Background: EF1143 from Enterococcus faecalis is a deoxyribonucleoside triphosphate (dNTP) triphosphohydrolase.
Results: Enzymatic studies and crystal structure of EF1143 bound to dNTPs show that dNTP binding at two adjacent allosteric sites can differentially modulate enzyme catalysis.
Conclusion: EF1143 enzyme catalysis is differentially regulated by four dNTPs.
Significance: Learning how the enzyme activity is regulated by substrates is fundamental for understanding enzyme catalysis and allostery.
Keywords: Allosteric regulation; Enzyme catalysis; Nucleoside nucleotide metabolism; Site directed mutagenesis; X-ray crystallography; dNTPase, dNTP, HD domain, triphosphohydrolase
Abstract
EF1143 from Enterococcus faecalis, a life-threatening pathogen that is resistant to common antibiotics, is a homo-tetrameric deoxyribonucleoside triphosphate (dNTP) triphosphohydrolase (dNTPase), converting dNTPs into the deoxyribonucleosides and triphosphate. The dNTPase activity of EF1143 is regulated by canonical dNTPs, which simultaneously act as substrates and activity modulators. Previous crystal structures of apo-EF1143 and the protein bound to both dGTP and dATP suggested allosteric regulation of its enzymatic activity by dGTP binding at four identical allosteric sites. However, whether and how other canonical dNTPs regulate the enzyme activity was not defined. Here, we present the crystal structure of EF1143 in complex with dGTP and dTTP. The new structure reveals that the tetrameric EF1143 contains four additional secondary allosteric sites adjacent to the previously identified dGTP-binding primary regulatory sites. Structural and enzyme kinetic studies indicate that dGTP binding to the first allosteric site, with nanomolar affinity, is a prerequisite for substrate docking and hydrolysis. Then, the presence of a particular dNTP in the second site either enhances or inhibits the dNTPase activity of EF1143. Our results provide the first mechanistic insight into dNTP-mediated regulation of dNTPase activity.
Introduction
DNA replication requires an appropriate balance of deoxyribonucleoside triphosphate (dNTP)4 pools to minimize mutations (1–3). The cellular levels of four canonical dNTPs (dATP, dGTP, dCTP, and dTTP) are tightly controlled by ribonucleoside diphosphate reductases and dNTP triphosphohydrolases (dNTPases) (4, 5). The former converts ribonucleoside diphosphate or ribonucleoside triphosphate (NTP) to deoxyribonucleoside diphosphate or dNTP (4), and the latter hydrolyzes dNTP into deoxyribonucleoside (dN) and triphosphate (6, 7). The dNTPases also play an important role in the cellular defense against pathogens. In primates, a dGTP-activated dNTPase, sterile alpha motif (SAM) and histidine-aspartate (HD) domain-containing protein 1 (SAMHD1), maintains dNTP pools at low levels, thereby blocking reverse transcription of retroviruses, including HIV and simian immunodeficiency virus, in some cell types (8–13). The virulence factor Vpx in HIV-2 and simian immunodeficiency virus antagonizes SAMHD1 by directing it for proteasome-dependent degradation (9, 10, 14). Similar host dNTPase-pathogen protein antagonist interplay is also observed in prokaryotes; the gene 1.2 product, encoded by bacteriophage T7, inhibits the Escherichia coli dGTPase, allowing productive infection (15, 16).
Enterococcus faecalis is a Gram-positive bacterium found in the intestinal tract of humans. It is a common, life-threatening, hospital-acquired infection, due to frequent mutations that result in the development of antibiotic resistance (17–20). Understanding the molecular mechanisms of E. faecalis pathogenicity and adaptation to antibiotics is important to prevent disease. Several virulence factors encoded by Enterococcus have been identified (21, 22). In this study, we focused on a dNTPase, which controls dNTP pools in the organism.
Biophysical characterization of several dNTPases indicates that they exist as homo-oligomers. For example, crystal structures of the HD domains of human SAMHD1 and bacterial EF1143, a dNTPase from E. faecalis, showed that they are dimer and tetramer, respectively (Protein Data Bank (PDB) IDs 3U1N and 3IRH, respectively) (6, 23). Similarly, 067745_AQUAE, a dNTPase from the prokaryotic organism Aquifex aeolicus, is thought to be active as a tetramer (24), whereas Tt-dNTPase from Thermus thermophilus HB8 is a hexamer (25), based on crystal structures (PDB IDs 2HEK and 2DQB, respectively).
The nature of the homo-oligomeric quaternary states of dNTPases suggests allosteric regulation of their enzymatic activity by effector molecules. Indeed, SAMHD1 undergoes dGTP-dependent tetramerization, which is the catalytically active form of the enzyme (26, 27), and extensive kinetic characterization of the Tt-dNTPase showed that its enzymatic activity was regulated by both dATP and dCTP (28). The most intriguing feature of the biochemically characterized dNTPases, including SAMHD1, Tt-dNTPase, and EF1143, is that dNTPs are both effectors and substrates. Structural comparison of the apo form of EF1143 with a dGTP effector and dATP substrate-bound form uncovered a few important features of its allosteric regulation and catalytic activity. Still, many mechanistic aspects of its complex regulation remained unclear. Here, we present the crystal structure of EF1143 in complex with dGTP and dTTP, bound at two distinctive regulatory sites. The residues at the primary allosteric site are configured to selectively bind only dGTP with high affinity. Detailed biochemical analyses, guided by the crystal structure, suggest that dTTP and dGTP function as negative regulators of the enzyme, whereas dATP and dCTP are positive allosteric effectors, binding to the same promiscuous secondary regulatory site adjacent to the dGTP-binding primary allosteric site. Taken together, these results reveal complex molecular mechanisms of substrate-mediated regulation of EF1143 catalysis.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Cloning of N-terminally His6-tagged E. faecalis HD-domain protein EF1143 was described previously (23). Site-specific mutants of EF1143 were prepared using QuikChange mutagenesis kits (Agilent). Wild-type and mutant EF1143 proteins were expressed in Ros2 (DE3) using auto-induction medium at 18 °C for 24 h (29). The recombinant proteins were purified by two-step liquid column chromatography: a 5-ml nickel-nitrilotriacetic acid column (GE Healthcare) and a Hi-Load Superdex200 16/60 column. Purified proteins were stored in a gel filtration column chromatography buffer containing 25 mm sodium phosphate, pH 7.5, 150 mm NaCl, 1 mm DTT, 10% glycerol, and 0.02% sodium azide.
Protein Crystallization and X-ray Data Collection
The protein stock solution for co-crystallization trials contained 5 mg/ml protein, 1 mm MgCl2, 1.8 mm dGTP, 1.8 mm dTTP, 3.6 mm Tris-HCl, and 250 mm NaCl. Crystal screening was done at room temperature in 96-well Corning crystallization plates. Protein stock was mixed with the crystallization conditions in 1:1 ratio. Diffraction quality crystals belonging to the orthogonal space group P212121 were obtained in 0.1 m buffer containing sodium malonate/imidazole/boric acid (at a 2:3:3 molar ratio), pH 5.0, and 25% w/v PEG 1500 in a 2-μl sitting drop at 22 °C. The crystals were flash-frozen in liquid nitrogen. Diffraction data were collected at 100 K at the LS-CAT (21-ID-F) beamline at the Advanced Photon Source, Argonne National Laboratory. All the data were processed with HKL2000 (30).
Structure Determination and Sequence Analysis
The structure of the EF1143 in complex with dGTP and dTTP was solved by molecular replacement using Phaser (31) and chain A from the EF1143-dGTP-dATP complex as a search model (PDB ID 3IRH). The structure was completed using Coot (32) and REFMAC (33). Crystallographic data and the refinement statistics are in Table 1. Graphical presentations were done with PyMOL (41). Sequence homology analysis was performed using BLAST (34) and ClustalW (35).
TABLE 1.
Data collection and refinement statistics for the E. faecalis EF1143 protein complex with Ni2+, dTTP, and dGTP
r.m.s.d., root mean square deviation.
| Complex with Ni2+/dGTP/dTTP | |
|---|---|
| Data collection | |
| Space group | P212121 |
| a, b, c (Å) | 91.54, 144.63, 155.56 |
| Unique reflection | 81,766 |
| Test set | 4304 |
| Resolution/last shell (Å) | 30-2.35/2.43-2.35 |
| I/σ(I) | 16.8/2.1 |
| Rmergea (%) | 0.07/0.43 |
| Completeness (%) | 99.4/94.9 |
| Overall redundancy | 4.2/2.6 |
| Refinement | |
| Rworkb | 0.184 |
| Rfreec | 0.235 |
| Biso (protein/ligands/water), Å2 | 46.1/43.9/46.4 |
| r.m.s.d. bond length, Å2 | 0.009 |
| r.m.s.d. bond angle, ° | 1.15 |
| Ramachandran distribution | |
| Most favored | 1482 (91.8%) |
| Allowed | 129 (8.0%) |
| Outside allowed | 4 (0.2%) |
| Water molecules | 734 |
| PDB ID | 4LRL |
a Rmerge = ΣhΣi|Ih,i − 〈Ih〉|/ΣhΣiIh,i, where Ih,i is the ith observation of the reflection h, whereas 〈Ih〉 is the mean intensity of reflections h.
b Rwork = Σ‖Fo|− |Fc‖/|Fo|.
c Rfree was calculated with a small fraction (5%) of randomly selected reflections.
dNTPase Assays
EF1143 enzyme activities were studied in a reaction buffer containing 20 mm Tris-HCl, pH 7.8, 50 mm NaCl, 2 mm MgCl2, 5% glycerol, and varying concentrations of dNTPs, as indicated in the figure legends, with 0.1 μm EF1143 and 5 μm BSA. The reactions were quenched with a final concentration of 50 mm EDTA after specific time intervals (2, 4, 6, and 8 min or 4, 8, 12, and 32 min). The products (dNs) and substrates (dNTPs) were separated by reversed phase HPLC using a CAPCELL PAK C18 column (Shiseido Fine Chemicals, 4.6 × 75 mm) and were quantified by peak integration of absorbance traces at 260 nm, as described previously (27).
Analysis of Kinetic Data
The kinetic data were fitted with either the Michaelis-Menten equation (Equation 1) or the allosteric sigmoidal equation (Equation 2)
where v0 is the initial velocity, Vmax is the maximum velocity, Km and Khalf are ligand concentrations at half-maximal velocity, and h is the Hill coefficient.
RESULTS
The Overall Structure of EF1143 in Complex with dGTP and dTTP
The new crystal structure of EF1143 in complex with dGTP and dTTP (PDB ID 4LRL) demonstrates the same tetrameric architecture as the previously reported EF1143 in complex with dGTP and dATP (PDB ID 3IRH, root mean square deviation between the two structures is 0.67 Å over 1631 Cα) and, likely, represents the biological assembly of EF1143 in solution (23). Each EF1143 monomer consists of a bowl-shaped catalytic HD domain (residues 1–329) and a C-terminal subdomain (residues 330–456), shown with pale tones in Fig. 1A. Four dGTP molecules in the 4LRL structure were found at the previously identified allosteric sites in the interfaces between the monomers. The detailed description of these dGTP-binding sites can be found elsewhere (23) and is reiterated in Fig. 1B. Guanine base forms five hydrogen-bonding interactions with the side chains of Asn-36, Gln-41, and Arg-44, providing base type specificity. The specificity for dGTP binding at this site was confirmed by kinetic studies, as described in a later section (see Fig. 5). Surprisingly, however, in the 4LRL structure, the electron density of dTTP was observed next to dGTP, indicating the existence of a secondary allosteric site (supplemental Fig. S1). This contrasts with the 3IRH structure, where dATP was found in the catalytic site.
FIGURE 1.
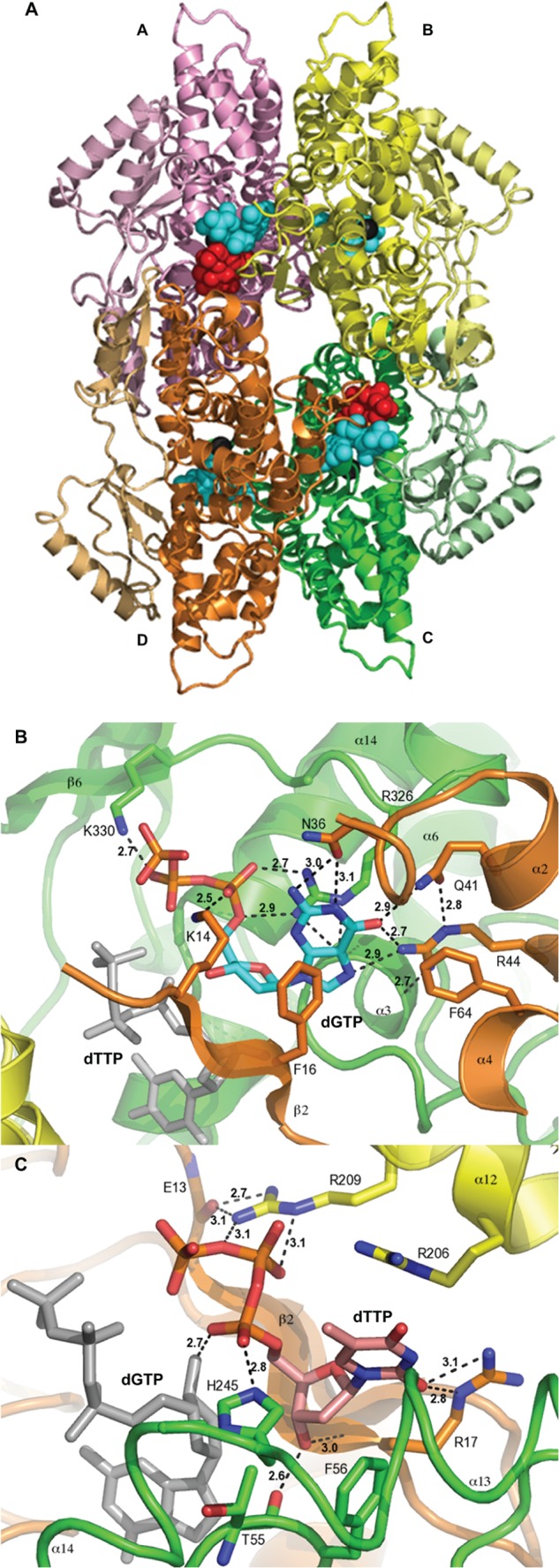
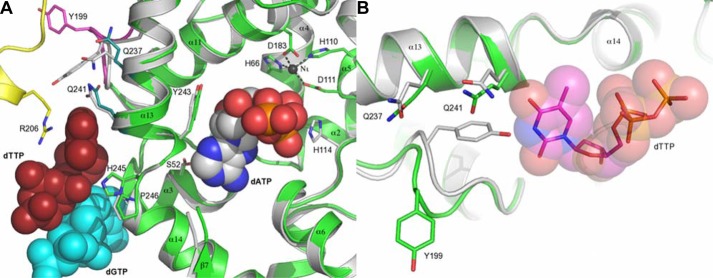
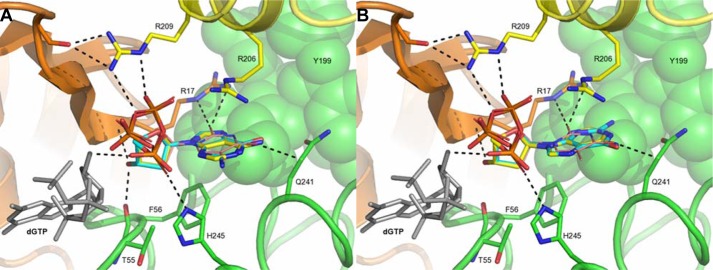
Crystal structure of EF1143 in complex with dGTP and dTTP. A, general view of the EF1143 homo-tetramer bound to dGTP and dTTP (PDB ID: 4LRL). Each monomer is identified with distinct colors and labeled as chains A–D. The N-terminal catalytic domain of each subunit is colored in brighter tones, whereas the C-terminal subdomains are shown in pale shades. Dication metal atoms in the catalytic centers are shown as black spheres. dGTP (cyan spheres) occupies each of four primary allosteric sites, and dTTP (red spheres) occupies two (those of chains A and C) of four secondary regulatory sites. B, dGTP (cyan carbon atoms) binding at the primary allosteric site in 4LRL. Chains B, C, and D are shown in yellow, green, and orange, respectively. dTTP is shown in gray stick representation. H-bonds are depicted by dashed lines, and H-bond lengths are shown. This binding site was also described in our previous publication (23). C, details of the dTTP (pink carbon atoms) interaction at the secondary allosteric site of 4LRL. Chains B, C, and D are depicted as in B. A dGTP molecule, in the primary allosteric site, is shown in gray. Selected side chains are shown and labeled. H-bond lengths are indicated.
FIGURE 5.
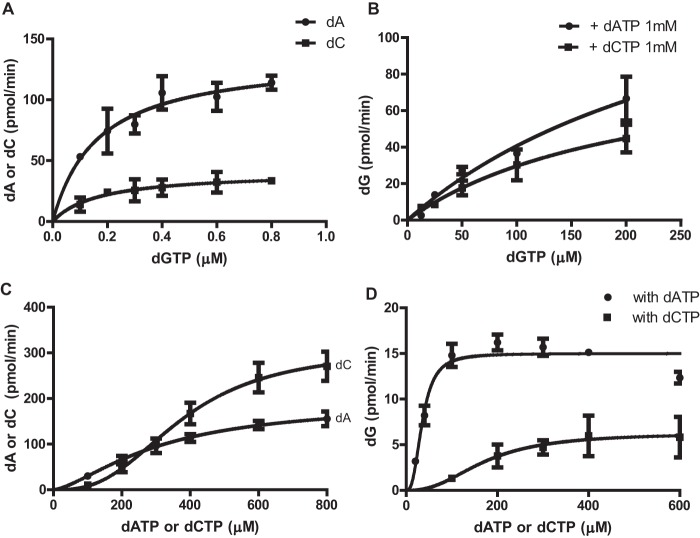
Enzyme kinetics of dGTP-, dATP-, and dCTP-dependent dNTPase activity of EF1143. A, the rate of dATP (circles) or dCTP (squares) conversion was determined in the presence of dGTP (0–0.8 μm), with an initial dATP or dCTP concentration of 1 mm. The data were fitted with the Michaelis-Menten model (Equation 1). Vmax and Km were determined to be 47.0 ± 3.4 nmol/nmol·min and 0.17 ± 0.05 μm, respectively, for dATP, and 11.5 ± 1.7 nmol/nmol·min and 0.17 ± 0.08 μm, respectively, for dCTP. B, the rate of dGTP conversion was determined with the initial dGTP concentration ranging from 0 to 200 μm and a fixed concentration (1 mm) of dATP (circles) or dCTP (squares). Vmax and Km were determined to be 47.3 ± 17.8 nmol/nmol·min and 315 ± 170 μm, respectively, for the dGTP/dATP mixture, and 25.9 ± 7.3 nmol/nmol·min and 213 ± 98 μm, respectively, for the dGTP/dCTP mixture when the data were fitted with the Michaelis-Menten model (Equation 1). C, the rate of dATP (circles) or dCTP (squares) conversion was determined in the presence of increasing concentrations of dATP (circles) or dCTP (squares) and a fixed concentration of dGTP (16 μm). The data were fitted with an allosteric sigmoidal model (Equation 2). Vmax, Khalf, and Hill coefficient (h) were determined to be 54.1 ± 7.0 nmol/nmol·min, 310 ± 63 μm, and 1.5 ± 0.3, respectively, for dATP. The kinetic values for dCTP were 86.2 ± 7.8 nmol/nmol·min, 376 ± 36 μm, and 2.7 ± 0.5, respectively. D, the rate of dG production was also determined from the experiments performed in C, and the data were fitted with an allosteric sigmoidal model. Vmax, Khalf, and h for dGTPase activity in the presence of dATP (circles) were determined to be 4.2 ± 0.1 nmol/nmol·min, 35 ± 4 μm, and 2.7 ± 0.7, respectively. In the presence of varying dCTP concentrations (squares), the kinetic values for dGTPase activity were 1.8 ± 0.3 nmol/nmol·min, 172 ± 46 μm, and 2.4 ± 1.4, respectively. Error bars, S.D.
There are four identical secondary allosteric sites per tetramer, but only two were occupied by dTTP in 4LRL. Several distinct features typify the site (Fig. 1C). First, residues belonging to three different chains (particularly, B, C, and D) form the binding pocket. Second, the site appears to be specific for the deoxyribose moiety, with the 3′-hydroxyl group interacting with the main chain atoms of Thr-55 (chain C) and Arg-17 (chain D). The substitution of the deoxyribose with the ribose moiety would result in structural clash between the 2′-hydroxyl group and Phe-56 (chain C). Third, the triphosphate moiety, specifically, oxygen atoms of the Pα and Pβ groups, is recognized by His-245 of chain C and Arg-209 of chain B. Interestingly, one of the Pα oxygen atoms also has an H-bond with the deoxyribose hydroxyl group of dGTP. Fourth, the guanidinium group of Arg-17 in chain D interacts with the ortho-carbonyl group of thymine. The fact that only one of the three functional groups of the dTTP base is involved in H-bonding suggests the promiscuous nature of the secondary site. This is in stark contrast to the specific dGTP binding mode at the primary allosteric site. The promiscuity of this site may play a key role in regulation of the hydrolase activity, depending on the presence of particular dNTPs as elaborated in later sections.
dTTP Binding Alters the Geometry of EF1143 near the Active Site
Superposition of chain B of 3IRH and chain C of 4LRL (Fig. 2A) reveals a potential structural mechanism for modulation of the dNTPase activity by dTTP binding to the secondary allosteric site. The dTTP-binding site is separated from the catalytic site by a helix-turn-helix fragment comprising α13 … α14. The phenol plane of residue Tyr-243, in the α13 … α14 loop, is in close proximity to the deoxyribose moiety of dATP. The position of this residue would regulate the location of the Pα group, a nucleophilic attack target of the hydroxide ion stabilized by His-114 (supplemental Fig. S2). The distance between Cγ of Tyr-243 and C1 of the dATP ribose in 3IRH is 3.72 Å, on average, whereas the same positions are 3.57 Å apart, on average, in the substrate-free forms when dATP is modeled in the structure. This suggests, first, that the α13 … α14 helix-turn-helix fragment is mobile and, second, that dATP binding tends to expand the binding site. In contrast to the dATP effect, dTTP binding would change the Cγ … C1 distance to an average of 3.32 Å. This 0.40 Å shortening could be directly linked to the dTTP steric contact with residue Gln-241 on the opposite side of α13, causing Tyr-243 to shift into the catalytic site. Thus, dTTP binding results in contraction of the catalytic site.
FIGURE 2.
Modeling of EF1143 activity regulation by dGTP/dTTP binding at the allosteric sites. A, superposition of chain C (green) from 4LRL (EF1143/dGTP/dTTP) with chain B (gray) of 3IRH (EF1143/dGTP/dATP) using the H … HD … D coordination motifs. Chain B (yellow), dGTP (cyan spheres), and dTTP (red spheres) in 4LRL, as well as dATP (gray/red/blue spheres), are shown. Selected side chains are shown and labeled. Note that chain B of 3IRH, rather than chain C, was used for the superposition because chain C does not contain dATP at the active site. B, superposition of chain C (green) from 4LRL with chain A (gray) of 3IRH. Binding of the dTTP molecule (represented by spheres and sticks) to chain C (4LRL) changes loop 196–202 and, particularly, the configuration of Tyr-199, which otherwise would clash with dTTP.
An additional structural change related to dTTP binding at the second allosteric site is the movement of Tyr-199 and the loop that includes residues 195–202 (Fig. 2B). In the absence of ligand, Tyr-199 protrudes into the allosteric site. Thus, in the dTTP-bound state, the 195–202 loop must change its conformation, resulting in an almost 6.9 Å movement of the Tyr-199 Cγ position out of the secondary allosteric site. This new loop conformation is acceptable but more energetically strenuous, as suggested by the Ramachandran plot distribution; in the dTTP-bound state, three residues move from the preferred into the allowed regions (data not shown). This observation suggests that Tyr-199 and the associated loop could play the role of a “spring” that ensures the reversibility of ligand binding to the secondary allosteric site.
Mutational Analysis Combined with Activity Assays Correlates with the Structural Features
Our initial biochemical analysis demonstrated that, in the presence of Mg2+, the hydrolytic activity of EF1143 depended on the presence of at least two different dNTPs, one of which had to be dGTP (23). In this study, we measured the dATPase activity of EF1143 in the presence of both dATP and dGTP, with a dATP:dGTP ratio of 1250:1. To confirm the importance of several critical residues revealed from the 3IRH and 4LRL structures, several mutants were generated and tested for dATPase activity (Fig. 3). First, analysis of 3IRH, with dATP at the active site, suggested that the catalytic residues comprise a Glu-122-His-129 dyad plus His-114, which together facilitate the in-line nucleophilic attack of a hydrolytic water molecule at the α-phosphate center of the dNTP substrate (supplemental Fig. S2). Negligible catalytic activity of the H114A and H129A mutants supports this hypothesis (Fig. 3). Second, dGTP binding at the primary allosteric site features two distinctive patterns (Fig. 1B): ionic interactions between the triphosphate and basic residues (Lys-14, Arg-326, and Lys-330) and hydrogen bonding between the guanine base and hydrophilic residues (Asn-36, Gln-41 and Arg-44). All these residues (except Arg-44) were mutated, individually, to Ala, and their dATPase activities were compared. With the exception of R326A, all the mutants were significantly defective in dNTP hydrolysis. Finally, residues observed to interact with dTTP in 4LRL, at the second allosteric site, were mutated. Although mutation at Arg-17, Tyr-199, or Gln-241 resulted in significant loss of dATPase activity, R206Q or R209S showed only a moderate decrease in hydrolysis. However, double mutation of Arg-206 and Arg-209 resulted in complete loss of the EF1143 catalytic activity. Taken together, this mutagenesis analysis confirms the importance of residues observed to be involved in ligand binding at the catalytic site and the two allosteric sites.
FIGURE 3.
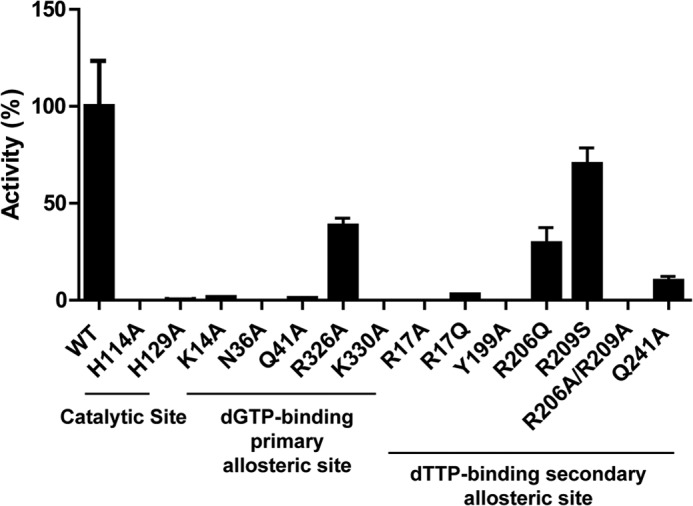
Mutational analysis of EF1143 validates the structure. Wild-type (WT) or mutant EF1143 (0.1 μm) was incubated in a reaction mixture containing 0.8 μm dGTP and 1 mm dATP. The rate of dATP conversion to dA was calculated for each mutant and is presented as the percentage of activity relative to WT. Error bars, S.D.
Survey of dNTPase Activity with Combinations of dNTPs
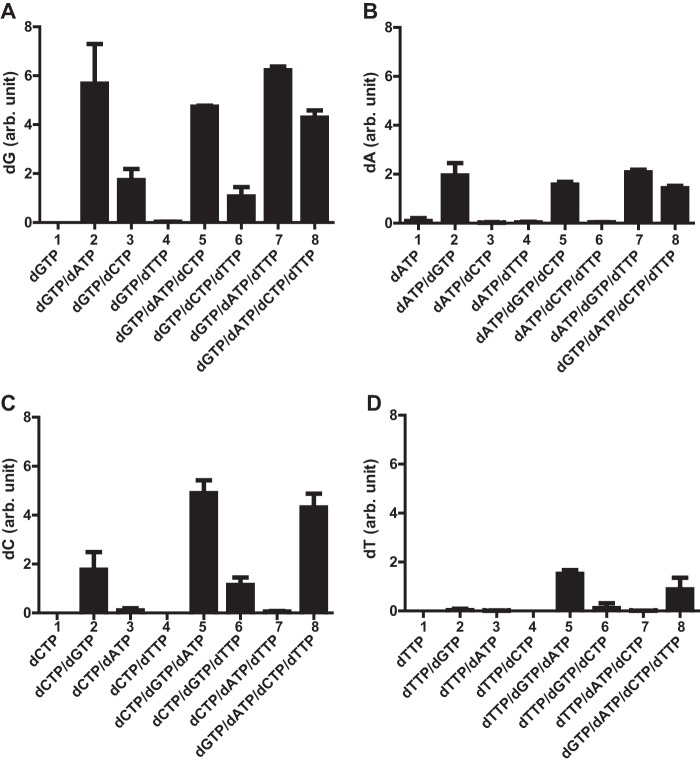
Our discovery of the promiscuous second allosteric site in 4LRL prompted us to speculate that the enzymatic activity of EF1143 is differentially regulated, depending on which dNTP occupies this regulatory site. To gain more insight into this, we first measured the dNTPase activity of EF1143 against each dNTP, in the absence or presence of one, two, or three additional dNTPs (Fig. 4). Consistent with our previous study (23), EF1143 failed to hydrolyze any of the dNTPs when a sole dNTP was present in the reaction mixture (Fig. 4, A–D, column 1 in each). Next, the data confirmed our previous results that, among the four dNTPs, only dGTP could function as an activator of dATP and dCTP hydrolysis (Fig. 4, B and C, columns 2–4). When the rate of dGTP hydrolysis was compared among reaction mixtures containing two dNTP types, dGTP hydrolysis was clearly enhanced by the presence of dATP or dCTP, but not dTTP, with dATP being the more potent activator (Fig. 4A, columns 2–4). Further, dTTP hydrolysis was virtually nonexistent when any one other dNTP was combined with dTTP in the reaction mixture (Fig. 4D, columns 2–4). This observation is consistent with 4LRL, where dTTP binding at the second allosteric site shrinks the catalytic site. However, dTTP hydrolysis did occur when both dGTP and dATP were present (Fig. 4D, column 5), suggesting that both dATP and dGTP are co-activators for hydrolysis of dTTP, with dGTP as the primary activator. In contrast, dTTP does not activate or enhance the enzyme's activity against any dNTP. For example, the addition of dTTP to reaction mixtures that contained dGTP and dATP or dCTP had no effect on the hydrolysis of dATP (Fig. 4B, column 2 versus column 7) or dCTP (Fig. 4C, column 2 versus column 6) at the initial concentrations tested. With all four dNTPs present at equal concentrations, the order of dNTP hydrolysis was dGTP > dCTP > dATP > dTTP (Fig. 4, A–D, compare column 8 from each panel). Based on all these comparisons, we conclude that dGTP is a primary activator because appreciable dNTP hydrolysis was not observed in any reaction mixture in the absence of dGTP. This conclusion is consistent with the crystal structures of EF1143, with dGTP at the primary allosteric site (Fig. 1). In addition, dATP is a stronger secondary activator than dCTP because the rates of dGTP and dTTP hydrolysis were higher with dATP in the reaction mixture as compared with those with dCTP.
FIGURE 4.
dNTPase activities of EF1143 in various mixtures of dNTPs. EF1143 (0.1 μm) was incubated with various combinations of dNTP (100 μm each), as indicated on the abscissa. A–D, the rates of dGTP conversion to dG (A), dATP to dA (B), dCTP to dC (C), and dTTP to dT (D) in each condition are shown on the ordinate. The individual rates were normalized to the dT conversion rate in the presence of all four dNTP (column 8 in D). Error bars, S.D. arb. unit, arbitrary unit.
Determination of Kinetic Constants of dGTP at the Allosteric Site and Catalytic Site
Analysis of the two crystal structures (3IRH and 4LRL), along with our dNTPase activity survey (Fig. 4), suggests that dGTP binding at the primary allosteric site is a prerequisite for the dATP- and dCTP-dependent activation of EF1143. To determine the binding constant for dGTP at the allosteric site, the rates of dATP and dCTP hydrolysis were determined in the presence of increasing concentrations of dGTP (Fig. 5A). Both sets of data were fitted with the Michaelis-Menten model (Equation 1), suggesting a simple model of dGTP binding at the primary allosteric site. Interestingly, the Km values were similar for both dATP and dCTP (0.17 ± 0.05 and 0.17 ± 0.08 μm, respectively). This suggests that dGTP binding at the allosteric site is independent of dATP and dCTP binding at the catalytic site. Further kinetic analyses, with higher dGTP concentrations, suggested that dGTP binding at the catalytic site also follows the Michaelis-Menten model (Equation 1) (Fig. 5B). The rates of dATP and dCTP hydrolysis were moderately constant over the range of dGTP concentrations examined (supplemental Fig. S3). Taken together, the data suggest that dGTP has a strong (∼2000-fold) preference for the allosteric site as compared with the catalytic site.
dATP and dCTP Allosterically Activate the dNTPase
To further understand the activation features of EF1143 by dATP and dCTP (Fig. 4), dNTPase assays were performed with increasing concentrations of dATP or dCTP in a reaction mixture containing a fixed concentration of dGTP, 16 μm, at which the primary allosteric site of the enzyme is fully occupied by dGTP (Kd of dGTP at the primary allosteric site is 0.17 μm). When the rates of dA and dC accumulation were plotted as a function of dATP and dCTP concentrations, respectively, the data were best fitted with sigmoidal curves (Equation 2) (Fig. 5C), indicating allostery. The rate of dC accumulation was more sensitive to dCTP concentration than the rate of dA accumulation was to dATP, resulting in a higher Hill coefficient value and Vmax for dCTP than dATP. When the rate of dG accumulation was plotted, dATP was far superior to dCTP in allosteric activation of dGTP hydrolysis (Fig. 5D). These data support the observation that dATP is a more potent activator of dGTP hydrolysis than dCTP, as seen when all dNTPs were present at 100 μm (Fig. 4A, columns 2 and 3). Further, at 100 μm, the rate of dA accumulation is slightly higher than that of dC accumulation (Fig. 5C), which also agrees with the data presented in Fig. 4. Taken together, the results show that dATP and dCTP allosterically activate the dNTPase with a different cooperativity for substrates. Our structural modeling suggests that both dATP and dCTP might bind at the secondary allosteric site, which is occupied by dTTP in 4LRL (see the section below).
dTTP and dGTP Inhibits dNTPase Activity at High Concentration
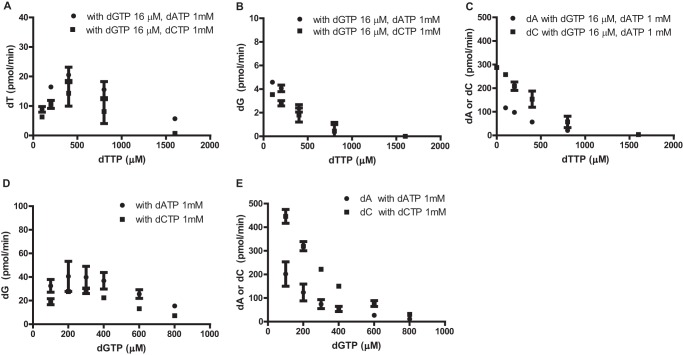
Comparison of crystal structures 3IRH and 4LRL (Fig. 2) suggests that dTTP binding at the secondary allosteric site could negatively influence EF1143 catalytic activity (Fig. 4). To further investigate this possibility, we measured the rate of dT accumulation in increasing concentrations of dTTP with a fixed concentration of dGTP and dATP or dCTP (Fig. 6A). Note that hydrolysis of dTTP can only be observed in the presence of both dGTP and dATP or dCTP (Fig. 4D). The rate of dTTP hydrolysis increased with increasing concentrations of dTTP, up to 400 μm, and then gradually declined at higher concentrations. Strikingly, no conversion of dTTP was observed at 1600 μm dTTP with dGTP and dCTP in the reaction mixture. Similar results were observed when the rates of dG, dA, and dC conversion were monitored with increasing concentrations of dTTP (Fig. 6, B and C).
FIGURE 6.
dTTP and dGTP at high concentrations inhibit dNTPase activity of EF1143. The dNTPase activity of EF1143 was determined in the presence of increasing concentrations of dTTP (0–1600 μm) and fixed concentrations of dGTP (16 μm) and dATP (1 mm) or dCTP (1 mm). A–C, the rates of dTTP (A), dGTP (B), and dATP and dCTP (C) conversion were plotted relative to dTTP concentration. D, the rate of dGTP triphosphohydrolase activity was determined in the presence of increasing concentrations of dGTP (100–800 μm) in the presence of either 1 mm dATP or 1 mm dCTP. E, the rate of dATP (circles) or dCTP (squares) conversion is plotted for the experiments performed in D. Error bars, S.D.
The 4LRL structure provides mechanistic insight into the inhibition mechanism by dTTP. The ligand interacts with residue Gln-241 of helix α13, shifting Tyr-243 toward the catalytic binding site by ∼0.4 Å (Fig. 2A). This movement could either prevent substrate binding or disrupt correct positioning of the dNTP for nucleophilic attack. Modeling of dGTP ligand binding at the second allosteric site (Fig. 7) suggests that it could exert a similar effect on helix α13/Gln-241, inhibiting enzyme catalysis. Consistent with this notion, at high dGTP concentrations (greater than 200 μm), the dNTPase was inhibited (Fig. 6, D and E). The dCTP molecule should be able to bind to the secondary allosteric site in a mode similar to that of dTTP. However, the deoxycytidine, in particular the amino group in para-position, would clash with Gln-241 if dCTP binds exactly in the same conformation as observed for dTTP in 4LRL. A similar observation could be made for dATP in either anti-conformation or syn-conformation (Fig. 7). Despite these caveats, the binding pocket appears to be large enough to allow cytosine/adenine plane rotation, for example, toward the Arg-236 residue that would prevent collision with Gln-241. The reduced, but still significant activity of the Q241A mutant in the dGTP/dATP mixture (Fig. 3) may be interpreted to support this hypothesis. Therefore, the steric difference exerted by the amino group in dCTP/dATP and the carbonyl group in the analogous position of dTTP/dGTP may differently affect the position of helix α13 and residue Tyr-243 in the catalytic site, thus, switching the enzyme between the inhibited and the activated states. Taken altogether, the kinetic data support a modulatory role of the secondary allosteric site upon binding to different dNTPs.
FIGURE 7.
Modeling of dNTP binding at the secondary allosteric site. A, modeling of the dGTP (cyan carbon atoms) and dATP (yellow carbon atoms) molecules in syn-conformation into the secondary allosteric site, formed by chains B (yellow), C (green), and D (orange) in 4LRL. The dGTP molecule in the primary allosteric site is shown as gray sticks, whereas the dTTP molecule in the second site is shown as pink/blue/red lines. H-bonding networks for the dTTP molecule are shown as black dashes, as well as the shortest distance (3.1 Å) between dTTP and Gln-241. In both syn-configuration and anti-configuration (see panel B), the dGTP molecule carbonyl group positions in the same place as the para-carbonyl in dTTP, whereas in the case of dATP, the bulkier amino group occupies this space. This suggests that dGTP might bind to the secondary allosteric site and exert the same inhibitory effect as dTTP. In contrast, dATP would, probably, rotate toward Arg-206 (to avoid the clash with Gln-241), which could weaken its influence on the active site. B, modeling of dGTP and dATP, in anti-conformation, into the secondary allosteric site. Color-coding is identical to panel A. H-bonding network and shortest distance to Gln-241 for the dTTP molecule are shown as black dashes within 3.1 Å distance.
DISCUSSION
The proper balance of cellular dNTP pools is critical for maintaining genome stability. The central role of ribonucleoside diphosphate reductases in this has been well documented (4). A recent study of SAMHD1, a mammalian dNTPase, suggested its essential role in balancing dNTP pools in concert with the ribonucleoside diphosphate reductase (5). We and others elucidated the structural basis of dGTP-dependent SAMHD1 tetramerization and allosteric activation (36, 37). In this study, we present a crystal structure of EF1143 in complex with dGTP and dTTP (4LRL) and provide insight into an elegant allosteric mechanism of dNTP-mediated inhibition of enzyme catalysis.
Comparison of 4LRL with the apo-EF1143 (2O6I) and dGTP/dATP bound forms (3IRH), along with extensive enzyme kinetic studies, presents a qualitative model for EF1143 allosteric regulation (Fig. 8). dGTP binding at the primary allosteric site is necessary for hydrolysis of the canonical dNTP substrates. Analysis of the N36A and Q41A mutants confirmed the critical importance of the guanine base-recognizing residues (Fig. 3). The enzyme kinetic analysis indicated high specificity of the primary allosteric site for dGTP (Fig. 5A). Taken together, binding of dGTP could be considered an enzyme preactivation step. Then, the enzyme can be further activated or inhibited by dNTPs binding to the secondary allosteric site, with different affinities. If the site is occupied by dATP or dCTP, the enzyme becomes catalytically active (Fig. 5, C and D). With dGTP or dTTP bound at the site, EF1143 is negatively regulated (Fig. 6). The physiological concentrations of dNTPs in prokaryotes are in the range of 100–300 μm and fluctuate several folds during bacteriophage T4 infection (38–40). At these concentrations, the primary allosteric site should be fully occupied with dGTP and substrate hydrolysis at the catalytic site could be modulated by different dNTPs binding at the secondary allosteric sites.
FIGURE 8.
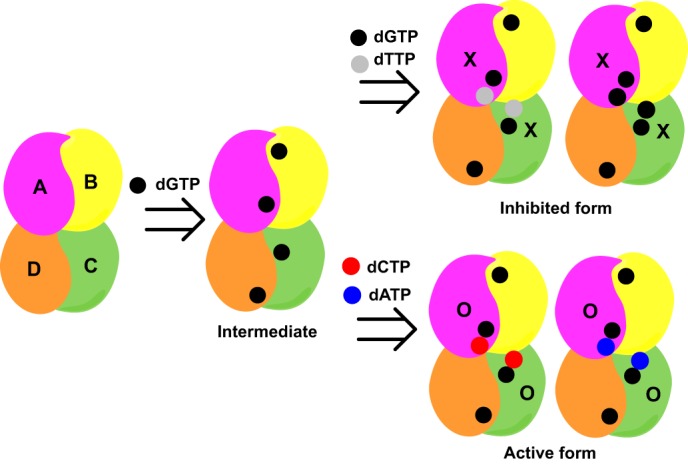
Model of dNTP-dependent allosteric regulation of EF1143 catalysis. dGTP binds to each of four primary allosteric sites (Intermediate). Binding of either dTTP or dGTP at the two secondary allosteric sites, adjacent to the primary site, results in an inhibited form, by closing the catalytic site (X), whereas binding of dATP or dCTP at the same secondary sites activates the enzyme (O).
To assess whether the proposed mechanism of EF1143 regulation by dNTPs could serve as a general model for homo-tetrameric dNTPases, we compared the amino acid sequences of EF1143 with 43 of its closest homologs, mostly from Enterococcus and Lactobacillus species (at least 97% coverage and 65% identity, supplemental Fig. S4). Among the closest sequence homologs, most of the critical residues in the catalytic and two allosteric sites are 100% conserved (supplemental Fig. S4, red boxes). We also carried out a structure-assisted comparison of EF1143 and SAMHD1 (supplemental Fig. S5). The human SAMHD1 dNTPase (sequence identity 36% over 274 residues) is a structural homolog of EF1143. For the conservation analysis, we superimposed SAMHD1 chain A from 4BZB (dGTP-bound tetrameric form) to chain C of EF1143 tetramer (supplemental Fig. S5A). Overall, all the dNTP-binding sites between EF1143 and SAMHD1 are highly conserved. At the dGTP-specific primary allosteric site, two critical residues for EF1143 preactivation, Arg-44 and Arg-326, have Arg-145 and Arg-451 as their counterparts in SAMHD1 (supplemental Fig. S5B). Of note, SAMHD1 R145A mutation resulted in less than 1% of wild-type dNTPase activity (6). Interestingly, hydrogen-bonding interactions between the base of deoxyribonucleoside triphosphate and surrounding residues in both enzymes suggest a promiscuity of the secondary allosteric site to other dNTPs (Fig. 1C) (36, 37). The catalytic residues Glu-122-His-129-His-114 in EF1143 correspond to Asp-218-His-233-His-210 in SAMHD1 (supplemental Fig. S5C). Thus, the hydrolysis in both enzymes may proceed in a similar manner, through the nucleophilic attack of the activated water species (hydroxide ion) on the Pα center.
Despite the structural similarities between EF1143 and SAMHD1, several important differences exist. Although both apo-bound and dGTP-bound EF1143 are tetramers in solution (23), SAMHD1 undergoes tetramerization upon dGTP binding (26, 27). Further, different dNTP binding at the secondary allosteric site of EF1143 can result in either active or inhibited forms, whereas no such regulation has been reported for SAMHD1.
A complete understanding of EF1143 regulation by dNTPs requires further work. Although our hypothesis that the switch between the active and inhibited modes could be regulated by binding of different ligands to the promiscuous secondary allosteric site agrees with the biochemical data, our suggestion is primarily based on modeling into the rigid protein structure. To prove that dGTP, dATP, and dCTP indeed bind to the secondary allosteric site, more crystallization experiments with different dNTP mixtures are needed. The issue also could be approached theoretically, using flexible molecular docking, molecular dynamics, or Monte-Carlo simulations, using our structures as a starting point. Furthermore, in 3IRH, only two out of four catalytic sites were occupied by dATP, and only two out of four secondary regulatory sites in 4LRL have dTTP. This might be an artifact of crystal packing but could also suggest communication between monomers in the tetrameric assembly. Finally, the function of the prominent and highly conserved C terminus of EF1143 remains unknown. These three structural aspects are currently under investigation.
Acknowledgment
We thank Dr. Teresa Brosenitsch for careful reading of the manuscript and editorial help.
This work was supported in part by National Institutes of Health Grant U54 GM094585 for the Midwest Center for Structural Genomics (to W. F. A.) and Grant P50GM082251 (to J. A.). This work was also supported by funds from the University of Pittsburgh School of Medicine (to J. A.).
This article was selected as a Paper of the Week.

This article contains supplemental Figs. S1–S5.
The atomic coordinates and structure factors (code 4LRL) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- dNTP
- deoxyribonucleoside triphosphate
- dNTPase
- deoxyribonucleoside triphosphate triphosphohydrolase
- dATPase
- dATP triphosphohydrolase
- SAMHD1
- sterile alpha motif and HD domain-containing protein 1
- HD domain
- histidine-aspartate domain.
REFERENCES
- 1. Kunkel T. A. (1992) DNA replication fidelity. J. Biol. Chem. 267, 18251–18254 [PubMed] [Google Scholar]
- 2. Kunz B. A., Kohalmi S. E., Kunkel T. A., Mathews C. K., McIntosh E. M., Reidy J. A. (1994) International Commission for Protection Against Environmental Mutagens and Carcinogens. Deoxyribonucleoside triphosphate levels: a critical factor in the maintenance of genetic stability. Mutat. Res. 318, 1–64 [DOI] [PubMed] [Google Scholar]
- 3. Kumar D., Abdulovic A. L., Viberg J., Nilsson A. K., Kunkel T. A., Chabes A. (2011) Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools. Nucleic Acids Res. 39, 1360–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nordlund P., Reichard P. (2006) Ribonucleotide reductases. Annu. Rev. Biochem. 75, 681–706 [DOI] [PubMed] [Google Scholar]
- 5. Franzolin E., Pontarin G., Rampazzo C., Miazzi C., Ferraro P., Palumbo E., Reichard P., Bianchi V. (2013) The deoxynucleotide triphosphohydrolase SAMHD1 is a major regulator of DNA precursor pools in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 110, 14272–14277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goldstone D. C., Ennis-Adeniran V., Hedden J. J., Groom H. C., Rice G. I., Christodoulou E., Walker P. A., Kelly G., Haire L. F., Yap M. W., de Carvalho L. P., Stoye J. P., Crow Y. J., Taylor I. A., Webb M. (2011) HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382 [DOI] [PubMed] [Google Scholar]
- 7. Powell R. D., Holland P. J., Hollis T., Perrino F. W. (2011) Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J. Biol. Chem. 286, 43596–43600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Berger A., Sommer A. F., Zwarg J., Hamdorf M., Welzel K., Esly N., Panitz S., Reuter A., Ramos I., Jatiani A., Mulder L. C., Fernandez-Sesma A., Rutsch F., Simon V., König R., Flory E. (2011) SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutieres syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 7, e1002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hrecka K., Hao C., Gierszewska M., Swanson S. K., Kesik-Brodacka M., Srivastava S., Florens L., Washburn M. P., Skowronski J. (2011) Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Laguette N., Sobhian B., Casartelli N., Ringeard M., Chable-Bessia C., Ségéral E., Yatim A., Emiliani S., Schwartz O., Benkirane M. (2011) SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baldauf H. M., Pan X., Erikson E., Schmidt S., Daddacha W., Burggraf M., Schenkova K., Ambiel I., Wabnitz G., Gramberg T., Panitz S., Flory E., Landau N. R., Sertel S., Rutsch F., Lasitschka F., Kim B., König R., Fackler O. T., Keppler O. T. (2012) SAMHD1 restricts HIV-1 infection in resting CD4+ T cells. Nat. Med. 18, 1682–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Descours B., Cribier A., Chable-Bessia C., Ayinde D., Rice G., Crow Y., Yatim A., Schwartz O., Laguette N., Benkirane M. (2012) SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4+ T-cells. Retrovirology 9, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. St Gelais C., de Silva S., Amie S. M., Coleman C. M., Hoy H., Hollenbaugh J. A., Kim B., Wu L. (2012) SAMHD1 restricts HIV-1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4+ T-lymphocytes cannot be upregulated by interferons. Retrovirology 9, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ahn J., Hao C., Yan J., DeLucia M., Mehrens J., Wang C., Gronenborn A. M., Skowronski J. (2012) HIV/simian immunodeficiency virus (SIV) accessory virulence factor Vpx loads the host cell restriction factor SAMHD1 onto the E3 ubiquitin ligase complex CRL4DCAF1. J. Biol. Chem. 287, 12550–12558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nakai H., Richardson C. C. (1990) The gene 1.2 protein of bacteriophage T7 interacts with the Escherichia coli dGTP triphosphohydrolase to form a GTP-binding protein. J. Biol. Chem. 265, 4411–4419 [PubMed] [Google Scholar]
- 16. Wurgler S. M., Richardson C. C. (1990) Structure and regulation of the gene for dGTP triphosphohydrolase from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 87, 2740–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wisplinghoff H., Seifert H., Wenzel R. P., Edmond M. B. (2003) Current trends in the epidemiology of nosocomial bloodstream infections in patients with hematological malignancies and solid neoplasms in hospitals in the United States. Clin. Infect. Dis. 36, 1103–1110 [DOI] [PubMed] [Google Scholar]
- 18. National Nosocomial Infections Surveillance System (2004) National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2004, issued October 2004. Am. J. Infect Control 32, 470–485 [DOI] [PubMed] [Google Scholar]
- 19. de Fátima Silva Lopes M., Ribeiro T., Abrantes M., Figueiredo Marques J. J., Tenreiro R., Crespo M. T. (2005) Antimicrobial resistance profiles of dairy and clinical isolates and type strains of enterococci. Int. J. Food Microbiol. 103, 191–198 [DOI] [PubMed] [Google Scholar]
- 20. Rosenthal V. D., Bijie H., Maki D. G., Mehta Y., Apisarnthanarak A., Medeiros E. A., Leblebicioglu H., Fisher D., Álvarez-Moreno C., Khader I. A., Del Rocío González Martínez M., Cuellar L. E., Navoa-Ng J. A., Abouqal R., Guanche Garcell H., Mitrev Z., Pirez García M. C., Hamdi A., Dueñas L., Cancel E., Gurskis V., Rasslan O., Ahmed A., Kanj S. S., Ugalde O. C., Mapp T., Raka L., Yuet Meng C., Thu le T. A., Ghazal S., Gikas A., Narváez L. P., Mejía N., Hadjieva N., Gamar Elanbya M. O., Guzmán Siritt M. E., Jayatilleke K., and INICC members (2012) International Nosocomial Infection Control Consortium (INICC) report, data summary of 36 countries, for 2004–2009. Am. J. Infect. Control 40, 396–407 [DOI] [PubMed] [Google Scholar]
- 21. Fisher K., Phillips C. (2009) The ecology, epidemiology and virulence of Enterococcus. Microbiology 155, 1749–1757 [DOI] [PubMed] [Google Scholar]
- 22. Sava I. G., Heikens E., Huebner J. (2010) Pathogenesis and immunity in enterococcal infections. Clin. Microbiol. Infect. 16, 533–540 [DOI] [PubMed] [Google Scholar]
- 23. Vorontsov I. I., Minasov G., Kiryukhina O., Brunzelle J. S., Shuvalova L., Anderson W. F. (2011) Characterization of the deoxynucleotide triphosphate triphosphohydrolase (dNTPase) activity of the EF1143 protein from Enterococcus faecalis and crystal structure of the activator-substrate complex. J. Biol. Chem. 286, 33158–33166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oganesyan V., Adams P. D., Jancarik J., Kim R., Kim S. H. (2007) Structure of O67745_AQUAE, a hypothetical protein from Aquifex aeolicus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63, 369–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kondo N., Nakagawa N., Ebihara A., Chen L., Liu Z. J., Wang B. C., Yokoyama S., Kuramitsu S., Masui R. (2007) Structure of dNTP-inducible dNTP triphosphohydrolase: insight into broad specificity for dNTPs and triphosphohydrolase-type hydrolysis. Acta Crystallogr. D Biol. Crystallogr. 63, 230–239 [DOI] [PubMed] [Google Scholar]
- 26. DeLucia M., Mehrens J., Wu Y., Ahn J. (2013) HIV-2 and SIVmac accessory virulence factor Vpx down-regulates SAMHD1 enzyme catalysis prior to proteasome-dependent degradation. J. Biol. Chem. 288, 19116–19126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yan J., Kaur S., DeLucia M., Hao C., Mehrens J., Wang C., Golczak M., Palczewski K., Gronenborn A. M., Ahn J., Skowronski J. (2013) Tetramerization of SAMHD1 is required for biological activity and inhibition of HIV infection. J. Biol. Chem. 288, 10406–10417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kondo N., Kuramitsu S., Masui R. (2004) Biochemical characterization of TT1383 from Thermus thermophilus identifies a novel dNTP triphosphohydrolase activity stimulated by dATP and dTTP. J. Biochem. 136, 221–231 [DOI] [PubMed] [Google Scholar]
- 29. Li Z., Kessler W., van den Heuvel J., Rinas U. (2011) Simple defined autoinduction medium for high-level recombinant protein production using T7-based Escherichia coli expression systems. Appl. Microbiol. Biotechnol. 91, 1203–1213 [DOI] [PubMed] [Google Scholar]
- 30. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 31. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 33. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 34. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 35. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 36. Zhu C., Gao W., Zhao K., Qin X., Zhang Y., Peng X., Zhang L., Dong Y., Zhang W., Li P., Wei W., Gong Y., Yu X. F. (2013) Structural insight into dGTP-dependent activation of tetrameric SAMHD1 deoxynucleoside triphosphate triphosphohydrolase. Nat. Commun. 4, 2722. [DOI] [PubMed] [Google Scholar]
- 37. Ji X., Wu Y., Yan J., Mehrens J., Yang H., Delucia M., Hao C., Gronenborn A. M., Skowronski J., Ahn J., Xiong Y. (2013) Mechanism of allosteric activation of SAMHD1 by dGTP. Nat. Struct. Mol. Biol. 20, 1304–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hopkins R. L., Goodman M. F. (1985) Ribonucleoside and deoxyribonucleoside triphosphate pools during 2-aminopurine mutagenesis in T4 mutator-, wild type-, and antimutator-infected Escherichia coli. J. Biol. Chem. 260, 6618–6622 [PubMed] [Google Scholar]
- 39. Sargent R. G., Mathews C. K. (1987) Imbalanced deoxyribonucleoside triphosphate pools and spontaneous mutation rates determined during dCMP deaminase-defective bacteriophage T4 infections. J. Biol. Chem. 262, 5546–5553 [PubMed] [Google Scholar]
- 40. Buckstein M. H., He J., Rubin H. (2008) Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J. Bacteriol. 190, 718–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]