfN44-e-3 were used to amplify the coding region of PfN44 without signal peptide. PCR products were cloned into the expression vector PET28a. Recombinant PfN44 with a His6 tag at the N terminus was overexpressed in Escherichia coli (Transsetta DE3) with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside at 10 °C for 24 h. Cells were homogenized and purified using a combination of His-trap HP columns and Superdex 75 10/300 GL column (GE) according to the manufacturer's instructions. Purified PfN44 was analyzed by SDS-PAGE and desalted by HiTrapTM Desalting Column (GE Healthcare) against 20 mm Na3PO4, 0.5 m NaCl buffer, pH 7.4. Anti-PfN44 polyclonal antibodies were raised in New Zealand rabbits following standard immunization procedures and were affinity-purified using Protein A+G-agarose (Beyotime) according to the manufacturer's instructions. EDTA-soluble matrix from different shell layers was prepared as described (46). The amount of extracted protein was determined using the BCA assay kit (Pierce) according to the manufacturer's instructions. The presence of PfN44 in different extracts of the shell was tested by Western blotting.
Immunolocalization of PfN44 in the Shell
Immunogold-labeling assays were conducted as described by Marin et al. (47) with some modifications. Anti-PfN44 was used as the primary antibody (1:200, v/v) and goat anti-rabbit coupled to 5-nm gold particles (Sigma) was the secondary antibody (1:400, v/v). Controls were performed without primary antibody or preimmune serum. Labeled samples were sputter-coated with carbon and scanned with an FEI Sirion2000 scanning electron microscope in the back scattering electron mode. Two pieces of shells from five oysters were analyzed in each experiment. Five images were randomly taken from each sample. Typical images were shown under “Results.”
Silencing of PfN44
RNAi assays was performed as described by Suzuki et al. (28) with some modifications. Primers RNAi-P1 and RNAi-P2 were used to amplify specific sequences from the PfN44 coding region. For GFP dsRNA synthesis, pEGFP-C1 (Clontech) was used as the template. PCR products were purified using TIANgel Midi Purification kits (TIANGEN). RiboMAXTM Large Scale RNA Production System (T7) kit (Promega) was used to synthesize dsRNA. RNase-free DNase I (TAKARA) was used to digest template DNA, and dsRNA was purified by RNAclean kits (TIANGEN). dsRNAs were diluted to 40 μg/100 μl or 80 μg/100 μl in PBS. Two-year-old oysters with 5–6-cm shell lengths were injected with dsRNAs or PBS into the adductor muscle. Each treatment group contained five oysters that were not manipulated in any way. Treated oysters were sacrificed 7 days after treatment. Shells were thoroughly washed with Milli-Q water, air-dried, and cut into sections. An FEI Quanta 200 scanning electron microscope was used to analyze the inner surface. All of the shells from the five oysters were examined. For each shell, at least 20 images were taken and examined, and the typical images were chosen to present the results.
Total RNA from mantle tissue was extracted and used to synthesize first-strand cDNA as described. Real-time quantitative PCR (qPCR) was conducted to quantify gene expression levels, with β-actin as an internal reference. qPCR was analyzed with a Mx3000PTM (Stratagene) using a SYBRH Premix ExTaqTMII kit (TAKARA). Parameters were: 95 °C for 30 s (1 cycle); 95 °C for 5 s, 55 °C for 30 s, 72 °C for 5 s (30 cycles); 72 °C for 30 s (see Table 1 for primers). Dissociation curves were investigated to determine product purity and amplification specificity. As a control, samples were normalized to 80 μg/100 μl of sample from the GFP dsRNA-injected group, which was assigned a relative value of 1.0. RNAi and other experiments were repeated once for reproducibility.
Shell Notching
Shell-notching assays were performed according to Mount (48) with some modifications. Close to the adductor muscle, a V-shaped notch was cut on the shell margin without disturbing the mantle tissue. Animals were randomly divided into eight groups with five animals each and returned to seawater tanks. At 0, 6, 12, 24, 48, 72, or 84 h after notching, oysters were sacrificed, and mantle tissues from the same group pooled and stored in liquid nitrogen. Mantle tissue from five oysters without shell notching was collected in the same way and used as controls. Total RNA extraction and first-strand cDNA synthesis were as described above. qPCR for gene expression was performed as described (27). Shell notching and qPCR experiments were repeated once. qPCR results show all replicates from all notching experiments.
Protein Labeling with Rhodamine and the Crystal Binding Assays
PfN44 and BSA were labeled with rhodamine using a HOOKTM (5/6) TAMRA-SE (Rhodamine) Labeling kit (G-Biosciences) according to the manufacturer's instructions. Protein crystal binding assays were as previously described (49). This experiment was repeated once to get a consistent result.
In Vitro Calcium Carbonate Crystallization Assay
Saturated calcium bicarbonate solution was prepared as described by Xu et al. (50). Aragonite crystallization was by the addition of different concentrations of magnesium chloride to saturated calcium bicarbonate solution. Crystallization experiments were performed by adding protein to freshly prepared solutions on siliconized glass slides. After 24 h, deposited crystals were washed with Milli-Q water. Air-dried crystals were characterized using an FEI Quanta 200 scanning electron microscope. Renishaw RM2000 Raman spectroscopy data, recorded at an excitation wavelength of 514 nm, were used to analyze crystal type. Spectra were scanned 3 times for 20 s in the range of 100–1400 cm−1. For each concentration of proteins, the crystallization experiments were done in five samples on the same glass slides. Each of the calcium carbonate crystallization experiments was done three times to get a consistent result. For the results of Raman spectroscopy analysis, at least 10 crystals of the same morphology were analyzed in one experiment. At least 15 crystals of the same morphology were analyzed by elemental analysis in each replicate experiment.
CD spectroscopy and Fluorescence Quenching
CD measurements were carried with a JASCO-600 spectropolarimeter using cuvettes with a 5-mm path length. PfN44 concentration was 200 μg/ml. Fluorescence spectra were collected using a Hitachi F-2500 spectrofluorometer, with a 1-cm path length cell and excitation wavelength of 280 nm. PfN44 concentration was 5 μm. The CD spectroscopy and fluorescence quenching analyses were done three times to get a consistent result.
Transition of ACC to Stable Crystals
50 mm CaCl2 together with 100 mm MgCl2 and 100 μg/ml PfN44 was prepared (solution A). 50 mm Na2CO3 was prepared (solution B). Solutions A and B were cooled on ice for 1 h. An equal volume of solution B was quickly added to solution A and mixed well at 4 °C. After 0, 24, or 48h, deposited crystals were washed by acetone, dried, and analyzed by x-ray diffraction and FTIR. This analysis was done by three times to get a consistent result.
RESULTS
Identification and Bioinformatics Analysis of PfN44
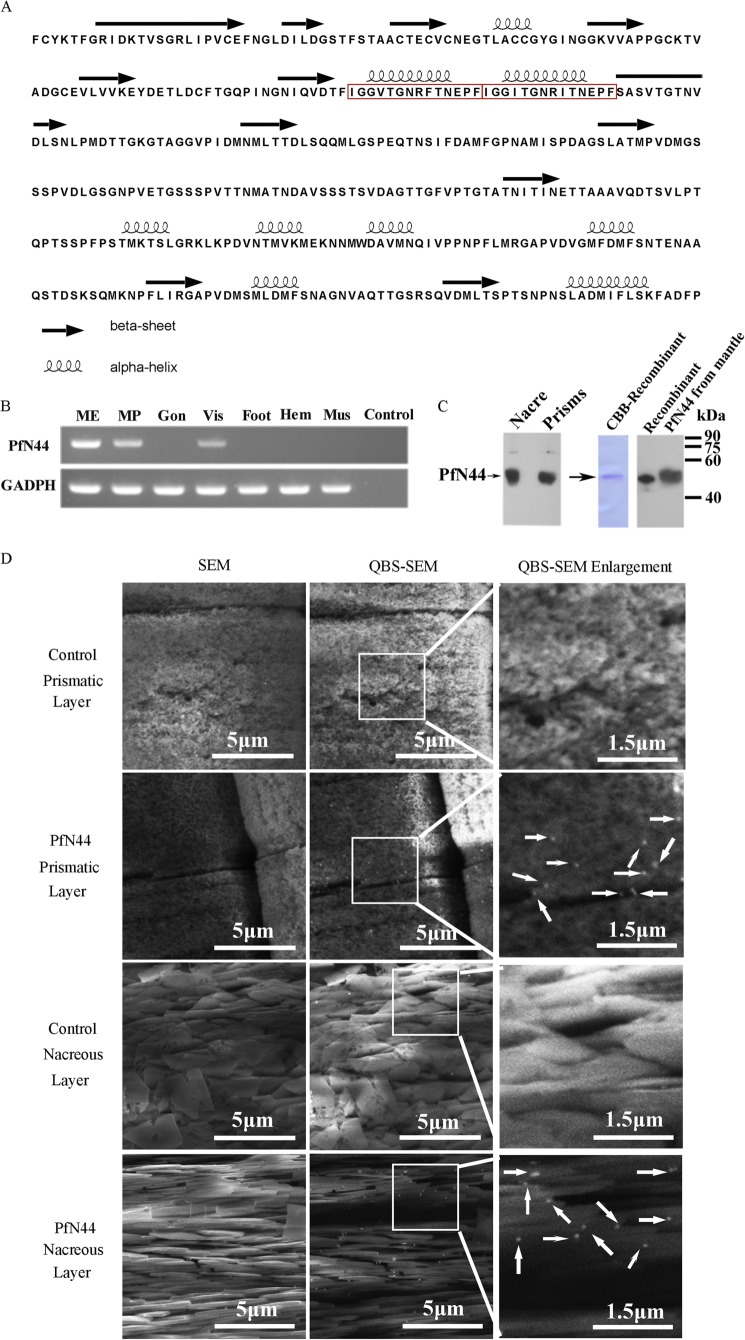
Based on the data in the suppression substrate hybridization library of D-shape stages (45), we searched the mantle tissue transcriptome and found a transcript in both the suppression substrate hybridization and transcriptome libraries. The transcript had a tandem repeat domain, which is suggested to be a feature of matrix proteins (51). We amplified this 1617-bp transcript (GenBankTM accession number KC238310) from mantle tissue cDNA. The product contained a 5′-untranslated region of 75 bp and a 3′-untranslated region of 222 bp. The open reading frame was 1320 bp and encoded a deduced 439-amino acid protein. By SignalP 3.0 prediction, the deduced protein contained a signal peptide of 17 amino acids. The mature protein was referred to as PfN44 based on the calculated molecular mass of 44.1 kDa. ProtScale analysis showed that PfN44 was an acidic protein with a theoretical isoelectric point of 4.25. A BLASTp search against the GenBankTM nr database indicated that PfN44 was similar to hypothetical protein CGI_10014775 of the Pacific oyster Crassostrea gigas with no similar genes in other species. However, hypothetical protein CGI_10014775 was not previously characterized, so the function of PfN44 was unknown. By Jpred prediction, PfN44 exhibited both the α helix and β sheet in its secondary structure (Fig. 1A).
FIGURE 1.
Detection of PfN44 as a matrix protein. A, secondary structure prediction for PfN44. Red box, repeated domain. B, tissue-specific gene expression of PfN44 by RT-PCR. Total RNA from mantle edge (ME), mantle pallial (MP), gonad (Gon), viscus (Vis), foot, hemocytes (Hem), and adductor muscle (Mus) was used. RT-PCR without template was the negative control (Control). Housekeeping gene GAPDH was the positive control. C, detection of PfN44 in the shell. Native PfN44 in EDTA-soluble shell matrix via Western blot. Recombinant protein and mantle tissue proteins were also tested. CBB, Coomassie Brilliant Blue. D, detection of native PfN44 in shells by immunogold staining. SEM of prismatic layer prisms and nacreous layer tablets with PfN44 in the prismatic and nacreous layers. Control, without primary antibody. Arrows, PfN44 in the shell. QBS-SEM, back-scattered electron mode scanning electron microscope.
Identification of PfN44 as a Matrix Protein in the P. fucata Shell
We analyzed the expression profile of PfN44 in different P. fucata tissues using RT-PCR. PfN44 expression was detected in the mantle pallial, mantle edge, and viscus (Fig. 1B). The mantle tissue is reported to be responsible for shell formation, whereas the mantle pallial is responsible for formation of the nacreous layer, and the mantle edge is responsible for prismatic layer formation (16). Although genes specifically expressed by the viscus tissue were not analyzed thoroughly, these results suggested that PfN44 could be involved in shell formation of both the nacreous layer and prismatic layer.
To further analyze whether PfN44 existed in the nacreous and prismatic layers, polyclonal antibodies against recombinant PfN44 were used for immunodetection of native PfN44. EDTA-soluble matrix from separated nacre and prisms was analyzed by Western blot. As shown in Fig. 1 C, PfN44 was detected in the EDTA-soluble matrix of both nacreous and prismatic layers. In controls, no signal was detected in the absence of the anti-PfN44 (data not shown). PfN44 protein can also be detected in the extract of viscus by Western blot (data not shown). We purified recombinant PfN44 and analyzed the purity by Coomassie Brilliant Blue staining (Fig. 1C). The recombinant protein was sufficiently pure for in vitro studies. Proteins expressed by P. fucata could be post-translationally modified by glycosylation or phosphorylation; therefore, we also compared the molecular weight of recombinant protein expressed by E. coli with proteins extracted from mantle tissue. As shown in Fig. 1C, recombinant and native proteins showed a similar molecular weight, indicating that PfN44 was not highly glycosylated or phosphorylated.
The microstructural distribution of PfN44 in shells was analyzed by immunogold labeling. Slightly notched prismatic and nacreous layers were incubated with anti-PfN44 as the primary antibody and 5-nm gold-labeled antibodies as the secondary antibody. The white spots detected by scanning electron microscopy in backscattered electron mode indicated gold elements. PfN44 was in the prisms of the prismatic layer and nacreous tablets of the nacreous layer (Fig. 1D). In controls, staining was not observed without anti-PfN44 (Fig. 1D) or with preimmune serum as the primary antibody (data not shown). These results together with expression profiles suggested that PfN44 participated in shell formation.
In Vivo Analysis of PfN44 Function during Shell Formation
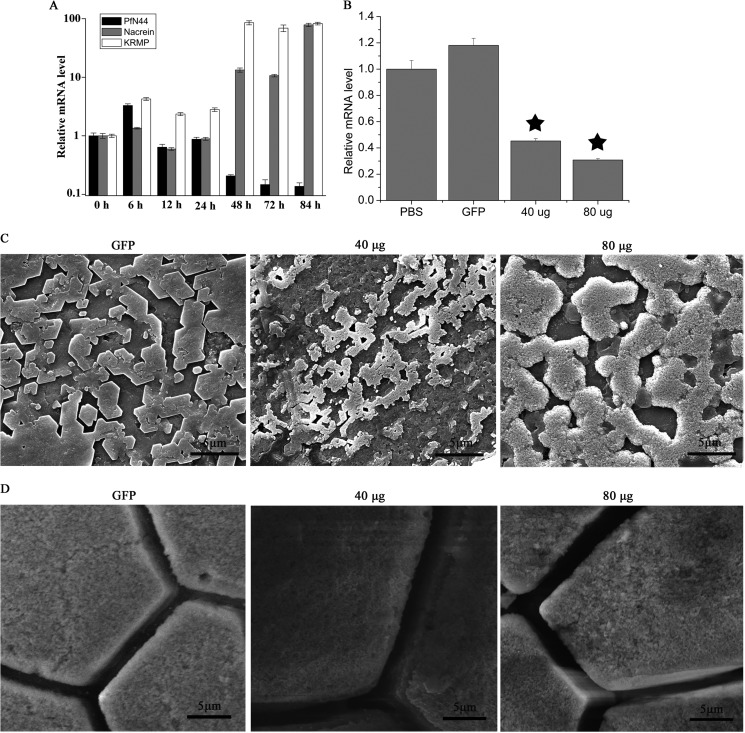
To investigate the PfN44 functions in shell formation, we conducted assays in which shells were slightly notched to induce shell formation. As shown in Fig. 2A, gene expression of PfN44 increased 6 h after shell notching and then decreased. At 48 h after shell notching, PfN44 expression decreased to approximate 10%. We also analyzed expression of two other matrix proteins, nacrein and lysine-rich matrix protein 3 (KRMP3). At 6 h after notching, nacrein expression had increased to 135%. In contrast, at 48 h after notching, nacrein expression had increased further. The expression pattern of KRMP3 was similar to nacrein after notching.
FIGURE 2.
In vivo functions of PfN44. A, PfN44 expression in shell notching. Expression of the unnotched groups (0 h) gave a relative value of 1.0. The y axis showed a log standard. B, PfN44 expression inhibited by RNAi. Control, PBS-injected groups gave a relative value of 1.0. The non-target control was GFP dsRNA. Star, significant (p < 0.001) difference compared with PBS-injected groups. C, SEM nacreous layer of inner surface. GFP dsRNA-injected group showed normal stair-like growth; 40 and 80 μg of PfN44 dsRNA-injected groups showed overgrowth. Scale bars, 5 μm. D, SEM prismatic layer of inner surface. Scale bars, 5 μm.
The function of PfN44 during shell formation was further analyzed by RNAi. To clarify the functions of PfN44 in shell formation, specifically designed dsRNAs of PfN44 were injected into oyster adductor muscles. Controls were injected with GFP dsRNA or PBS only. Seven days after injection, relative mRNA levels were analyzed. PfN44 expression in the GFP dsRNA-injected group was similar to the PBS-injected group. Compared with the PBS-injected control group, PfN44 expression decreased to approximate 45% in the group injected with 40 μg of dsRNA and 30% in the group injected with 80 μg of dsRNA (Fig. 2B). The surfaces of both the nacreous and prismatic layers of the treated groups were observed under SEM. In the GFP dsRNA-injected group, the surface of the nacreous layer showed a normal flat nacreous tablet with a stair-like growth pattern (Fig. 2C). The surface of the nacreous layer of the group was injected with 40 μg of PfN44 dsRNA, and the deposited tablets were porous with the overgrowth of crystals where the normal flat nacreous tablets were absent. In addition, these nacreous tablets were smaller than those that were normal. These changes in the nacreous tablets were more obvious in the 80-μg injection group, and the deposited tablets were larger than those that were normal. When the expression of PfN44 was depleted even more, the abnormal deposition of calcium carbonate was out of the control of the feedback system, so that in the 80 μg of dsRNA-treated tablets the deposited crystals were larger than the normal tablets. The surface of the prismatic layer was not substantially affected by the interference of PfN44 (Fig. 2D).
Binding Properties of PfN44 with Main Shell Components
To analyze the mechanisms of how PfN44 affected the shell formation, we investigated the binding properties of PfN44 with the main mineral components of the shell, i.e. calcite and aragonite. PfN44 was labeled with rhodamine and incubated with calcite or aragonite. Calcite and aragonite surfaces were labeled with rhodamine, indicating binding between calcium carbonate crystals and PfN44 (Fig. 3).
FIGURE 3.
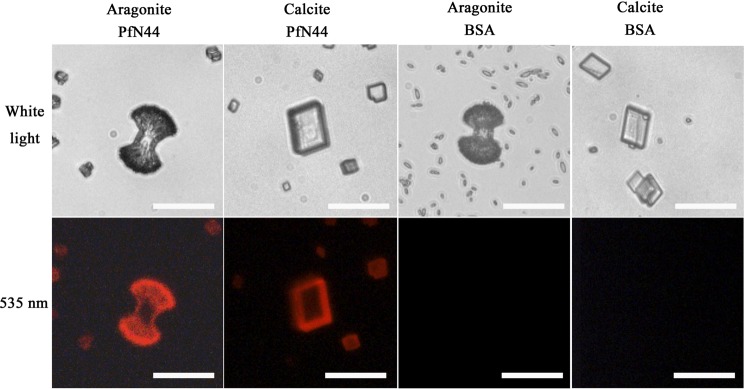
Binding profiles of PfN44. PfN44 was labeled with rhodamine and then incubated with calcite or aragonite. The rhodamine labeled BSA was used as a control. Scale bar, 20 μm. 109 crystals were examined, and 105 crystals showed the same pattern in the calcite binding assay. 124 crystals were examined, and 114 crystals showed the same pattern as shown in the aragonite binding assay.
Calcium Carbonate Crystallization in the Presence of PfN44
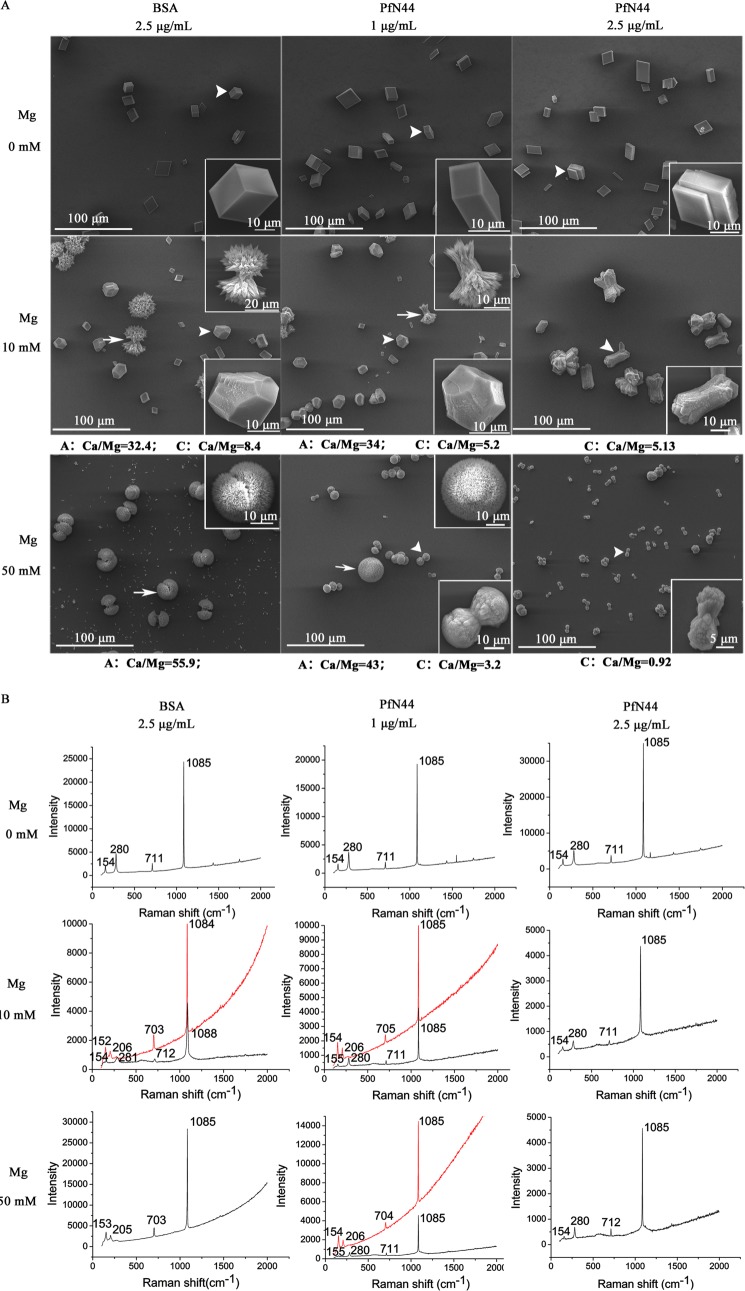
In vitro calcite and aragonite crystallization was used to investigate the function of PfN44 in calcium carbonate crystallization. In the in vitro crystallization assays, the functions of proteins were analyzed. If the proteins could induce the crystallization of calcium carbonate, the addition of protein would induce the overgrowth of some axis in the deposited crystals. If the protein could inhibit the crystallization of calcium carbonate, the addition of protein would inhibit the growth of some axis or specific deposition of certain kind of crystals. A calcite crystallization system free of magnesium was used to analyze the effects of PfN44 on calcite formation. With 2.5 μg/ml BSA as a control, crystals deposited from saturated calcium bicarbonate were typical calcite rhombohedra (Fig. 4A). With 1 or 2.5 μg/ml PfN44, deposited crystal morphology did not noticeably change. Raman spectra analysis showed that all deposited crystals were calcite (Fig. 4B).
FIGURE 4.
Effects of PfN44 on calcite and aragonite growth. A, SEM of deposited crystals. Enlarged images show crystals indicated by arrows and arrowheads. The control was 2.5 μg/ml BSA. Without magnesium, deposited crystals were all calcite. PfN44 did not change morphology. With 10 or 50 mm magnesium, PfN44 induced calcite-like crystals and inhibited aragonite deposition. Elemental compositions of crystals were analyzed by energy-dispersive x-ray spectroscopy. A, aragonite; C, calcite. At least images of 50 crystals in each spot were taken. The images shown in A are typical of these results. B, Raman spectra of deposited crystals in A. Red lines, crystals are indicated by arrows; black lines, crystals indicated by arrowheads.
To analyze the function of PfN44 in aragonite deposition, we conducted in vitro crystallization assays with 10 mm magnesium to induce deposition of both calcite and aragonite. With 10 mm magnesium and BSA as the control, deposited crystals were mixed with needle-like spindles and slightly notched calcite-like crystals (Fig. 4A). Raman spectra analysis showed that the two kinds of crystals were aragonite and calcite (Fig. 4B). With 1 μg/ml PfN44, fewer aragonite crystals were seen, and the size of the aragonite crystal decreased compared with the BSA control. With 2.5 μg/ml PfN44, deposited crystals were all calcite, as identified by Raman spectra analysis. This result suggested that PfN44 inhibited aragonite deposition. To address whether PfN44 inhibited aragonite crystallization, we conducted in vitro crystallization assays with 50 mm magnesium, which was sufficient to turn all deposited crystals to aragonite. As shown in Fig. 4A, when 2.5 μg/ml BSA was used as the control, deposited crystals were all aragonite with needle-like spindles forming a round morphology. With the addition of 1 μg/ml PfN44, two kinds of crystals formed; one was the same as the aragonite deposited with the BSA control, and the other was dumbbell-shaped calcite, as confirmed by Raman spectra. Deposited crystals were all dumbbell-shaped calcites in the presence of 2.5 μg/ml PfN44. These results suggested that aragonite deposition was inhibited by PfN44.
We also analyzed the elemental composition of deposited crystals using energy-dispersive x-ray spectroscopy. The ratio of Ca2+/Mg2+ decreased in deposited calcite when PfN44 was present. In the presence of 10 mm magnesium, the ratio of Ca2+/Mg2+ was 8.4 in calcite, decreasing to 5.2 with 1 μg/ml and 5.13 with 2.5 μg/ml PfN44. In the presence of 50 mm magnesium, the ratio of Ca2+/Mg2+ in deposited calcite was 3.2 with 1 μg/ml PfN44, decreasing to 0.92 with 2.5 μg/ml PfN44. The increase in magnesium content in calcite suggested that PfN44 stabilized magnesium calcite to control calcium carbonate crystallization.
Interactions between PfN44 and Magnesium
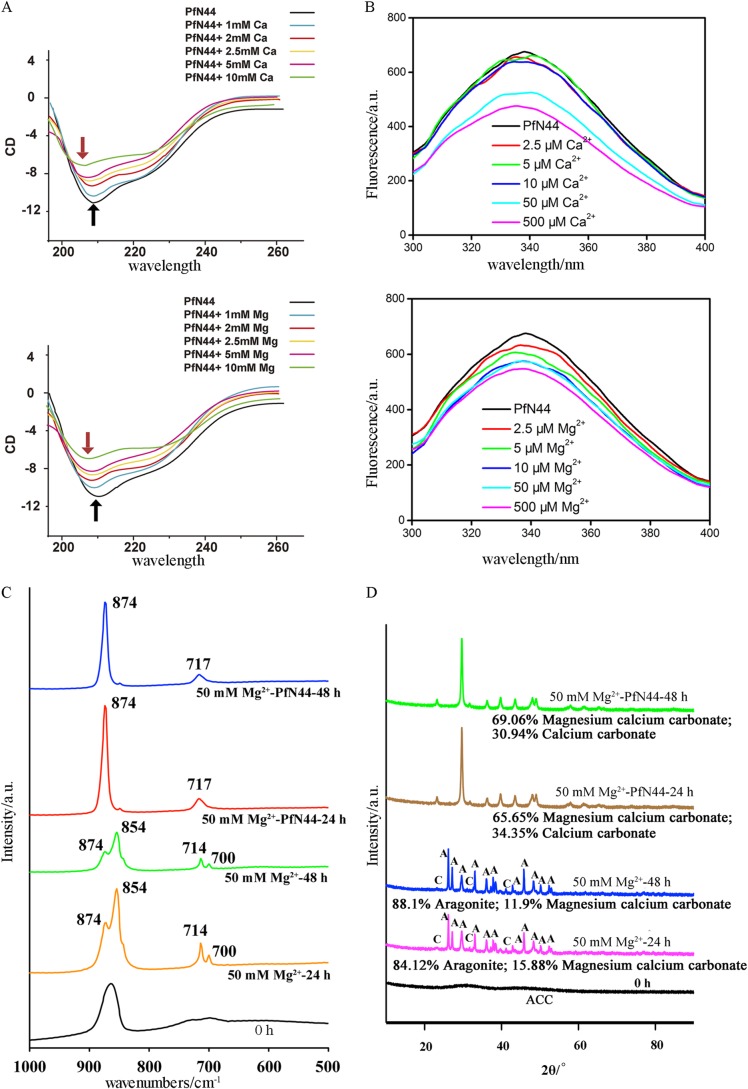
To address whether PfN44 interacted with calcium to control calcium carbonate formation, we analyzed near-UV CD spectra of PfN44 in the presence or absence of Ca2+. In the absence of calcium (1 mm EDTA), PfN44 showed typical mixed α helix and β sheet peaks consistent with the secondary structure prediction in Fig. 1A (Fig. 5A). In the presence of 1 mm calcium, the peak of the α helix did not change. When PfN44 was incubated with 2.5 mm calcium, the peak of the α helix at 208 nm shifted to a shorter wavelength. With higher concentrations of calcium, this CD spectrum shift became more pronounced. This result suggested that, by inducing a more flexible α helix structure, calcium increased the hydrophobicity of the PfN44 structure. Carbonate deposition in Mollusca is also affected by magnesium (52). Therefore, we analyzed the near-UV CD spectra of PfN44 in the presence of magnesium. With 1 mm or higher concentrations of magnesium, the α helix peak shifted to a shorter wavelength in a concentration-dependent manner (Fig. 5A). Mg2+ could induce the CD shift at only 1 mm, but Ca2+ could not make the shift happen at 2.5 mm or more. The shift caused by magnesium could be observed in the presence of magnesium at lower concentrations than that in the presence of calcium, suggesting a higher affinity between magnesium and PfN44.
FIGURE 5.
PfN44 interacts with magnesium to stabilize magnesium calcite. A, near-UV CD spectra for PfN44 in the absence (1 mm EDTA) or presence of calcium or magnesium. Arrows, α helix peaks at 208 nm. Black arrows, native protein; red, proteins with calcium or magnesium. B, intrinsic fluorescence of PfN44 in the absence (1 mm EDTA) or presence of calcium or magnesium. Shown are IR (C) and x-ray diffraction spectra (D) of precipitates from solutions with the indicated concentrations of magnesium and crystallization times.
To confirm the PfN44 interaction with Ca2+ and Mg2+, we analyzed the intrinsic fluorescence of PfN44 with or without Ca2+ and Mg2+. The addition of 2.5, 5, or 10 μm calcium quenched the intrinsic tryptophan fluorescence of PfN44 by ∼5.4%; however, with 50 μm calcium, quenching was 22.3% and with 500 μm calcium, 29.5% (Fig. 5B). As shown in Fig. 5B, quenching by 2.5, 5, or 10 μm magnesium was higher than quenching by calcium. Quenching by 10 μm magnesium was 14.8% higher than quenching by calcium. However, when PfN44 was incubated with 50 or 500 μm magnesium, intrinsic fluorescence was not further decreased. Quenching caused by magnesium was saturated at a low concentration so that intrinsic fluorescence was not further decreased. The substantial different quenching levels suggested that PfN44 bound magnesium more strongly than calcium.
Effects of PfN44 on Transition of ACC to Stable Crystals
ACC is the precursor of calcium carbonate biominerals in Mollusca (38). To find out how PfN44 inhibited aragonite deposition, we analyzed the function of PfN44 in the transition of ACC to stable calcium carbonate crystals. We first analyzed the effect of PfN44 on the transition of ACC to the stable crystals by FTIR. With 50 mm magnesium, ACC transformed to aragonite and calcite after 24 or 48 h of crystallization. Specific peaks at 700 and 854 nm showed deposited aragonite, and peaks at 714 and 874 nm showed the calcite. With 50 μg/ml PfN44, the FITR spectrum of deposited crystals showed only the specific peaks at 717 and 874 nm, indicating that the deposited crystals were all calcite (Fig. 5C).
To further characterize the polymorphs of deposited crystals, we analyzed the deposited crystals by x-ray diffraction. With 50 mm magnesium, ACC transformed to aragonite and Mg2+-bearing calcite after 24 or 48 h crystallization (Fig. 5D). When 50 μg/ml PfN44 was added to the crystallization system, aragonite was excluded in deposited crystals. Instead, ACC transformed to monoclinic calcium carbonate without magnesium and Mg2+-bearing calcite rhombohedral crystals. Compared with crystals deposited without PfN44, the amount of Mg2+-bearing calcite crystals increased more than 2-fold. In addition, with the 48 h crystallization time, the content of Mg2+-bearing calcite decreased from 15.88 ± 0.4% to 11.9 ± 0.2% without PfN44 (p value = 0.001), consistent with the hypothesis that Mg2+-bearing calcite is thermodynamically unstable. From 24 to 48 h, the content of deposited Mg2+-bearing calcite increased from 65.7 ± 1.0% to 69.1 ± 0.7% with PfN44 (p value = 0.0089), indicating that PfN44 stabilized the Mg2+-bearing calcite and increased its content. These results suggested that, in the transition of ACC to the stable crystals, PfN44 stabilized the Mg2+-bearing calcite to inhibit aragonite formation.
DISCUSSION
Biogenetic solid minerals known as biominerals are different from inorganic crystals. Biominerals have extraordinary physical and mechanical properties and can be synthesized in an ambient environment. In P. fucata, biominerals were deposited under the control of matrix proteins, specifically acidic proteins (44, 54–57). We analyzed the in vivo and in vitro functions of the novel acidic matrix protein PfN44.
In shell-notching experiments, the expression of PfN44, nacrein, and KRMP3 increased 6 h after notching, which might have been caused by the notching procedure. Notching disturbed the growth of the oyster, so the expression of matrix proteins might also have been disturbed after notching. Shell repair processes are similar to creating new shell. Calcium carbonate crystals accumulated on the matrix. Nacrein, which contains two carbonic anhydrase domains that are important in biomineralization, catalyzes the formation of hydrogen carbonate from water and CO2 (6). Although nacrein inhibits calcification (58), it must accumulate in the shell to provide sufficient hydrogen carbonate for crystal formation. KRMP participates in the framework formation of the prismatic layer. During shell repair, KRMP3 accumulates in the shell to facilitate this framework formation. In contrast to nacrein and KRMP, PfN44 expression decreased during shell repair. We found that PfN44 inhibited aragonite deposition in in vitro calcium carbonate crystallization and ACC transition. Therefore, PfN44 might be a “safety guard” that slows nacreous layer formation. During this process, some other matrix proteins, like MSI31, which formed the framework of nacreous layer and MSI7 which could induce the nucleation of aragonite, participated to control the formation of nacreous tablets. Repression of PfN44 would facilitate the formation of the nacreous layer during repair. Our results were consistent with the hypothesis that PfN44 inhibited aragonite formation to participate in shell formation. When PfN44 function was disturbed, only the nacreous layer was affected with a disordered deposition of crystals on the surface; the prismatic layer was not obviously affected. Although PfN44 was found in both the prismatic and nacreous layer, it had small effects on calcite deposition. As shown by in vitro crystallization, PfN44 clearly affected aragonite deposition in the presence of magnesium but had little effect on calcite deposition in the absence of magnesium. Disturbing PfN44 function affected the nacreous layer more than the prismatic layer. Shell formation is under control of different matrix proteins; more than one could induce the formation of the microstructure of shell. In the formation of prismatic layer, PfN44 might need to interact with other matrix proteins to control calcite deposition. Analyzing the functions of PfN44 together with other matrix proteins such as Prisilkin 39 and Shematrin would be interesting. In addition, it is also possible that PfN44 could perform some function in the prismatic layer that is not revealed by the RNAi studies because PfN44 mRNA expression is not sufficiently reduced. As PfN44 exists in the prismatic layer and could bind to the calcite, it may also exhibit some effects in the formation of prismatic layer. To reveal the functions of PfN44 in the prismatic layer, it would be useful to use bioinformatics analysis and yeast two-hybrid system to screen for the potential interacting proteins.
Magnesium is strongly adsorbed onto the surface of precipitated calcite because it is more strongly hydrated than calcium. In addition, magnesium might be dehydrated when it is incorporated into calcite, generating a barrier to calcite growth. However, magnesium is not incorporated into the aragonite structure to inhibit the aragonite formation. Therefore, when magnesium is added to a crystallization system, it incorporates into the calcite to form magnesium calcite. Thermodynamically unstable magnesium calcite disrupts the formation of calcite and induces aragonite deposition. Until now, only a few small molecules have been found to be involved in magnesium calcite formation, such as carboxylated molecules that increase the magnesium content in the Mg2+-bearing calcite (33) and polyacrylic acid sodium salt and dextran sulfate sodium salt, which can promote the deposition of high magnesium calcite under ambient conditions (53). Matrix proteins might control calcium carbonate crystallization in pearl oyster shell formation based on evidence showing magnesium calcite in the shell (36). The matrix proteins that control magnesium content are, therefore, important in shell formation. However, no direct evidence shows that matrix proteins participate in the formation of calcium carbonate crystals by interacting with magnesium. Here we found that the matrix protein PfN44 interacted with magnesium and stabilized the formation of magnesium calcite to inhibit aragonite deposition. Studies on biomineralization have analyzed the relationship between matrix proteins and calcium. Magnesium is important in the crystallization of calcium carbonate. The relationship between matrix proteins and magnesium could provide information about the mechanism of biomineralization. Studying interactions between matrix proteins and magnesium in biomineralization could shed light on biomineralization and facilitate the synthesis of new functional materials.
In conclusion, the acidic protein PfN44 participated in the shell formation by inhibiting aragonite deposition. Further research on the relationship between the sequence of shell formation and functions of PfN44 would provide more information about the molecular mechanism of shell formation.
This work was supported by National Basic Research Program of China Grant 2010CB126405, National High Technology Research and Development Program of China Grant 2010AA09Z405, National Natural Science Foundation of China Grants 31172382 and U0831001 (Joint Fund with Guangdong), and Independent Research Projects of Tsinghua University Grant 20111080964. Grant 2011ZX08011-006 is a Major Project of Ministry of Agriculture.
- ACC
- amorphous calcium carbonate
- RACE
- rapid amplification of cDNA ends
- SEM
- scanning electron microscope
- dsRNA
- double-stranded RNA
- qPCR
- quantitative PCR.
REFERENCES
- 1. Wilkinson B. H. (1979) Biomineralization, paleoceanography, and the evolution of calcareous marine organisms. Geology 7, 524–527 [Google Scholar]
- 2. Simkiss K. (1977) Bio-mineralization and detoxification. Calcif. Tissue Res. 24, 199–200 [DOI] [PubMed] [Google Scholar]
- 3. Paine M. L., White S. N., Luo W., Fong H., Sarikaya M., Snead M. L. (2001) Regulated gene expression dictates enamel structure and tooth function. Matrix Biol. 20, 273–292 [DOI] [PubMed] [Google Scholar]
- 4. Radha A. V., Forbes T. Z., Killian C. E., Gilbert P. U., Navrotsky A. (2010) Transformation and crystallization energetics of synthetic and biogenic amorphous calcium carbonate. Proc. Natl. Acad. Sci. U.S.A. 107, 16438–16443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lowenstam H. A., Abbott D. P. (1975) Vaterite. Mineralization product of hard tissues of a marine organism (Ascidiacea). Science 188, 363–365 [DOI] [PubMed] [Google Scholar]
- 6. Miyamoto H., Miyashita T., Okushima M., Nakano S., Morita T., Matsushiro A. (1996) A carbonic anhydrase from the nacreous layer in oyster pearls. Proc. Natl. Acad. Sci. U.S.A. 93, 9657–9660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aizenberg J., Addadi L., Weiner S., Lambert G. (1996) Stabilization of amorphous calcium carbonate by specialized macromolecules in biological and synthetic precipitates. Adv. Mater. 8, 222–226 [Google Scholar]
- 8. Muramoto K., Yako H., Murakami K., Odo S., Kamiya H. (1994) Inhibition of the growth of calcium carbonate crystals by multiple lectins in the celomic fluid of the acorn barnacle Megabalanus rosa. Comp. Biochem. Physiol. B 107, 401–409 [Google Scholar]
- 9. Borbas J. E., Wheeler A. P., Sikes C. S. (1991) molluscan shell matrix phosphoproteins. Correlation of degree of phosphorylation to shell mineral microstructure and to in vitro regulation of mineralization. J. Exp. Zool. 258, 1–13 [Google Scholar]
- 10. Maruyama K., Mikawa T., Ebashi S. (1984) Detection of calcium-binding proteins by Ca-45 autoradiography on nitrocellulose membrane after sodium dodecyl sulfate gel electrophoresis. J. Biochem. 95, 511–519 [DOI] [PubMed] [Google Scholar]
- 11. Ueno M. (1980) Calcium transport in crayfish gastrolith disk. Morphology of gastrolith disk and ultrahistochemical demonstration of calcium. J. Exp. Zool. 213, 161–171 [Google Scholar]
- 12. Addadi L., Weiner S. (1997) Biomineralization. A pavement of pearl. Nature 389, 912–915 [Google Scholar]
- 13. Weiner S. (2008) Biomineralization. A structural perspective. J. Struct. Biol. 163, 229–234 [DOI] [PubMed] [Google Scholar]
- 14. Furuhashi T., Schwarzinger C., Miksik I., Smrz M., Beran A. (2009) Molluscan shell evolution with review of shell calcification hypothesis. Comp. Biochem. Physiol. B 154, 351–371 [DOI] [PubMed] [Google Scholar]
- 15. Bonucci E. (2009) Calcification and silicification. A comparative survey of the early stages of biomineralization. J. Bone Miner. Metab. 27, 255–264 [DOI] [PubMed] [Google Scholar]
- 16. Marin F., Luquet G., Marie B., Medakovic D. (2008) Molluscan shell proteins. Primary structure, origin, and evolution. Curr. Top. Dev. Biol. 80, 209–276 [DOI] [PubMed] [Google Scholar]
- 17. Almeida M. J., Milet C., Peduzzi J., Pereira L., Haigle J., Barthelemy M., Lopez E. (2000) Effect of water-soluble matrix fraction extracted from the nacre of Pinctada maxima on the alkaline phosphatase activity of cultured fibroblasts. J. Exp. Zool. 288, 327–334 [DOI] [PubMed] [Google Scholar]
- 18. Liu X., Li J., Xiang L., Sun J., Zheng G., Zhang G., Wang H., Xie L., Zhang R. (2012) The role of matrix proteins in the control of nacreous layer deposition during pearl formation. Proc. Biol. Sci. 279, 1000–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou Y., He Z., Huang J., Gong N., Yan Z., Liu X., Sun J., Wang H.., Zhang G., Xie L., Zhang R. (2010) Cloning and characterization of the activin-like receptor 1 homolog (Pf-Alr1) in the pearl oyster, Pinctada fucata. Comp. Biochem. Physiol. B 156, 158–167 [DOI] [PubMed] [Google Scholar]
- 20. Miyazaki Y., Nishida T., Aoki H., Samata T. (2010) Expression of genes responsible for biomineralization of Pinctada fucata during development. Comp. Biochem. Physiol. B 155, 241–248 [DOI] [PubMed] [Google Scholar]
- 21. Glazer L., Shechter A., Tom M., Yudkovski Y., Weil S., Aflalo E. D., Pamuru R. R., Khalaila I., Bentov S., Berman A., Sagi A. (2010) A protein involved in the assembly of an extracellular calcium storage matrix. J. Biol. Chem. 285, 12831–12839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tsukamoto D., Sarashina I., Endo K. (2004) Structure and expression of an unusually acidic matrix protein of pearl oyster shells. Biochem. Biophys. Res. Commun. 320, 1175–1180 [DOI] [PubMed] [Google Scholar]
- 23. Zhang C., Xie L., Huang J., Liu X., Zhang R. (2006) A novel matrix protein family participating in the prismatic layer framework formation of pearl oyster, Pinctada fucata. Biochem. Biophys. Res. Commun. 344, 735–740 [DOI] [PubMed] [Google Scholar]
- 24. Zhang Y., Xie L., Meng Q., Jiang T., Pu R., Chen L., Zhang R. (2003) A novel matrix protein participating in the nacre framework formation of pearl oyster, Pinctada fucata. Comp. Biochem. Physiol. B 135, 565–573 [DOI] [PubMed] [Google Scholar]
- 25. Sudo S., Fujikawa T., Nagakura T., Ohkubo T., Sakaguchi K., Tanaka M., Nakashima K., Takahashi T. (1997) Structures of mollusc shell framework proteins. Nature 387, 563–564 [DOI] [PubMed] [Google Scholar]
- 26. Samata T., Hayashi N., Kono M., Hasegawa K., Horita C., Akera S. (1999) A new matrix protein family related to the nacreous layer formation of Pinctada fucata. FEBS Lett. 462, 225–229 [DOI] [PubMed] [Google Scholar]
- 27. Yano M., Nagai K., Morimoto K., Miyamoto H. (2007) A novel nacre protein N19 in the pearl oyster Pinctada fucata. Biochem. Biophys. Res. Commun. 362, 158–163 [DOI] [PubMed] [Google Scholar]
- 28. Suzuki M., Saruwatari K., Kogure T., Yamamoto Y., Nishimura T., Kato T., Nagasawa H. (2009) An acidic matrix protein, Pif, is a key macromolecule for nacre formation. Science 325, 1388–1390 [DOI] [PubMed] [Google Scholar]
- 29. Suzuki M., Murayama E., Inoue H., Ozaki N., Tohse H., Kogure T., Nagasawa H. (2004) Characterization of prismalin-14, a novel matrix protein from the prismatic layer of the Japanese pearl oyster (Pinctada fucata). Biochem. J. 382, 205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weiner S. (1979) Aspartic acid-rich proteins. Major components of the soluble organic matrix of mollusk shells. Calcif. Tissue Int. 29, 163–167 [DOI] [PubMed] [Google Scholar]
- 31. Fujimura T., Wada K., Iwaki T. (1995) Development and morphology of the pearl oyster larvae, Pinctada fucata. Venus, the Japanese Journal of Malacology 54, 25–48 [Google Scholar]
- 32. Kono M., Hayashi N., Samata T. (2000) Molecular mechanism of the nacreous layer formation in Pinctada maxima. Biochem. Biophys. Res. Commun. 269, 213–218 [DOI] [PubMed] [Google Scholar]
- 33. Wang D., Wallace A. F., De Yoreo J. J., Dove P. M. (2009) Carboxylated molecules regulate magnesium content of amorphous calcium carbonates during calcification. Proc. Natl. Acad. Sci. U.S.A. 106, 21511–21516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Weiner S., Levi-Kalisman Y., Raz S., Addadi L. (2003) Biologically formed amorphous calcium carbonate. Connect Tissue Res. 44, 214–218 [PubMed] [Google Scholar]
- 35. Raz S., Weiner S., Addadi L. (2000) Formation of high magnesian calcites via an amorphous precursor phase. Possible biological Implications. Adv. Mater. 12, 38–42 [Google Scholar]
- 36. Bischoff W. D., Mackenzie F. T., Bishop F. C. (1987) Stabilities of synthetic magnesian calcites in aqueous solution. Comparison with biogenic materials. Geochim. Cosmochim. Acta 51, 1413–1423 [Google Scholar]
- 37. Weiner S., Addadi L. (2011) Crystallization pathways in biomineralization. Annu. Rev. Mater. Res. 41, 21–40 [Google Scholar]
- 38. Weiner S., Sagi I., Addadi L. (2005) Choosing the crystallization path less traveled. Science 309, 1027–1028 [DOI] [PubMed] [Google Scholar]
- 39. Mouriès L. P., Almeida M. J., Milet C., Berland S., Lopez E. (2002) Bioactivity of nacre water-soluble organic matrix from the bivalve mollusk Pinctada maxima in three mammalian cell types: fibroblasts, bone marrow stromal cells, and osteoblasts. Comp. Biochem. Physiol. B 132, 217–229 [DOI] [PubMed] [Google Scholar]
- 40. Sud D., Doumenc D., Lopez E., Milet C. (2001) Role of water-soluble matrix fraction, extracted from the nacre of Pinctada maxima, in the regulation of cell activity in abalone mantle cell culture (Haliotis Tuberculata). Tissue Cell 33, 154–160 [DOI] [PubMed] [Google Scholar]
- 41. Albeck S., Weiner S., Addadi L. (1996) Polysaccharides of intracrystalline glycoproteins modulate calcite crystal growth in Vitro. Chem. Eur. J. 2, 278–284 [Google Scholar]
- 42. Fu G., Valiyaveettil S., Wopenka B., Morse D. E. (2005) Caco3 Biomineralization. Acidic 8-kDa proteins isolated from aragonitic abalone shell nacre can specifically modify calcite crystal morphology. Biomacromolecules 6, 1289–1298 [DOI] [PubMed] [Google Scholar]
- 43. Michenfelder M., Fu G., Lawrence C., Weaver J. C., Wustman B. A., Taranto L., Evans J. S., Morse D. E. (2003) Characterization of two molluscan crystal-modulating biomineralization proteins and identification of putative mineral binding domains. Biopolymers 70, 522–533 [DOI] [PubMed] [Google Scholar]
- 44. Gotliv B. A., Addadi L., Weiner S. (2003) Mollusk shell acidic proteins. In search of individual functions. Chembiochem 4, 522–529 [DOI] [PubMed] [Google Scholar]
- 45. Fang D., Xu G., Hu Y., Pan C., Xie L., Zhang R. (2011) Identification of genes directly involved in shell formation and their functions in pearl oyster, Pinctada fucata. Plos ONE 6, e21860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fang D., Pan C., Lin H., Lin Y., Zhang G., Wang H., He M., Xie L., Zhang R. (2012) Novel basic protein, Pfn23, functions as key macromolecule during nacre formation. J. Biol. Chem. 287, 15776–15785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Marin F., Pokroy B., Luquet G., Layrolle P., De Groot K. (2007) Protein mapping of calcium carbonate biominerals by immunogold. Biomaterials 28, 2368–2377 [DOI] [PubMed] [Google Scholar]
- 48. Mount A. S. (2004) Hemocyte-mediated shell mineralization in the eastern oyster. Science 304, 297–300 [DOI] [PubMed] [Google Scholar]
- 49. Ma Z., Huang J., Sun J., Wang G., Li C., Xie L., Zhang R. (2007) A novel extrapallial fluid protein controls the morphology of nacre lamellae in the pearl oyster, Pinctada fucata. J. Biol. Chem. 282, 23253–23263 [DOI] [PubMed] [Google Scholar]
- 50. Xu G. F., Yao N., Aksay I. A., Groves J. T. (1998) Biomimetic synthesis of macroscopic-scale calcium carbonate thin films. Evidence for a multistep assembly process. J. Am. Chem. Soc. 120, 11977–11985 [Google Scholar]
- 51. Marin F., Luquet G. (2004) Molluscan shell proteins. Comptes. Rendus. Palevol. 3, 469–492 [Google Scholar]
- 52. Wilbur K. M., Bernhardt A. M. (1984) Effects of amino acids, magnesium, and molluscan extrapallial fluid on crystallization of calcium carbonate. In vitro experiments. Biol. Bull. 166, 251–259 [Google Scholar]
- 53. Long X., Ma Y., Qi L. (2011) In vitro synthesis of high Mg2+ calcite under ambient conditions and its implication for biomineralization process. Cryst. Growth Des. 11, 2866–2873 [Google Scholar]
- 54. Marie B., Marin F., Marie A., Bédouet L., Dubost L., Alcaraz G., Milet C., Luquet G. (2009) Evolution of nacre. Biochemistry and proteomics of the shell organic matrix of the cephalopod Nautilus macromphalus. Chembiochem. 10, 1495–1506 [DOI] [PubMed] [Google Scholar]
- 55. Marie B., Luquet G., Pais De Barros J.-P., Guichard N., Morel S., Alcaraz G., Bollache L., Marin F. (2007) The shell matrix of the freshwater mussel unio pictorum (Paleoheterodonta, Unionoida). FEBS J. 274, 2933–2945 [DOI] [PubMed] [Google Scholar]
- 56. Nudelman F., Shimoni E., Klein E., Rousseau M., Bourrat X., Lopez E., Addadi L., Weiner S. (2008) Forming nacreous layer of the shells of the bivalves Atrina rigida and Pinctada margaritifera. An environmental- and cryo-scanning electron microscopy study. J. Struct. Biol. 162, 290–300 [DOI] [PubMed] [Google Scholar]
- 57. Pereira-Mouriès L., Almeida M.-J., Ribeiro C., Peduzzi J., Barthélemy M., Milet C., Lopez E. (2002) Soluble silk-like organic matrix in the nacreous layer of the bivalve Pinctada maxima. Eur. J. Biochem. 269, 4994–5003 [DOI] [PubMed] [Google Scholar]
- 58. Miyamoto H., Miyoshi F., Kohno J. (2005) The carbonic anhydrase domain protein nacrein is expressed in the epithelial cells of the mantle and acts as a negative regulator in calcification in the mollusc Pinctada fucata. Zool. Sci. 22, 311–315 [DOI] [PubMed] [Google Scholar]